Key message
A protocol for the isolation of egg apparatus cells from the basal angiosperm Amborella trichopoda to generate RNA-seq data for evolutionary studies of fertilization-associated genes.
Abstract
Sexual reproduction is particularly complex in flowering plants (angiosperms). Studies in eudicot and monocot model species have significantly contributed to our knowledge on cell fate specification of gametophytic cells and on the numerous cellular communication events necessary to deliver the two sperm cells into the embryo sac and to accomplish double fertilization. However, for a deeper understanding of the evolution of these processes, morphological, genomic and gene expression studies in extant basal angiosperms are inevitable. The basal angiosperm Amborella trichopoda is of special importance for evolutionary studies, as it is likely sister to all other living angiosperms. Here, we report about a method to isolate Amborella egg apparatus cells and on genome-wide gene expression profiles in these cells. Our transcriptomics data revealed Amborella-specific genes and genes conserved in eudicots and monocots. Gene products include secreted proteins, such as small cysteine-rich proteins previously reported to act as extracellular signaling molecules with important roles during double fertilization. The detection of transcripts encoding EGG CELL 1 (EC1) and related prolamin-like family proteins in Amborella egg cells demonstrates the potential of the generated data set to study conserved molecular mechanisms and the evolution of fertilization-related genes and their encoded proteins.
Similar content being viewed by others
Introduction
Molecular and fossil evidence indicates that flowering plants (angiosperms) arose abruptly in the late Jurassic/early Cretaceous from a not yet identified ancestral lineage. Since then, angiosperms rapidly diversified to form over 350,000 species alive today (Bell et al. 2005; Doyle 2012; Willis and McElwain 2013; Scutt 2018) and represent the most abundant and ecologically successful group of plants on earth. Amborella trichopoda, a woody shrub endemic to New Caledonia, holds a key position in the angiosperm evolutionary tree as the most basal angiosperm. It is the sole living species of the Amborellaceae family which is strongly supported as the sister lineage of all other extant flowering plants (Amborella Genome Project 2013). For this reason, Amborella has been the focus of many phylogenetic, genomic and reproductive biology studies, providing a deeper understanding regarding the evolution of flowering plants (for review, see Scutt 2018).
Among the defining features of angiosperms are carpels, which enclose and protect the ovules, and the process of double fertilization involving two pairs of male and female gametes, respectively (Dresselhaus et al. 2016; Scutt 2018). The highly reduced female gametophyte (embryo sac) develops within the ovule and contains the egg cell and central cell. Both cells become fertilized and give rise to the embryo and the embryo-nourishing endosperm of the seed, respectively. A large majority of extant angiosperms produce the monosporic Polygonum-type embryo sac, which was first described in Polygonum divaricatum (Strasburger 1879). This originates from a single haploid spore (the functional megaspore), which undergoes three incomplete mitotic division cycles to develop into an eight-nucleate syncytium. After cellularization, the Polygonum-type embryo sac is seven celled but eight nucleate, with three cells at each pole and a large binucleate central cell. The egg apparatus is located close to the micropylar entrance point of the pollen tube and comprises one egg cell and two accessory synergid cells, which are required for pollen tube attraction and reception (for review, see Higashiyama and Yang 2017). Three antipodal cells form at the opposite chalazal end of the embryo sac, but little is known about their function (Sprunck and Groß-Hardt 2011).
Although the Polygonum-type embryo sac is most prevalent in flowering plants, embryo sac formation is highly diverse across different plant taxa, suggesting an extensive degree of developmental experimentation during angiosperm evolution (Friedman 2006). Embryo sacs can also develop from two or four functional megaspores (bisporic and tetrasporic development, respectively), and/or from a varying number of mitotic divisions following meiosis. Within the embryo sac, the cells can furthermore occupy different positions (Maheshwari 1950; Huang and Russell 1992).
Notably, none of the members of the earliest extant angiosperm clades produces a Polygonum-type embryo sac (Friedman 2006). The four-celled Nuphar/Schisandra-type embryo sac comprising one egg cell, two synergid cells and a uninucleate central cell appears to be universal among extant members of the Nymphaeales and occurs also in Austrobaileyales (e.g., Williams and Friedman 2004; Tobe et al. 2007; Friedman 2008; Rudall et al. 2008). According to the present knowledge, the nine-nucleate, eight-celled Amborella-type female gametophyte is restricted to Amborella (Friedman and Ryerson 2009) and might represent a critical link between angiosperms and gymnosperms (Friedman 2006). Detailed structural analyses revealed that Amborella embryo sac development initially parallels that of the Polygonum-type, since immature embryo sacs are eight nucleate and seven celled, with each three cells at the chalazal and the micropylar pole. However, prior to embryo sac maturation, one of the three cells at the micropylar pole undergoes a final mitotic and cytokinetic division to produce two daughter cells: a third synergid cell and the egg cell. At maturity, the Amborella embryo sac is therefore eight celled, with three antipodal cells, a large binucleate central cell and a unique four-celled egg apparatus comprising three synergid cells and the egg cell which is the lineal sister cell of a synergid cell (Friedman 2006).
From studies in model plants such as Arabidopsis and maize, we know that cell fate specification processes in the developing embryo sac are tightly controlled and a few of the genes involved in these have been identified (for review, see Tekleyohans et al. 2017; Zhou et al. 2017). Previous studies furthermore showed that synergid cells, egg cells and central cells fulfill unique functions during flowering plant reproduction, which are reflected by distinct gene expression profiles (Márton et al. 2005; Okuda et al. 2009; Wuest et al. 2010; Anderson et al. 2013; Chen et al. 2017). These include molecules mediating pollen tube guidance to the ovule, induction of pollen tube burst, gamete interaction, activation and fusion (Okuda et al. 2009; Amien et al. 2010; Márton et al. 2012; Sprunck et al. 2012; Takeuchi and Higashiyama 2012; Ge et al. 2017). Nevertheless, a deeper molecular understanding of the evolution of flowering plant embryo sac formation is missing, and so far, the transcriptomes of individual embryo sac cells from basal angiosperms remained unexplored.
Here, we describe a method to manually isolate viable Amborella egg apparatus cells. Their morphology was investigated, and they were subsequently used to generate genome-wide gene expression profiles from a low number of cells for bioinformatics analysis. In this pioneer study, we identified Amborella-specific genes preferentially expressed in egg apparatus cells and first candidate genes for comparative evolutionary and functional studies.
Materials and methods
Plant material and growth conditions
Flowers were harvested from a female Amborella plant (one flowering period per year), grown in the greenhouse under controlled conditions of 12 h of light, 16.2 °C and constant humidity of 66%. Control tissues of tepals, leaves and roots were harvested and immediately frozen in liquid nitrogen.
Isolation of egg apparatus cells
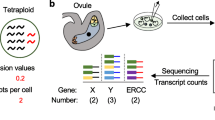
The isolation procedure was performed only on the most recently fully opened flowers with visible wet stigmas, harvested in the morning (8:00 am). Flowers were harvested in a glass petri dish on wet filter paper, to keep high humidity until dissection. All following steps were performed at room temperature, without any breaks and in an expeditious manner (approximately 1.5 h for processing ten carpels). Under a Nikon ZM645 stereo microscope, individual carpels were gently removed from the inflorescence using disposable hypodermic needles (0.4 mm diameter, 13 mm length) and placed in a new glass petri dish. Subsequently, carpels were cross-sectioned using a scalpel (No. 11 sterile carbon steel surgical blade). Cutting angle and position of cross section is indicated in Fig. 1b. The lower portion of dissected carpels is immediately transferred to a microscopy Imaging Dish GC 1.5 (130-098-284, Miltenyi Biotec GmbH), filled with 0.3 M filter-sterilized mannitol solution. Subsequently, hypodermic needles (0.4 mm diameter, 13 mm length) were used to further dissect the lower portion of the carpel in mannitol solution. The inner integument was separated from the outer integument, and the inner part of the ovule was gently lifted out of the carpel. The remains of the outer ovary sections were removed. Subsequently, the release of egg apparatus cells from the ovule tips was performed under a Nikon Eclipse TE2000-S inverted microscope equipped with a 20× objective (Plan Fluor ELWD 20×/0.45, DIC L/N1). For the isolation procedure, fine-tipped glass needles were prepared from borosilicate glass rods (4 mm diameter, 10 cm length, Hilgenberg GmbH) by using a heating device (Bunsen burner). To release egg apparatus cells from the ovule tip, one glass needle was used to hold the inner integument in place. The cells were then released by gently tapping on the outer side of the ovule with the second glass needle. Once egg apparatus cells were released, and their diameter was recorded by using a 40× objective (Plan Fluor ELWD 40×/0.60, DIC L/N1, Nikon). Cells were immediately collected using a CellTram® Oil (Eppendorf) equipped with a Ringcaps® glass 50-micron capillary (Hirschmann Instruments™ 9,600,150). The fine tip of the glass capillary was prepared using a micropipette puller as described elsewhere (Englhart et al. 2017). Individual cells were transferred to 0.5 ml LoBind reaction tubes (Sigma Aldrich) in a minimal amount of mannitol solution (max. 3–5 µl) and immediately frozen in liquid nitrogen for further analyses. Frozen cells were stored at − 80 °C.
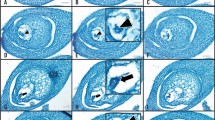
Female flower of A. trichopoda and isolation procedure of egg apparatus cells. a Female flower at anthesis. b Longitudinal cut of a mature carpel (side view). Note that the abaxial side of the carpel is more curved, while the adaxial side is flat. The red dashed line shows the angle of the cross section through the carpel. The position of the egg apparatus cells (asterisk) within the embryo sac (dotted line) is indicated. c Scheme of the Amborella embryo sac. Note that the cross section (red dotted line) traverses the lumen of the central cell. d Lower portion of the dissected carpel in mannitol solution. The dashed lines border the ovules outer integument. The arrowhead marks the position where the inner integument is manually separated from the outer integument and lifted out of the carpel. e Characteristic appearance of manually dissected ovule tip with the lower portion of the embryo sac (dotted line), surrounded by nucellus and inner integument. f Dissected ovule tip releasing an egg apparatus cell. g Morphology of released egg apparatus cell. The fluorescent dyes SYBR Green I and FM 4–64 were applied to stain the nucleus (green) and the plasma membrane (red), respectively. AC, antipodal cells; ad, adaxial side of carpel; CC, central cell; Cp, carpel; EAC, egg apparatus cell; EC, egg cell; ES, embryo sac; M, micropyle; N, nucleus; Ov, ovule; PM, plasma membrane; PN, polar nuclei; S, stigma; St, staminode; Sy, synergids; T, tepal; V, vacuole. Scale bars in a, b: 1 mm; in d, e: 100 μm; in f: 25 μm; in g: 10 μm
RNA isolation, library preparation and sequencing
For control tissues, total RNA was isolated from three biological replicates of tepals, leaves and roots with a Spectrum™ Plant Total RNA Kit (Sigma Aldrich) according to manufacturer’s instructions. RNA quality was assessed on the Agilent 2100 Bioanalyzer with the RNA 6000 Pico LabChip reagent set (Agilent) (Supplementary Figure 1). RNA integrity (RIN) values ranged from 6.7 to 9.4. Using 250 ng of total RNA, library preparation and RNA-seq were carried out as described in the Illumina TruSeq Stranded mRNA Sample Preparation Guide, the Illumina HiSeq 1000 System User Guide (Illumina, Inc.) and the KAPA Library Quantification Kit—Illumina/ABI Prism User Guide (Kapa Biosystems, Inc.).
Total RNA from egg apparatus cells was extracted according to the “Purification of total RNA from animal and human cells” protocol of the RNeasy Plus Micro Kit (Qiagen). Twenty-eight cells of L-(diameter from 25.85 to 17.96 µm) and M-(diameter from 17.65 to 16.35 µm) size categories and 26 cells of the S-size category (diameter from 16.17 to 13.13 µm) were used for RNA extraction. Finally, total RNA was eluted in 12 μl of nuclease-free water. RNA quality control measurements on the Bioanalyzer were unsuitable, since the RNA quantities obtained from the low number of egg apparatus cells were below the assay sensitivity. The SMARTer Ultra Low Input RNA Kit for Sequencing v4 (Clontech Laboratories, Inc.) was used to generate first-strand cDNA from 500 pg total RNA. Double-stranded cDNA was amplified by LD PCR (14 cycles) and purified via magnetic bead cleanup. Library preparation was carried out as described in the Illumina Nextera XT Sample Preparation Guide (Illumina, Inc.). 150 pg of input cDNA was tagmented (tagged and fragmented) by the Nextera XT transposome. Products were purified and amplified via a limited-cycle PCR program to generate multiplexed sequencing libraries. For the PCR step, 1:5 dilutions of index 1 (i7) and index 2 (i5) primers were used.
Libraries from egg apparatus cells and control tissues were quantified using the KAPA SYBR FAST ABI Prism Library Quantification Kit. Equimolar amounts of each library were used for cluster generation on the cBot (TruSeq SR Cluster Kit v3). Sequencing runs were performed on a HiSeq 1000 instrument using the indexed, 2 × 100 cycles paired end (PE) protocol and the TruSeq SBS v3 Kit. Image analysis and base calling resulted in “.bcl” files, which were converted into “.fastq” files using the CASAVA1.8.2 software. RNA extraction of egg apparatus cells and Illumina deep sequencing were carried out at the genomics core facility of the University of Regensburg (Center for Fluorescent Bioanalytics KFB; www.kfb-regensburg.de).
RNA-seq data analysis
RNA-seq data were processed using Kallisto (Bray et al. 2016). Reads were mapped to the Amborella genome v1.0 (Amborella Genome Project 2013), obtained from Ensembl Plants (Kersey et al. 2016). Gene expression values were determined using Transcripts Per Million (TPM) (Conesa et al. 2016) and clustered using Python Seaborn software package with “Euclidean” distance and “complete” method, which was also used to generate the heat map shown in Fig. 3c. A comprehensive overview on normalized expression data in Amborella egg apparatus cells, leaves, roots and tepals is shown in Supplementary Table 1. RNA-seq data will be publicly available in the open source web server CoNekT (Proost and Mutwil 2018).
Results and discussion
Isolation of Amborella egg apparatus cells
We report here an isolation procedure for Amborella egg apparatus cells by manually dissecting carpels of female flowers, without treating ovules with cell wall-degrading enzymes. Enzyme-free methods have been successfully established mainly for the isolation of egg cells from angiosperm species such as Brassica napus, Hordeum vulgare, Oryza sativa, Plumbago zeylanica and Triticum aestivum (Table 1). After gently removing carpels from female flowers of Amborella (Fig. 1a), their characteristic shape with a flat adaxial and round abaxial side became apparent. Carpels were placed laterally to perform a cross section through ovaries as indicated in Fig. 1b. It was very clear in our studies that position and angle of cross sections determined later success of releasing egg apparatus cells. Cross sections traverse the lumen of central cells (Fig. 1c), which will be destroyed and, thus, cannot be isolated by the described protocol. Immediate transfer of the lower part of carpels into 0.3 M mannitol solution is essential to prevent osmotic stress and to avoid the irreversible collapse of egg apparatus cells by drying out (Fig. 1d). In pilot experiments, we tested different mannitol molarities and determined 0.3 M mannitol to be optimal to maintain the shape and size of released egg apparatus cells. Similar mannitol concentrations have been reported for the isolation of egg apparatus cells from rice and spider lily (Uchiumi et al. 2006; Ohshika and Ikeda 1994). By contrast, egg apparatus cells from other flowering plant species require higher osmolarities, such as 0.55 M mannitol for wheat (Kovács et al. 1994), or even 0.7 M mannitol for Plumbago zeylanica and rapeseed (Cao and Russell 1997, Katoh et al. 1997). We used one hypodermic needle to hold the outer part of the carpel in place. With a second hypodermic needle, we gently separated the inner integument from the outer integument and lifted ovule tips out of carpel sections (Fig. 1e). After removing remains of carpels from the mannitol solution, release of egg apparatus cells was performed and observed under an inverted microscope. Two fine-tipped glass needles were used to hold one ovule tip and to gently push the egg apparatus cells out of the tip, respectively. One egg apparatus cell, just being released from an ovule tip, is shown in Fig. 1f. Subsequently, egg apparatus cells were imaged, transferred into a 0.5 ml reaction tube by micropipette aspiration and immediately frozen in liquid nitrogen. Approximately 5–10 apparently intact egg apparatus cells could be isolated from 50 carpels.
Morphology and identity of isolated egg apparatus cells
Depending on the plant species, either egg or synergid cells were reported to be slightly larger in diameter (Table 1). Only rice and petunia egg apparatus cells were reported to be rather similar in size (Van Went and Kwee 1990; Uchiumi et al. 2006). Furthermore, egg cells and synergid cells isolated from Arabidopsis, or rice, showed distinct cellular morphologies (Englhart et al. 2017; Ohnishi et al. 2011). To be able to distinguish Amborella egg cells and synergid cells without the help of molecular markers, we studied the morphology of isolated egg apparatus cells in more detail. Typically, intact cells exhibited a plasma membrane, a vacuole of variable size and a nucleus, which could be stained by SYBR Green I (Fig. 1g). Comparable to previous reports on Plumbago, wheat, barley and rice egg cells that were isolated by non-enzymatic cell isolation techniques (e.g., Holm et al. 1994; Cao and Russell 1997; Sprunck et al. 2005; Leljak-Levanic et al. 2013; Ohnishi et al. 2011), egg apparatus cells from Amborella became immediately round during the isolation process, suggesting that either a very thin primary cell wall or no cell wall was present.
Eventually, a group of spherical cells was simultaneously released from one ovary tip (Fig. 2). These cells exhibited slightly different morphologies in terms of cell size and granular, contrast-rich structures in their cytoplasm (Fig. 2a). The diameter of “large” cells ranged from 18 to 25 µm (± 2.23), and they were frequently attached to either one, two or three smaller cells (Fig. 2a, b, d). The cytoplasm of these “large” cells, which we considered as egg cells (see below), contained relatively few granular particles and a large vacuole that pushed the nucleus (with its clearly recognizable nucleolus) to the cell periphery (Fig. 2d). Diameters of “small” egg apparatus cells ranged from 13 to 15 µm (± 1.28). Furthermore, these cells featured less vacuoles, more granular particles in the cytoplasm and were sometimes released in a group of three (Fig. 2c). Based on their similar morphology and the fact that the female gametophyte of A. trichopoda contains three synergid cells (Friedman and Ryerson 2009), we determined the cells with diameters ranging from 13 to 15 µm as synergid cells.
Morphology of isolated egg apparatus cells from A. trichopoda. a Four spherical cells are released from the dissected embryo sac. Three cells appear slightly darker and smaller (arrowheads). b Group of one larger and two smaller cells that remain together after isolation. Two cells exhibit a clearly visible vacuole (V). c Group of three cells with similar size and morphology. d Two released cells showing different features in terms of size and cell morphology. The larger cell exhibits a clearly visible nucleolus (N) and a vacuole (V), while the smaller cell lacks a large vacuole and contains dark granules in the cytoplasm. Scale bars: 10 μm
Eventually, we observed clusters of egg apparatus cells where one of the three synergid cells was larger than the other two (Fig. 2a). Furthermore, a substantial number of isolated egg apparatus cells did not exhibit unambiguous morphological features and cell diameters, which prevented us from classifying them either as egg cells, or as synergid cells. However, the Amborella egg cell is unique in that it differentiates very late, only after a terminal cell division of one of the three micropylar cells of the seven-celled, eight-nucleate immature embryo sac present in pre-anthesis floral buds (Friedman 2006). The Amborella egg cell is therefore a lineal sister of the third synergid cell, while in all other angiosperm embryo sacs, the egg cell is the mitotic sister of a polar nucleus of the central cell (Huang and Russell 1992; Friedman 2006). Considering this uniqueness, the close relation between the egg cell and the third synergid cell of Amborella might be reflected by a more similar morphology, especially when the two daughter cells are isolated rather short after the final cell division. For these reasons, we assumed that the “intermediate-sized” and less vacuolated group of egg apparatus cells include young egg cells and their mitotic sister synergid cells, which cannot be unequivocally distinguished by their size and/or morphology. To obtain high-quality RNA-seq data, we had to pool at least 25 cells per replicate. We therefore assigned three egg apparatus cell size categories, based on the cell diameter (Fig. 3a). We pooled 28 cells with a diameter ranging from 17.96 to 25.85 µm as “Large” (L) group predominantly comprising egg cells. A group of 28 cells with diameters of 16.35 to 17.65 µm was classified as “Medium” (M) group enriched in young egg cells and third synergid cells. 26 cells with a diameter ranging from 13.13 to 16.17 µm were assigned as “Small” (S) group predominantly consisting of synergid cells.
Size categories of A. trichopoda egg apparatus cells and overview of RNA-seq data generated. a According to their diameter, 82 isolated egg apparatus cells were pooled into groups of large (L), medium (M) and small (S) cells. b Number of expressed genes (TPM ≥ 1) in each cell each size category and overlap (%) of the total number of expressed genes (15,409) among the three categories. c Heat map comparing the top 6000 genes expressed in each cell size category. Genes are present in rows, while samples are present in columns. Z-score normalized values are shown, ranging from 0 (mean expression, in white) to 1 (maximum standard deviation away from the mean of expression, in red)
RNA-seq analysis of isolated embryo sac cells
After RNA extraction, library preparation and Illumina sequencing, we obtained a total of 57.58 million (M) reads for the three groups of egg apparatus cells that mapped to Amborella genome v1 (Amborella Genome 2013). We also generated RNA-seq data from sporophytic control tissues (three biological replicates of leaves, roots and tepals, respectively) and obtained a total number of 22.62 M mapped reads for leaves, 19.07 M for roots and 50.38 M for tepals. Supplementary Table 1 provides Amborella gene symbols and normalized transcripts per million (TPM) values obtained for the three egg apparatus cell samples and the sporophytic control tissues.
A principal component analysis (PCA) performed on the expression data revealed that the three egg apparatus samples group closely together and are clearly separated from the clusters formed by the biological replicates of the three control tissues (Supplementary Figure 2). Among the egg apparatus cell size categories, L- and M-size cell types grouped together more closely.
We employed a threshold level of ≥ 1 TPM to estimate the number of expressed genes in the three groups of egg apparatus cells. Thereby, we detected 11,841 expressed genes in the category of large (L) cells, 11,808 expressed genes in the intermediate-sized cell category (M) and 12,501 expressed genes in the group of small (S) egg apparatus cells (Fig. 3b). Overall, 15,409 genes were expressed in the Amborella egg apparatus cells. Considering a total number of 27,313 coding genes in the genome of A. trichopoda (Ensembl Plants database release 41; September 2018), the number of egg apparatus-expressed genes comprises 56.42% of all Amborella genes.
When comparing gene expression patterns in the three cell size categories, we detected 8737 genes to be conjointly expressed in large, medium and small egg apparatus cells (Fig. 3b and Supplementary Table 2). Despite this rather large overlap (56.7% of 15,409 egg apparatus-expressed genes), a comparison of the top 6000 genes expressed in each cell size category identified prominent gene clusters with differential expression in either the L-, M- or S-group of egg apparatus cells (Fig. 3c). These distinct transcriptome signatures could reflect a cell type-dependent accumulation of transcripts necessary for the egg cells and synergid cells to fulfill their specialized roles during double fertilization. However, more biological replicates are needed to analyze the differential gene expression in Amborella egg apparatus cell types in detail, as only single replicates of each cell size category have been generated during this pioneer study.
Egg apparatus-enriched genes encode CRPs, novel proteins and proteins with homologs in eudicots and/or monocots
We next analyzed the strongest expressed genes that were specifically enriched in the respective size category of egg apparatus cells, but were not expressed in the sporophytic control tissues. Therefore, we excluded all genes with a TPM ≥ 3 in roots, leaves and tepals. In Table 2, we summarize the ten strongest expressed Amborella genes enriched in each cell size category and their best match in sequence similarity searches. Table 2 also provides information on three more plant species annotated for subsequent BLASTP hits. The division of the best BLAST matches to Amborella sequences, approximately equally between eudicots and monocots, is in agreement with Amborella’s phylogenetic position as a basal angiosperm, equally distantly related to these two groups (Amborella Genome 2013). Notably, six of the 30 genes shown in Table 2 and Supplementary Table 3 were annotated to encode “hypothetical proteins” in A. trichopoda and did not reveal significant sequence similarities to any protein from other flowering plant species.
More than one-third of proteins encoded by the 30 most abundantly enriched transcripts in Amborella egg apparatus cells were predicted to be directed to the secretory pathway, or to localize in the extracellular space. These include a potential Amborella-specific small secreted cysteine-rich protein (CRP) of 109 amino acids (encoded by AMTR_s00059p00189800) and another small secreted CRP with eight conserved cysteine residues (AMTR_s00162p00083730) showing sequence similarity to non-specific lipid transfer proteins (nsLTPs), which are a part of the prolamin superfamily and abundant in liverworts, mosses and all other investigated land plants (Edstam et al. 2011).
A defensin-like (DEFL) protein and a putative RALF (RAPID ALKALINIZATION FACTOR), encoded by AMTR_s00067p00126450 and AMTR_s00067p00138490, respectively, were detected in the S-group (enriched in synergid cells). RALFs are CRPs of around 5-kDa that have been identified as extracellular ligands of the Catharanthus roseus RLK1-like (CrRLK1L) family of receptor-like kinases, known to regulate cell expansion, pollen tube cell integrity and burst (Haruta et al. 2014; Ge et al. 2017). The Amborella genome was reported to encode nine RALF family proteins belonging to the major clades II, III and IV (Campbell and Turner 2017). Multiple sequence alignment of S-group-enriched clade IV-C AMTR_s00067p00138490 with other clade IV RALFs from maize, rice and Arabidopsis revealed that it contains the four cysteine residues typically conserved in RALF proteins, but lacks the YISY motif conserved in clade I-III RALFs (Supplementary Figure 3). When we investigated all nine RALFs for their expression in Amborella sporophytic tissues and egg apparatus cells, the second clade IV-C RALF-related AMTR_s00067p00137560 also showed weak, but selective expression in egg apparatus cells, while clade II-B (AMTR_s00045p00202280) and clade III-C RALFs (AMTR_s00017p00211590) were almost ubiquitously expressed. Strikingly, clade IV-B RALF-related AMTR_s00045p00207290 exhibited extremely strong expression values in the synergid-enriched group of egg apparatus cells (Fig. 4). It therefore seems likely that the respective RALF-related protein holds an important function as extracellular ligand in Amborella synergid cells.
Expression pattern of Amborella genes encoding RALFs. The classification of RALF family proteins into four major clades is according to Campbell and Turner (2017). Nine genes encoding RALF and RALF-related proteins were identified in the Amborella genome, belonging to clades II-B, III-B to III-D and IV-B and IV-C, respectively. Clade IV-B RALF-related AMTR_s00045p00207290 is expressed in the control tissues (tepals, leaves, roots), but highly enriched in S-group egg apparatus cells (synergids) when compared with all other RALF and RALF-related genes. Clade IV-C RALF-related genes AMTR_s00067p00138490 and AMTR_s00067p00137560 are selectively present in egg apparatus cells, without detectable expression in the control tissues
The above mentioned DEFL did not reveal any similarity to synergid-expressed DEFL family proteins with known functions such as LUREs, which act as species-specific pollen tube attractants in Torenia and Arabidopsis (Okuda et al. 2009; Takeuchi and Higashiyama 2012), or maize ES1-4 (EMBRYO SAC1-4), which induce pollen tube tip burst (Amien et al. 2010). However, related functions cannot be excluded as DEFLs are highly polymorphic proteins, making it difficult to determine exact relationships of gene orthology (Higashiyama and Takeuchi 2015). Furthermore, other DEFLs with enriched expression in Amborella synergid cells might be present among the genes comprising egg apparatus-expressed genes with transcripts in roots, leaves or tepals (Supplementary Table 4).
Amborella egg cells express EGG CELL 1 and other DUF784/DUF1278 gene family members
Importantly, we also identified four genes encoding CRPs with a prolamin-like domain (PF05617) to be specifically expressed in Amborella egg apparatus cells (Supplementary Table 3). These include two of the strongest expressed genes in egg cells (L-group: AMTR_s00067p00156690 and AMTR_s00032p00113070), the strongest expressed gene in the M-group (AMTR_s00017p00240840) and one gene in the S-group (AMTR_s00067p00155100). A fifth CRP with prolamin-like characteristics was detected when we manually inspected the predicted protein sequence encoded by AMTR_s00058p00172040, which is the strongest expressed genes in egg cells (L-group) (Table 2).
Prolamin-like CRPs comprise Domain of Unknown Function 784 (DUF784) and DUF1278 proteins, which are often collectively termed as ECA1 gametogenesis-related proteins (Zhang 2009). In Arabidopsis, DUF784 and DUF1278 proteins are encoded by embryo sac-specific gene families (Jones-Rhoades et al. 2007). So far, a function for prolamin-like CRPs has only been assigned for five Arabidopsis DUF1278 family members termed EGG CELL 1 (EC1). EC1 proteins are secreted by the egg cell upon sperm cell arrival and essential for successful gamete interactions during double fertilization (Sprunck et al. 2012; Rademacher and Sprunck 2013). Notably, sequence similarity searches using both AMTR_s00067p00156690 and AMTR_s00017p00240840 resulted in matches to proteins annotated as Egg cell-secreted protein 1.4 (Table 2). To investigate Amborella prolamin-like proteins in more detail, we performed multiple sequence alignments and maximum likelihood phylogenies (Fig. 5). For sequence comparisons, we included EC1 proteins with validated expression in wheat, Arabidopsis, maize and rice egg cells (Sprunck et al. 2005; Ohnishi et al. 2011; Sprunck et al. 2012; Chen et al. 2017; Resentini et al. 2017), but also Arabidopsis DUF784 and DUF1278 proteins with validated expression in synergid cells (Jones-Rhoades et al. 2007; Steffen et al. 2007). The phylogenetic tree in Fig. 5 shows individual clades formed by EC1 proteins, synergid-expressed DUF1278 proteins and DUF784 proteins, respectively. Four Amborella prolamin-like proteins do not locate to these clades. However, the protein encoded by AMTR_s00017p00240840 forms a subclade with EC1 proteins from maize and rice suggesting that it represents a true Amborella EC1 ortholog (Fig. 5). Although a similar function for Amborella EC1 during double fertilization remains to be experimentally confirmed, our findings strongly suggest that the molecular mechanism of EC1-mediated sperm activation by the egg cell, as previously reported for Arabidopsis EC1 (Sprunck et al. 2012; Rademacher and Sprunck 2013), existed already in the most recent common ancestor of all extant flowering plants.
Phylogeny of EC1 and other prolamin-like proteins. Five Amborella prolamin-like proteins (in red) were aligned with egg cell-expressed EC1 proteins from Arabidopsis, wheat, maize and rice and with other DUF1278 and DUF784 family proteins from Arabidopsis that were shown to be expressed in synergid cells. The unrooted tree was constructed with MEGA7 (Kumar et al. 2016) using the neighbor-joining method, 1000 bootstrap replicates and Poisson correction. Proteins are labeled with previously published names and accession numbers, or with their respective gene identifiers. Abbreviations: ECR, EC1-related DUF1278 protein (Sprunck et al. 2014); MYB98dep, expression depends on the transcription factor Myb98 (Jones-Rhoades et al. 2007); SY, expressed in synergid cells, verified by reporter studies (Jones-Rhoades et al. 2007; Steffen et al. 2007)
Conclusions
We have shown that the described method for the isolation of living egg apparatus cells from the basal angiosperm Amborella is suitable for subsequent investigations including omics approaches, but it will also be valuable for in vitro fertilization experiments to generate zygote and embryo stages. A limited number of Amborella egg apparatus cells was used to generate RNA-Seq data and to identify cell type-enriched transcripts. The presented data set revealed first candidate genes for comparative evolutionary and functional studies in angiosperms, and more genes remain to be identified. It will also allow the search for molecular footprints of non-flowering seed plants, for example genes expressed in archegonia of gymnosperms. For future transcriptome studies of Amborella embryo sac cells, it is important to generate more biological replicates. A detailed comparison of gene expression programs in egg cells and synergid cells from Amborella with those from eudicot and monocot egg cells and synergid cells will contribute to the identification of evolutionary conserved molecular mechanisms involved in their specific functions during double fertilization. Future transcriptome studies should also include the Amborella central cell. This will require technical adaptions of the presented method to avoid the destruction of this large and highly vacuolated cell that occupies most of the volume of the embryo sac. Furthermore, the unambiguous classification of Amborella egg cells and synergid cells will be essential to prevent cross-contamination between pooled cell populations. Nevertheless, single-cell RNA sequencing technologies are rapidly evolving and will soon allow the profiling and classification of individual embryo sac cells, isolated from Amborella and other angiosperms.
Author contribution statement
SS and TD designed the study, MFT established the cell isolation protocol and performed experiments, SP and MM processed raw RNA-seq data to enable data analyses by MFT, MM and SS. CS provided the female Amborella plant used for cell isolation. SS wrote the manuscript, with input from TD, MFT and CS. All authors approved the manuscript.
Change history
25 February 2019
The article Transcriptomics of manually isolated <Emphasis Type="Italic">Amborella trichopoda</Emphasis> egg apparatus cells, written by María Flores-Tornero, Sebastian Proost, Marek Mutwil, Charles P. Scutt, Thomas Dresselhaus, Stefanie Sprunck, was originally published electronically on the publisher’s internet portal (currently SpringerLink) on 1 February 2019 without open access
References
Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342:1241089. https://doi.org/10.1126/science.1241089
Amien S, Kliwer I, Márton ML, Debener T, Geiger D, Becker D, Dresselhaus T (2010) Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol 8:e1000388
Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V (2013) Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76:729–741. https://doi.org/10.1111/tpj.12336
Bell CD, Soltis DE, Soltis PS (2005) The age of the angiosperms: a molecular timescale without a clock. Evolution 59:1245–1258. https://doi.org/10.1111/j.0014-3820.2005.tb01775.x
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525. https://doi.org/10.1038/nbt.3519
Campbell L, Turner SR (2017) A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front Plant Sci 8:37. https://doi.org/10.3389/fpls.2017.00037
Cao Y, Russell SD (1997) Mechanical isolation and ultrastructural characterization of viable egg cells in Plumbago zeylanica. Sex Plant Reprod 10:368–373. https://doi.org/10.1007/s004970050111
Chen SH, Yang YH, Liao JP, Kuang AX, Tian HQ (2008) Isolation of egg cells and zygotes of Torenia fournieri L. and determination of their surface charge. Zygote 16:179–186. https://doi.org/10.1017/S0967199408004693
Chen J, Strieder N, Krohn NG, Cyprys P, Sprunck S, Engelmann JC, Dresselhaus T (2017) Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29:2106–2125
Conesa A et al (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17:1–13. https://doi.org/10.1186/s13059-016-0881-8
Doyle JA (2012) Molecular and fossil evidence on the origin of angiosperms. In: Jeanloz R (ed) Annual review of earth and planetary sciences, vol 40. Department of Evolution and Ecology, University of California, Davis, pp 301–326
Dresselhaus T, Sprunck S, Wessel GM (2016) Fertilization mechanisms in flowering plants. Curr Biol 26:R125–R139. https://doi.org/10.1016/j.cub.2015.12.032
Edstam MM, Viitanen L, Salminen TA, Edqvist J (2011) Evolutionary history of the non-specific lipid transfer proteins. Mol Plant 4:947–964. https://doi.org/10.1093/mp/ssr019
Englhart M, Šoljić L, Sprunck S (2017) Manual isolation of living cells from the Arabidopsis thaliana female gametophyte by micromanipulation. In: Schmidt A (ed) Plant germline development: methods and protocols. Springer, New York, pp 221–234. https://doi.org/10.1007/978-1-4939-7286-9_18
Friedman WE (2006) Embryological evidence for developmental lability during early angiosperm evolution. Nature 441:337–340
Friedman WE (2008) Hydatellaceae are water lilies with gymnospermous tendencies. Nature 453:94–97
Friedman WE, Ryerson KC (2009) Reconstructing the ancestral female gametophyte of angiosperms: insights from Amborella and other ancient lineages of flowering plants. Am J Bot 96:129–143. https://doi.org/10.3732/ajb.0800311
Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, Hou S, Huang Q, Lai L, Moura DS, Gu H, Dong J, Wu HM, Dresselhaus T, Xiao J, Cheung AY, Qu LJ (2017) Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358:1596–1600. https://doi.org/10.1126/science.aao3642
Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343:408–411
He E-M, Wang Y-Y, Liu H-H, Zhu X-Y, Tian H (2012) Egg cell isolation in Datura stramonium (Solanaceae). Ann Bot Fenn 49:7–12. https://doi.org/10.5735/085.049.0102
Higashiyama T, Takeuchi H (2015) The mechanism and key molecules involved in pollen tube guidance. Annu Rev Plant Biol 66:393–413
Higashiyama T, Yang W-c (2017) Gametophytic pollen tube guidance: attractant peptides, gametic controls, and receptors. Plant Physiol 173:112–121
Holm PB, Knudsen S, Mouritzen P, Negri D, Olsen FL, Roue C (1994) Regeneration of fertile barley plants from mechanically isolated protoplasts of the fertilized egg cell. Plant Cell 6:531–543. https://doi.org/10.1105/tpc.6.4.531
Hoshino Y, Murata N, Shinoda K (2006) Isolation of individual egg cells and zygotes in Alstroemeria followed by manual selection with a microcapillary-connected micropump. Ann Bot 97:1139–1144. https://doi.org/10.1093/aob/mcl072
Hu S-Y, L-g Li, Zhu C (1985) Isolation of viable embryo sacs and their protoplasts of Nicotiana tabacum. Acta Bot Sin 27:343–347
Huang B-Q, Russell SD (1992) Female germ unit: organization, isolation, and function. Int Rev Cytol 140:233–293
Huang B-Q, Pierson ES, Russell SD, Tiezzi A, Cresti M (1992) Video microscopic observations of living, isolated embryo sacs of Nicotiana and their component cells. Sex Plant Rep 5:156–162. https://doi.org/10.1007/bf00194876
Jones-Rhoades MW, Borevitz JO, Preuss D (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3:1848–1861
Katoh N, Lorz H, Kranz E (1997) Isolation of viable egg cells of rape (Brassica napus L.). Zygote 5:31–33
Kersey PJ et al (2016) Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res 44:D574–D580. https://doi.org/10.1093/nar/gkv1209
Kovács M, Barnabás B, Kranz E (1994) The isolation of viable egg cells of wheat (Triticum aestivum L.). Sex Plant Reprod 7:311–312. https://doi.org/10.1007/bf00227715
Kranz E, Bautor J, Lörz H (1991) In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sex Plant Reprod 4:12–16. https://doi.org/10.1007/bf00194565
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Leljak-Levanić D, Juranić M, Sprunck S (2013) De novo zygotic transcription in wheat (Triticum aestivum L.) includes genes encoding small putative secreted peptides and a protein involved in proteasomal degradation. Plant Reprod 26:267–285. https://doi.org/10.1007/s00497-013-0229-4
Lin M-Z, Chen L, Zhu X-Y, Tian H-Q, Teixeira da Silva JA (2012) Isolation of eggs and synergids in Ceiba speciosa. Ann Bot Fenn 49:229–233. https://doi.org/10.5735/085.049.0402
Maheshwari P (1950) An introduction to the embryology of angiosperms. McGraw-Hill, New York
Márton ML, Cordts S, Broadhvest J, Dresselhaus T (2005) Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307:573. https://doi.org/10.1126/science.1104954
Márton ML, Fastner A, Uebler S, Dresselhaus T (2012) Overcoming hybridization barriers by the secretion of the maize pollen tube attractant ZmEA1 from Arabidopsis ovules. Curr Biol 22:1194–1198
Mól R (1986) Isolation of protoplasts from female gametophytes of Torenia fournieri. Plant Cell Rep 5:202–206. https://doi.org/10.1007/bf00269119
Ohnishi T, Takanashi H, Mogi M, Takahashi H, Kikuchi S, Yano K, Okamoto T, Fujita M, Kurata N, Tsutsumi N (2011) Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol 155:881–891. https://doi.org/10.1104/pp.110.167502
Ohshika K, Ikeda H (1994) Isolation and preservation of the living embryo sac of Crinum asiaticum L. var. japonicum baker. J Plant Res 107:17–21. https://doi.org/10.1007/bf02344525
Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, Kawano N, Sakakibara T, Namiki S, Itoh K, Otsuka K, Matsuzaki M, Nozaki H, Kuroiwa T, Nakano A, Kanaoka MM, Dresselhaus T, Sasaki N, Higashiyama T (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458:357–361. https://doi.org/10.1038/nature07882
Proost S, Mutwil M (2018) CoNekT: an open-source framework for comparative genomic and transcriptomic network analyses. Nucleic Acids Res 46:W133–W140. https://doi.org/10.1093/nar/gky336
Rademacher S, Sprunck S (2013) Downregulation of egg cell-secreted EC1 is accompanied with delayed gamete fusion and polytubey. Plant Signal Behav 8:e27377
Resentini F et al (2017) SUPPRESSOR OF FRIGIDA (SUF4) supports gamete fusion via regulating Arabidopsis EC1 gene expression. Plant Physiol 173:155–166. https://doi.org/10.1104/pp.16.01024
Rudall PJ, Remizowa MV, Beer AS, Bradshaw E, Stevenson DW, Macfarlane TD, Tuckett RE et al (2008) Comparative ovule and megagametophyte development in Hydatellaceae and water lilies reveal a mosaic of features among the earliest angiosperms. Ann Bot 101:941–956
Scutt CP (2018) The origin of angiosperms. In: Nuño de la Rosa L, Müller GB (eds) Evolutionary developmental biology. Springer, Berlin. https://doi.org/10.1007/978-3-319-33038-9_60-1
Sprunck S, Groß-Hardt R (2011) Nuclear behavior, cell polarity and cell specification in the female gametophyte. Sex Plant Reprod 24:123–136
Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41:660–672. https://doi.org/10.1111/j.1365-313X.2005.02332.x
Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338:1093–1097
Sprunck S, Hackenberg T, Englhart M, Vogler F (2014) Same same but different: sperm-activating EC1 and ECA1 gametogenesis-related family proteins. Biochem Soc Trans 42:401–407. https://doi.org/10.1042/BST20140039
Steffen JG, Kang IH, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51:281–292. https://doi.org/10.1111/j.1365-313X.2007.03137.x
Strasburger E (1879) Die Angiospermen und die Gymnospermen. Fischer, Jena, p 1879
Takeuchi H, Higashiyama T (2012) A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol 10:e1001449. https://doi.org/10.1371/journal.pbio.1001449
Tekleyohans DG, Nakel T, Groß-Hardt R (2017) Patterning the female gametophyte of flowering plants. Plant Physiol 173:122–129. https://doi.org/10.1104/pp.16.01472
Tian HQ, Russell SD (1997) Micromanipulation of male and female gametes of Nicotiana tabacum: II. Preliminary attempts for in vitro fertilization and egg cell culture. Plant Cell Rep 16:657–661. https://doi.org/10.1007/bf01275510
Tobe H, Kimoto Y, Prakash N (2007) Development and structure of the female gametophyte in Austrobaileya scandens (Austrobaileyaceae). J Plant Res 120:431–436
Uchiumi T, Komatsu S, Koshiba T, Okamoto T (2006) Isolation of gametes and central cells from Oryza sativa L. Sex Plant Rep 19:37–45. https://doi.org/10.1007/s00497-006-0020-x
van der Maas HM, Zaal MACM, de Jong ER, Krens FA, Van Went JL (1993) Isolation of viable egg cells of perennial ryegrass (Lolium perenne L.). Protoplasma 173:86–89. https://doi.org/10.1007/bf01378865
Van Went JL, Kwee H-S (1990) Enzymatic isolation of living embryo sacs of Petunia. Sex Plant Rep 3:257–262. https://doi.org/10.1007/bf00202883
Williams JH, Friedman WE (2004) The four-celled female gametophyte of Illicium (Illiciaceae; Austrobaileyales): implications for understanding the origin and early evolution of monocots, eumagnoliids, and eudicots. Am J Bot 91:332–351
Willis K, McElwain J (2013) The evolution of plants, 2nd edn. Oxford University Press, Oxford, ISBN: 9780199292233
Wuest SE et al (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20:506–512. https://doi.org/10.1016/j.cub.2010.01.051
Yang SJ, Mei Wei D, Tian H (2015) Isolation of sperm cells, egg cells, synergids and central cells from Solanum verbascifolium L. J Plant Biochem Biot 24:400–407. https://doi.org/10.1007/s13562-014-0290-6
Zhang D (2009) Homology between DUF784, DUF1278 domains and the plant prolamin superfamily typifies evolutionary changes of disulfide bonding patterns. Cell Cycle 8:3428–3430. https://doi.org/10.4161/cc.8.20.9674
Zhou L-Z, Juranic M, Dresselhaus T (2017) Germline development and fertilization mechanisms in maize. Mol Plant 10:389–401
Acknowledgements
We are grateful to Maximilian Weigend, Cornelia Löhne and Bernhard Reinken (Botanical Garden of the University of Bonn, Germany) for providing Amborella plant material. We thank Maria Lindemeier for her support in single-cell collection. Illumina deep sequencing was carried out at a genomics core facility: Center of Excellence for Fluorescent Bioanalytics (KFB, University of Regensburg, Germany). This work was supported by the ERA-CAPS Grant EVOREPRO (DR 334/12-1) to SS and TD, funded by the Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Dolf Weijers.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised due to a retrospective Open Access order.
A contribution to the special issue ‘Cellular Omics Methods in Plant Reproduction Research’.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure
1 Electropherograms of total RNA samples isolated from tepals, leaves and roots. (PDF 1115 kb)
Supplementary Figure
2 Principal Component Analysis (PCA). (PDF 809 kb)
Supplementary Figure
3 (PDF 305 kb)
Supplementary Table
1 Normalized gene expression values in egg apparatus cells of large (L)-, medium (M)- and small (S)-size categories in comparison with tepals, leaves and roots. (XLSX 7581 kb)
Supplementary Table
2 Genes expressed in large (L), medium (M) and small (S) egg apparatus cell size categories (TPM ≥ 1) and their overlap. (PDF 2206 kb)
Supplementary Table
3 Top ten list of A. trichopoda genes enriched in large (L), medium (M), or small (S) cell size categories, respectively, but not expressed in sepals, leaves and roots (average TPM < 3). (XLSX 22 kb)
Supplementary Table
4Amborella trichopoda expressed genes (TPM ≥ 1) in large (L), medium (M), and small (S) egg apparatus cell size categories and average expression values in the control tissues. (XLSX 2753 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Flores-Tornero, M., Proost, S., Mutwil, M. et al. Transcriptomics of manually isolated Amborella trichopoda egg apparatus cells. Plant Reprod 32, 15–27 (2019). https://doi.org/10.1007/s00497-019-00361-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-019-00361-0