Abstract
Apomixis, a type of asexual reproduction in angiosperms, results in progenies that are genetically identical to the mother plant. It is a highly desirable trait in agriculture due to its potential to preserve heterosis of F1 hybrids through subsequent generations. However, no major crops are apomictic. Deciphering mechanisms underlying apomixis becomes one of the alternatives to engineer self-reproducing capability into major crops. Parthenogenesis, a major component of apomixis, commonly described as the ability to initiate embryo formation from the egg cell without fertilization, also can be valuable in plant breeding for doubled haploid production. A deeper understanding of transcriptional differences between parthenogenetic and sexual or non-parthenogenetic eggs can assist with pathway engineering. By conducting laser capture microdissection-based RNA-seq on sexual and parthenogenetic egg cells on the day of anthesis, a de novo transcriptome for the Cenchrus ciliaris egg cells was created, transcriptional profiles that distinguish the parthenogenetic egg from its sexual counterpart were identified, and functional roles for a few transcription factors in promoting natural parthenogenesis were suggested. These transcriptome data expand upon previous gene expression studies and will be a resource for future research on the transcriptome of egg cells in parthenogenetic and sexual genotypes.
Similar content being viewed by others
Introduction
Apomixis is commonly defined as a reproductive phenomenon in angiosperms where embryos form from maternal cells in the ovule without meiosis and syngamy, resulting in asexual seed formation1,2,3. Apomixis is widespread in flowering plants and has now been described in 148 genera as adventitious embryony where the embryo arises from somatic cells in the ovule, 110 genera as aposporous and 68 genera as diplosporous where the embryo develops from the unreduced egg in a nucellar or megaspore mother cell-derived embryo sac, respectively4. While apomicts are by nature clonal and can produce seeds of identical genotype to the mother plant, no major crops have this self-reproducing capability. Elucidating molecular mechanisms of apomixis will significantly enhance the plant breeding toolbox for preserving genetic composition of elite hybrid cultivars.
Significant strides have been made in deciphering the genetic control of apomixis in a wide range of natural apomicts3, although genes underlying the multiple components comprising apomixis are still not fully elucidated. In the natural apomict Pennisetum squamulatum (syn. Cenchrus squamulatus), mapping in a F1 population derived from a cross between Pennisetum glaucum (syn. C. americanus; pearl millet) x P. squamulatum segregating for apospory and sexuality revealed 12 apospory-linked markers, which defined a contiguous apospory-specific genomic region (ASGR)5. The linkage of many of these markers to apospory was further shown in C. ciliaris6. The physical size of the ASGR in P. squamulatum was determined to encompass ~ 50 Mb based on fluorescent in situ hybridization mapping of multiple ASGR-linked and one ASGR-recombinant BAC clones. The physical size of the ASGR in C. ciliaris could not be determined due to a lack of identified ASGR-recombinant BAC clones; however, the co-linearity of shared ASGR-linked BAC clones between the two species suggested that the two apomictic species share a similar physical size7,8,9. Sequencing of ASGR-linked bacterial artificial chromosome clones from P. squamulatum and C. ciliaris led to the identification of genes within the region including an AP2-domain containing transcription factor (ASGR-BABY BOOM-like, ASGR-BBML)10,11. The PsASGR-BBML transgene was shown to induce parthenogenesis in sexual pearl millet as well as in maize and rice, but not in Arabidopsis12,13.
Comparative genomics and transcriptomics studies of apomictic plants and their sexual relatives or siblings often can help unlock the functional molecular components of apomixis that have not been genetically tractable. However, genomes of most natural apomicts remain unsequenced, and targeting the critical tissues where components of apomixis are expressed for RNA-seq has been biologically and technically challenging14. Recent expression studies have focused on and advanced our understanding of the first component in the apomixis pathway, termed apomeiosis, by investigating differentially expressed molecular signatures between sexual and apomictic reproductive tissues. Apomeiosis encompasses the initiation of mitotic divisions leading to the production of unreduced embryo sacs derived either from a somatic cell of the ovule nucellus (apospory) or from a megaspore mother cell (diplospory). Differential expression in whole ovules, each containing one to a few apomeiotic cells, to more targeted cells isolated using laser capture microdissection (LCM) has been investigated15,16,17,21,22,23. Gene expression activity within monocots prior to and after fertilization has been interrogated in isolated egg/zygote cells of maize and rice, with an emphasis on zygotic genome activation24,25. An important missing component in current expression studies is the transcriptomic comparison between sexual and parthenogenetic egg cells. Parthenogenesis, embryo development of the egg cell without fertilization, is the second component of the apomixis pathway, with transcriptomic analysis providing an approach to expose pathways underlying natural parthenogenesis.
Discovery of genes driving parthenogenesis in the natural apomict C. ciliaris has been challenging due to limited genomic resources and technical difficulties in accessing the egg cell expressing parthenogenesis. Droplet-based single cell RNA-seq recently has been applied to acquire gene expression profiles for complex tissues such as Arabidopsis roots and developing ears of maize26,27. Yet it remains challenging for the current technology to capture extremely rare transcripts underlying core traits of interest in sparse and spatially-restricted cells like eggs. By conducting laser capture microdissection (LCM)-based RNA-seq on unfertilized sexual and parthenogenetic eggs on the day of anthesis and deep sequencing, combined with de novo transcriptome assembly and computational analyses, we created a de novo transcriptome with sequence information for C. ciliaris eggs. Transcriptional profiles that distinguish the parthenogenetic egg from its sexual counterpart were identified, and suggested functional roles for a few key transcription factors and pathways in promoting natural parthenogenesis. Our transcriptome data complement previous gene expression studies and will be an important resource for research on natural parthenogenesis.
Results and discussion
Parthenogenetic embryo frequency
Twelve percent of unpollinated ovaries from obligately apomictic genotype B-12–9, two days after anthesis, were shown to contain parthenogenetic embryos (Supplementary Fig. S1; Table 1) as evidenced by the presence of a multicellular embryo without endosperm development deduced from retention of the polar nuclei. This frequency of parthenogenesis in unpollinated ovaries of apomictic C. ciliaris is consistent with observations of Veille-Calzada28 who reported 7–27% proembryos in unpollinated ovaries of different genotypes. No parthenogenetic embryos were observed in the sexual genotype B-2s. These data confirm the apomictic and sexual phenotypes of the plants used in this experiment and the obligate nature of sexuality for B-2s.
Egg identification and collection
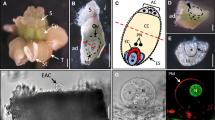
Serial sections of sexual C. ciliaris genotype B-2s showed a single sexual embryo sac containing the egg apparatus at the micropylar end, two polar nuclei, and antipodals at the chalazal end (Fig. 1A–F). In aposporous genotype B-12–9, an egg apparatus and polar nuclei, but no antipodals, were observed in the aposporous embryo sac nearest to the micropylar end of the ovule (Fig. 1G–L). An additional aposporous embryo sac also was frequently detected. This is consistent with previous descriptions of aposporous embryo sac development in C. ciliaris29.
Buffelgrass ovary sections stained with Safranin & FastGreen. (A–F) Serial sections of sexual genotype B-2s showing a single sexual embryo sac. (G–L) Serial sections of aposporous genotype B-12–9 showing multiple aposporous embryo sacs. Egg cells are indicated by arrows and polar nuclei are indicated by triangles, scale bar = 50 μm. Enlarged insets have been provided for some images.
De novo egg, ovule, and ovule plus egg transcriptome assemblies
De novo assembly and annotation of the C. ciliaris egg transcriptome was challenging for several reasons. (1) No annotated reference C. ciliaris genome sequence is available to guide transcriptome assembly. (2) The ultra-low input quantity of egg cell RNA required multiple cycles of cDNA amplification for downstream library preparation, potentially introducing PCR-generated nucleotide errors within the sequencing reads resulting in an excess of allelic variants. (3) The inevitable compromised quality of LCM-derived RNA and a poly-A based cDNA synthesis strategy resulting in the loss of 5′mRNA ends30.
To partially address these challenges, high-quality, abundant ovule RNA was used for amplification-free cDNA library construction. The resulting reads were used to improve the assembly and annotation of LCM egg reads based on the hypothesis that the egg, as a biological subset of ovule tissues, will be represented in the whole ovule transcriptome with more complete transcripts than in the LCM egg transcriptome. Therefore, three de novo assemblies were constructed using Trinity: ovule only, LCM egg, and LCM egg plus ovule. Statistics, metrics for completeness, and number of unique plant annotations for each assembly were compared (Table 2).
As expected, a significantly longer N50 (1747 vs 433 bases) and higher overall alignment rate (96.1% vs 77.6%) were achieved in the ovule assembly compared with the LCM egg, while the combined LCM egg plus ovule assembly had a slightly lower N50 than ovule alone. By subjecting our transcriptome assemblies to BUSCO analysis31, the ovule transcriptome was found to be near-complete, the LCM egg transcriptome highly fragmented and the combined LCM egg plus ovule transcriptome slightly less complete than ovule alone. A BLASTN (e-value < 1e−10) search against the NCBI nt database found the greatest number of unique plant hit descriptions (36,861) in the LCM egg plus ovule assembly with slightly fewer (33,826) in the ovule assembly. The fewest unique plant hit descriptions (23,049) were found in the LCM egg assembly. These observations indicate that most LCM egg transcripts are represented in the ovule transcriptome, and that the LCM egg plus ovule assembly not only properly captured the diversity from both individual assemblies but also captured transcripts that failed to be assembled and annotated solely by LCM egg or ovule data alone. For example, CcASGR-BBML reads were detected in the apomictic ovule read data but no contig was present in the ovule assembly, whereas a CcASGR-BBML contig (TRINITY_DN28916_c0_g1_i1) was identified in the LCM egg plus ovule assembly.
The number of contigs in the LCM egg assembly and in the LCM egg plus ovule assembly with detectable egg cell expression (hit count > 0 across all LCM egg libraries) was examined (Table 3). The LCM egg plus ovule assembly showed an additional 14,831 contigs with LCM egg expression (a 9.8% increase) and a 26.8% increase in the number of annotations identified. These results support the combination of reads generated from the LCM egg and ovule to improve both assembly and annotation of egg cell sequences. The LCM egg plus ovule assembly was used for the downstream analyses.
Cell-type specific expression patterns
The top 50 most abundantly expressed LCM egg transcripts accounted for 12.7% of total LCM egg reads and included constitutively expressed transcripts from mitochondrion, chloroplast, and ribosomal protein genes (Supplementary Table S2). Among the 165,998 transcripts expressed in the LCM egg, 91,730 (55.26%) had hits in the NCBI nt database (e-value < 1e−10), and 71,631 of those (78.1%) to a close relative, Setaria italica (foxtail millet), consistent with the known evolutionary relationship between these species32,33,34.
To further check the fidelity of our transcriptome data, the expression of egg cell-specific gene EC1 (egg cell 1)35, synergid predominant gene MYB9836 and a previously experimentally verified parthenogenesis gene ASGR-BBM-Like12 was examined. Three potential EC1 orthologs (EC1.2-like, EC1.3 and EC1.4) were expressed in all parthenogenetic and sexual LCM egg libraries (Fig. 2). RNA in situ hybridization experiments using a ssRNA probe designed from the CcEC1.3 transcript (TRINITY_DN34886_c2_g1_i5) further confirmed its egg cell expression specificity in both sexual and apomictic ovary sections (Fig. 3A,B). MYB98, identified as one of the top five synergid-enriched transcripts in rice [Log2 (synergid/egg cell) = 7.2337], was detected at extremely low levels in the LCM egg reads, showing that egg cells likely were preferentially captured during LCM or that synergids in the egg apparatus from apomicts differed in their gene expression pattern from those in sexuals (Fig. 2). CcASGR-BBM-like expression was detected in all parthenogenetic LCM egg libraries, and was completely absent from the sexual LCM egg libraries, indicating correctly captured eggs with or without parthenogenesis expression. RNA in situ hybridization experiments with an ASGR-BBML ssRNA probe also confirmed expression specifically in the parthenogenetic egg cell (Fig. 3C,D). The expression patterns of EC1, MYB98-like and CcASGR-BBM-like genes in our data, based on TPM and RNA in situs, correlate with previously published expression patterns12,37 and support the highly enriched egg-cell specificity of our LCM egg reads.
RNA in situ hybridization of sexual (B-2S) and parthenogenetic (B-12–9) ovary sections at day of anthesis. (A) A sexual ovary section displaying egg-cell specific signal with an EC1.3 probe. (B) A parthenogenetic ovary section with multiple aposporous embryo sacs, displaying two egg-cell signals with an EC1.3 probe. (C) A sexual embryo sac displaying observed background/sense signal. (D) A parthenogenetic ovary section with a single aposporous embryo sac displaying egg-cell CcASGR-BBML signal. Black outlines define both sexual and aposporous embryo sacs. Sexual embryo sacs are denoted with brackets defining the antipodals (ANT) which do not develop in aposporous embryo sacs. The red arrow denotes the egg cell.
Differentially expressed embryonic transcription factors reveal distinctions between parthenogenetic and sexual eggs
The expression profiles across all samples were clustered through principle component analyses with excellent correlation among biological replicates clustering tightly according to genotype and tissue type (Fig. 4). Pairwise comparisons of parthenogenetic LCM egg reads to sexual LCM egg reads were performed to identify differentially expressed genes (DEGs) as those with log2FC > 2, false discovery rate (FDR) < 0.05 using DESeq238. Of the 4,625 differentially expressed Trinity transcripts (Supplementary Table S3), 2,571 were up-regulated in the parthenogenetic egg, illustrated in a heat map that includes all the differentially expressed transcripts in parthenogenetic eggs relative to sexual eggs (Fig. 5). Several embryogenesis-related transcription factors were significantly expressed in the parthenogenetic eggs compared to sexual eggs (Fig. 6). Parthenogenetic up-regulated DEGs were further subjected to gene ontology enrichment analyses which identified 175 over-represented GO terms (Supplementary Table S4) including terms such as embryo development (GO: 0,009,790), reproductive process (GO: 0,022,414), transcription factor activity and transcription factor binding (GO: 0,000,989). In a similar enrichment of GO-terms for up-regulated genes in a comparison of parthenogenetic eggs from Boechera and sexual eggs from Arabidopsis39, the only category in common with our results was for regulation of transcription, DNA-templated (GO:0,006,355). This difference could be attributed to developmental differences between apomictic eudicot Boechera and the apomictic monocot Cenchrus where parthenogenesis in Boechera is repressed in the absence of central cell fertilization40.
To assess major regulatory distinctions between parthenogenetic and sexual egg transcripts, LCM egg plus ovule transcripts were subjected to a BLASTX (e-value < 1e-10) search against the plantTFDB (Plant Transcription Factor Database41). The search identified 72 potential transcription factors spanning 27 families de novo expressed in parthenogenetic eggs and 21 potential transcription factors spanning 11 families de novo expressed in sexual eggs (Fig. 7; Supplementary Table S5). The AP2 and WOX TF families were among 16 TF families that were exclusively de novo expressed in parthenogenetic eggs. These gene families have been shown to play major roles in embryogenesis and embryo development in monocots and eudicots42,43. Using microarrays, a fertilization-induced WOX gene was identified in rice zygotes 2-3 h post-pollination44. From RNA-seq data, a comparison of transcription factor families up-regulated in maize zygotes 12 h after pollination compared with maize eggs showed considerable overlap with those up-regulated in C. ciliaris parthenogenetic versus sexual eggs, namely AP2, bHLH, bZIP, C3H, GRAS, Homeobox (WOX), MADS, MYB, NAC, Trihelix, WRKY, YABBY, and ZF-HD25. At least for transcriptional regulation, this supports our hypothesis that parthenogenetic eggs resemble early stage zygotes that have initiated the maternal to zygotic transition25,45.
As expected, an ASGR-BABY BOOM-like (ASGR-BBML) gene was de novo expressed in parthenogenetic eggs. Expression of this apomixis-locus derived gene induces parthenogenesis in unfertilized eggs of sexual pearl millet, rice and maize in the absence of pollination12,13. Ectopic overexpression of Brassica BBM induces somatic embryogenesis in Arabidopsis46 and the rice BBM1 (LOC_Os11g19060.1) transgene, expressed from an egg-cell-specific promoter, induces embryogenesis in rice eggs without fertilization47. Native OsBBM1 expression in rice was detected in zygotes at 2.5 h after pollination (HAP) (corresponding to karyogamy) in a male-origin-specific manner, and ectopic expression in egg cells under an egg-cell-specific promoter induced parthenogenesis. Differential gene expression analysis also provided evidence for de novo expression of a C. ciliaris BBM2-like gene (TRINITY_DN31800_c0_g1_i1) in unfertilized parthenogenetic LCM eggs. The parthenogenesis gene CcASGR-BBML was de novo expressed at very low levels (average TPM = 3.7) in parthenogenetic eggs, while the CcBBM2-like gene was also de novo expressed in parthenogenetic eggs with relatively abundant expression (average TPM = 44.6). No read counts were identified for the CcBBM2-like transcript in sexual egg sequences. Based on the observation that BBM has a transcriptional auto-activation nature48, we speculate that CcBBM2 expression may be due to CcASGR-BBML activation and CcBBM2 auto-activation. The parthenogenesis pathway may be initiated by CcASGR-BBML and proceed through its activation of CcBBM2 together with CcBBM2 auto-activation to further promote cell proliferation and embryogenesis.
C. ciliaris WUSCHEL-related homeobox 7 (WOX7) genes (TRINITY_DN42296_c3_g1_i3 and TRINITY_DN42296_c3_g1_i2) showed de novo and up-regulated expression in parthenogenetic eggs, respectively. Over-expression of maize BBM (orthologous to the C. ciliaris BBM2-like gene) and WUSCHEL2 in somatic tissues of maize, sorghum, rice and sugarcane greatly increases embryogenic potential thereby enhancing transformation frequency49. It is possible that the de novo-expressed and up regulated WUSCHEL from parthenogenetic eggs works in concert with BBM2 to promote parthenogenesis. It is noteworthy that BBM and WUSCHEL were de novo expressed in rice zygotes, 2.5 h after pollination at karyogamy24 suggesting a role in promoting zygotic embryogenesis. WUSCHEL is thought to be required for maintaining meristem identity50 and its overexpression can induce organogenesis and somatic embryogenesis in shoot and root tissues42,51. Several genes overlapped between our DEGs and WUSCHEL-target genes52 (Supplementary Table S6) but none of them plays a central role in embryogenesis suggesting that some of the WUSCHEL-mediated transcriptional change may be ancillary to regulating parthenogenesis or that another set of embryogenesis-related targets is specific to CcWOX7.
Besides CcBBM2 and CcWOX7, embryonic factor CcAINTEGUMENTA-LIKE 5 (AIL5) gene (TRINITY_DN44016_c2_g3_i3 and TRINITY_DN44016_c2_g3_i4) showed de novo and up-regulated expression in parthenogenetic LCM egg. Ectopic expression of Arabidopsis AIL5 can induce somatic embryo formation in Arabidopsis53. The identification in parthenogenetic eggs of up-regulated and de novo expressed embryonic transcription factors that previously have been shown to function in parthenogenesis, somatic embryogenesis, or zygotic embryogenesis, demonstrated the utility of our data and confirmed that embryogenic potential is one of the major functional distinctions between parthenogenetic and sexual eggs.
Transcriptional change in parthenogenetic eggs may confer the core parthenogenesis pathway
Based on prior research that the transcription factor ASGR-BBML is the only experimentally verified apomict-derived parthenogenesis gene12,13, and that BBM transcription factors are known to function with other proteins to control cell proliferation and somatic embryogenesis48,54,55, we hypothesize that the core natural parthenogenesis pathway initiated by ASGR-BBML may advance through interactions with other TFs followed by up- or down-regulation of their target genes.
Candidate BBM-target genes that were directly activated by BBM expression in Arabidopsis seedlings and those with DNA sites bound by BBM in Arabidopsis somatic embryos through chromatin immunoprecipitation sequencing (ChIP-seq)48,54,55 were examined. Overlap between these previously reported genes and our differentially expressed genes was found. Putative orthologs were identified through BLASTN description annotation (NCBI nt database) similarities and/or sequence similarities through BLASTX search using full length contigs (their corresponding best hit in NCBI nt database) against Araport11_genes.201606.pep (1e−5) (Supplementary Table S7). These included BBM and AIL5 described above. In addition, potential orthologs (TRINITY_DN34773_c0_g1_i2 nuclear transcription factor Y subunit A-1 and TRINITY_DN30545_c0_g1_i1 nuclear transcription factor Y subunit A-10) of CcNUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT A-9, similar to another BBM target gene thought to play a role in inducing embryogenesis when overexpressed in Arabidopsis56, were upregulated in parthenogenetic eggs suggesting that BBM may also interact with NF-YA to control embryogenesis.
Apart from the parthenogenetic egg up-regulated transcripts with similarity to BBM-target genes that correspond to genes with known function to induce embryogenesis, those that may play roles in transcription, signaling, protein–protein interaction and cytoskeleton organization that function in embryogenesis also were identified. For example, two members of a well-studied gene family ACTIN DEPOLYMERIZING FACTOR (ADF), ADF2 (TRINITY_DN36541_c0_g1_i1, Log2FC = 6.34, FDR = 0.03, average TPM in parthenogenetic egg = 40.03) and ADF5 (TRINITY_DN37217_c0_g3_i4, Log2FC = 8.75, FDR = 0.001, average TPM = 12.35), thought to control actin dynamics57 and reorganize the actin cytoskeleton under BBM activation48, were up-regulated in parthenogenetic eggs.
Histone modification and chromatin remodeling factors are differentially expressed in parthenogenetic vs sexual eggs
The chromosomal regions associated with apomixis in C. ciliaris are largely heterochromatic8, suggesting potential for epigenetic regulation. Comparative small RNA-seq between sexual and apomictic spikelets of Paspalum notatum provided evidence that RNA-dependent regulation of auxin pathways may be important for the expression of apomixis20. A comparative transcriptomics study between sexual and apomictic ovule nucellus at a pre-meiotic developmental stage in Hypericum perforatum L. showed that RNA-mediated DNA methylation, histone and chromatin modifications were associated with aposporous apomixis expression19. Moreover, a methylation status analysis of the apomixis-specific region in Paspalum spp. suggests a possible epigenetic control of parthenogenesis, where the factors controlling repression of parthenogenesis might be inactivated in apomictic Paspalum by DNA methylation58.
Chromatin states are mainly determined via two mechanisms: covalent modifications of histones and DNA methylation59. Major histone modification events can be categorized into acetylation, methylation, phosphorylation and ubiquitination60, which affect chromatin states resulting in activation or repression of gene expression61,62,63. In plants, studies have shown that histone acetylation/deacetylation and histone methylation play a fundamental role in regulating plant development.
Among DEGs between parthenogenetic and sexual eggs, multiple histone deacetylase genes (TRINITY_DN41443_c1_g1_i1, TRINITY_DN39020_c3_g1_i2, TRINITY_DN37516_c2_g2_i6, and TRINITY_DN42464_c1_g1_i7) were exclusively up-regulated in parthenogenetic eggs while a single putative MYST histone acetyltransferase gene (TRINITY_DN38730_c1_g1_i7) was up-regulated in sexual eggs. Hypoacetylation is known to be associated with chromatin condensation and transcriptional suppression63, which suggests that there are some genomic regions in parthenogenetic eggs that are hypoacetylated and genes within those regions may be transcriptionally silent. Alternatively, evidence is emerging in plants for a balance of acetylation/deacetylation for gene activation64. The short fruit phenotype in cucumber results from a mutation in Histone deacetylase complex1 (HDC1; SF2 in cucumber), a gene whose product interacts with multiple histone deacetylases to enhance expression of cell cycle, DNA replication, and chromatin assembly genes and promote cell division, but also to repress expression of suppressors of auxin, gibberellin, and cytokinin biosynthesis and responses. Limited histone 3 acetylation in gene bodies and properly acetylated promoters and enhancers were shown to promote transcriptional elongation, at least in mammalian cells65.
Another potential role for histone deacetylases is to allow methylation of the H3K9 residue66,67 and interaction with DNA methyltransferase to repress gene expression68. Multiple histone methylation factors were differentially expressed, most being up-regulated in parthenogenetic eggs. Among these were Cchistone-lysine N-methyltransferase ATXR4 (TRINITY_DN34478_c0_g1_i3), Cchistone-lysine N-methyltransferase TRX1 (TRINITY_DN46041_c2_g1_i2), CcH3 lysine-9 specific SUVH1 (TRINITY_DN42267_c0_g1_i1), and CcH3 lysine-9 specific SUVH4 (KRYPTONITE) (TRINITY_DN46013_c1_g1_i6). SUVH4 encodes a protein that methylates H3K9, is required for the maintenance of cytosine methylation in the CpNpG context, and represses retrotransposon activity in Arabidopsis69. Another parthenogenetic up-regulated histone-lysine N-methyltransferase gene, CcH3 lysine-9 specific SUVH1 shows similarity with a gene in Arabidopsis that is associated with heterochromatic H3K9 dimethylation, and may play a role in heterochromatin gene silencing70. Apart from histone modification, DNA methylation is also involved in determining chromatin states. One of the best characterized classes of DNA methylation genes in Arabidopsis, DOMAINS REARRANGED METHYL TRANSFERASE2 (DRM2), responsible for de novo DNA methylation in all sequence contexts and mediation of transgene silencing71, was represented by one CcDRM2 gene (TRINITY_DN36171_c0_g1_i3), de novo expressed in parthenogenetic eggs. Another CcDRM2 gene (TRINITY_DN46376_c0_g4_i1) was up-regulated in sexual eggs. The de novo expression of one CcDRM2 gene in parthenogenetic eggs and the up-regulation of another in sexual eggs implies that increased de novo DNA methylation may be common to both parthenogenetic eggs with parthenogenetic fate and sexual eggs with embryogenesis fate upon fertilization. This is consistent with the observation that embryogenesis is characterized by increased de novo DNA methylation72. However, it is not known if these two CcDRM2 genes function somehow to differentiate sexual and parthenogenetic egg cell fate through de novo DNA methylation.
Hypoacetylation and H3K9 methylation activities suggest enhancement of repressive chromatin states in parthenogenetic eggs even though they are transitioning to an active stage of cell division. Specific chromatin regions may be repressed in parthenogenetic eggs while others are derepressed in order to release totipotency and achieve competency to initiate parthenogenesis. Day0 unpollinated parthenogenetic eggs are biologically active in terms of having the capacity to form embryos two days after anthesis and are transcriptionally active as indicated by GO enrichment analyses reflecting the active side of chromatin states. Several Jumonji-like proteins were up-regulated in parthenogenetic eggs including JMJ25-like (TRINITY_DN44176_c1_g1_i3; TRINITY_DN46670_c1_g4_i1) which demethylates H3K9, regulating gene expression through epigenetic modifications73,74. Histone 3 lysine 9 methylation homeostasis may be regulated by the interplay of Jumonji and SUVH proteins. Histone 3 lysine 4 methylation, positively associated with actively transcribed genes75, was indicated by a COMPASS-like H3K4 histone methylase component WDR5B gene and a H3K4 histone-lysine N-methyltransferase CcTRX1 gene, both up-regulated in parthenogenetic eggs. Similar genes were shown to activate flowering under long day conditions76,77. A rice ortholog was shown to promote flowering by interacting with a DNA-binding C2H2 zinc finger protein SIP1 to activate EARLY HEADING DATE 1, a B-type response regulator78. Although the histone-lysine N-methyltransferase ATXR4 was up-regulated in parthenogenetic eggs, no direct evidence is available to infer its function. In Arabidopsis ATX1, ATX2, ATXR3, and ATXR7 positively regulate FLC via H3K4 methylation79,80,81,82 while ATXR5 and ATXR6 control the heterochromatin condensation and heterochromatic elements silencing via methylation of H3K2783. Taken together, the differential expression of histone deacetylases and histone methylases as well as a de novo expressed DNA methyltransferase is likely to both repress and activate a spectrum of genes that characterize parthenogenetic eggs assuming a parthenogenesis fate.
Cell cycle
The idealized cell cycle is comprised of four successive phases: G1 (Gap 1), S (Synthesis), G2 (Gap 2) and M (Mitosis), through which parthenogenetic egg cells must traverse as they become competent for cell division. The expression profiles of some key cell-cycle regulators were examined in an attempt to identify the cell cycle stages for Day 0 parthenogenetic and sexual egg cells.
A CcBREAST CANCER SUSCEPTIBILITY 1 (BRCA1) transcript (TRINITY_DN29461_c0_g1_i1) was up-regulated in parthenogenetic eggs. This gene plays a role in the G2/M checkpoint, particularly upon detection of DNA damage, limiting the proliferation of cells containing replication defects84,85. Similarly, another gene, CcSUPPRESSOR OF GAMMA RESPONSE 1 (SOG1) (TRINITY_DN39490_c0_g1_i2), that responds to DNA damage signals, and whose phosphorylated form controls several cell cycle and DNA damage repair genes86, was up-regulated in parthenogenetic eggs. BRCA1 has been identified as a target of SOG1, but was uniquely up-regulated in parthenogenetic eggs compared with other common targets. Since the expression of BRCA1 was barely detectable in the sexual eggs, the parthenogenetic egg might have reached the G2/M checkpoint while the sexual egg remained at a stage prior to G2. Aside from a potential cell cycle marker, it is tempting to speculate that an apomict undergoing many cycles of clonal reproduction through seeds may have adapted gene expression to ensure DNA repair in somatic cell-derived eggs destined to undergo parthenogenesis.
Auxin and calcium
Auxin plays a role in virtually every aspect of plant growth and development including early embryogenesis87 and gamete specification in the female gametophyte88. In relation to apomixis, auxin treatment of the inflorescence of apomictic Poa pratensis was used to rapidly and reliably test whether the egg cell was competent for parthenogenesis, showing that exogenously supplied auxin promoted the expression of apomictic parthenogenesis89.
Among DEGs, an auxin receptor CcTRANSPORT INHIBITOR RESPONSE 1 (TIR1) gene (TRINITY_DN40331_c0_g1_i1)90,91,92 was up-regulated in parthenogenetic eggs indicating that the parthenogenetic egg may differ from the sexual egg in auxin perception. Interestingly, auxin negative regulators CcAUX/IAA genes (IAA17, TRINITY_DN37013_c0_g1_i2; IAA2, TRINITY_DN44579_c0_g3_i1; IAA31, TRINITY_DN39140_c0_g1_i3; IAA29, TRINITY_DN32631_c0_g1_i2) were exclusively up-regulated in parthenogenetic eggs. AUX/IAAs are thought to interact with SCFTIR1 and lead to ubiquitination and degradation through the 26S proteasome in the presence of high auxin concentrations, derepressing auxin response factors (ARFs) to allow ARFs to transcriptionally activate or repress downstream auxin responsive genes93,94. Interestingly, 10 BTB/POZ-domain transcripts were up-regulated in parthenogenetic eggs versus only one in sexual eggs, suggesting an active proteasomal ubiquitin-mediated degradation pathway in parthenogenetic eggs. At least one of these BTB/POZ-domain transcripts contains an ankyrin repeat domain similar to BLADE ON PETIOLE (BOP) lateral organ boundary genes95. The ankyrin repeat interacts with basic Leu repeat transcription factors96 for which six transcripts were up-regulated in parthenogenetic eggs. These up-regulated genes suggest a regulatory network in parthenogenetic eggs involving a dynamic transcriptional and protein turnover response to hormones shaping growth and development. Induction of auxin pathway genes in maize zygotes 12 h after pollination was also observed25.
Crosstalk between auxin and ethylene signaling pathways leading to cooperative regulation of developmental processes has been documented97. ETHYLENE INSENSITIVE 2 (EIN2; TRINITY_DN47673_c2_g1_i1) along with 14 ethylene responsive transcription factors were up-regulated in parthenogenetic eggs versus only one in sexual eggs. Among these transcription factors were the embryogenesis related genes already discussed, BBM2 and AIL5, but also other AP2-domain containing, ethylene responsive genes CYTOKININ RESPONSE FACTOR (CRF1; TRINITY_DN40560_c0_g1_i1), ABA INSENSITIVE (ABI4; TRINITY_DN49291_c1_g1_i1), RELATED TO APETALA2 (RAP2; TRINITY_DN39096_c2_g1_i2; TRINITY_DN46047_c0_g3_i2), ERF61 (TRINITY_DN42510_c2_g1_i1), ERF73 (TRINITY_DN40591_c1_g2_i1), and WAX INDUCER 1 (WIN1; TRINITY_DN42753_c2_g1_i3). These transcription factors may integrate multiple hormone responses.
Calcium ionophore is reported to stimulate parthenogenesis in mouse oocytes98 but calcium alone is not sufficient to induce parthenogenesis in plants. As for plants, Ca2+ changes in egg cells are widely thought to be associated with successful fertilization and egg activation99,100,101. In our data, intracellular calcium receptor protein genes, calmodulin (TRINITY_DN44663_c0_g1_i2; TRINITY DN33744_c3_g1_i1; TRINITY_DN34327_c1_g1_i3)102 and its effector calcium/calmodulin-dependent protein kinases (TRINITY_DN44558_c1_g2_i1; TRINITY_DN40595_c1_g3_i1)103, were exclusively up-regulated in parthenogenetic eggs compared to the sexual eggs, suggesting the presence of an internal Ca2+ increase and calcium-triggered signaling pathway in parthenogenetic eggs. This may reflect some aspects of parthenogenetic egg activation but its correlation to parthenogenesis is still unknown.
With the differentially expressed genes and their annotations generated in this study, the potential core pathways diagnostic of natural parthenogenesis in an apomictic grass species and pathways that may play an ancillary role in promoting parthenogenesis based on sequence similarities to previously identified genes and pathways were discussed. However, to experimentally test our hypotheses, chromatin immunoprecipitation-seq (ChiP-seq) on transcription factor-bound DNA would be essential to study target genes or protein-DNA interactions. Yeast two hybrid experiments may be needed to experimentally investigate physical interactions between proteins encoded by differentially expressed genes or other embryogenic factors identified in this study. Bisulfite- or ATAC(Assay for Transposase Accessible Chromatin)-sequencing could potentially be used to ultimately check the chromatin state differences between sexual and parthenogenetic egg cells, but both would be challenging at a single cell level in plants. Progress recently has been made to apply single nucleus Assay for Transposase Accessible Chromatin sequencing (sNucATAC-seq) technologies to decipher cell-type-specific pattern of chromatin accessibility in Arabidopsis roots104. As an initial step, this study has provided numerous sequence and computational analyses for sexual and parthenogenetic eggs, which are valuable for developing testable hypotheses to further explore natural parthenogenesis in grasses.
Materials and methods
Plant material and floret collection
The C. ciliaris plants used as source materials were vegetatively propagated tillers of the sexual genotype B-2s and natural apomictic genotype B-12–9105. Plants were grown in the greenhouse (24–30 °C) for head collection from July to September in 2016. Heads were bagged prior to stigma exsertion and stigmas were manually removed with tweezers upon appearance. The heads remained bagged until the day of anther exsertion (anthesis) at which time florets were collected from 8:00–11:00 am with fresh anthers half or fully exserted and immediately fixed in Farmer’s fixative (ethanol:glacial acetic acid, 3:1)106 and stored at 4 °C for overnight. Fixed florets were then transferred and stored in 70% ethanol (DEPC-treated water) at 4 °C.
Ovary clearing and microscopic observation
For ovary clearing, stigma-free heads were collected on the day of (Day0) and two days after (Day2) anthesis and immediately fixed in Farmer’s fixative. Ovaries were dissected from florets, and dehydrated in a graded ethanol series for two hours in each step as follows: 70% ethanol, 85% ethanol, and 100% ethanol, followed by another 100% ethanol incubation overnight. Ovaries were transferred to a methyl salicylate series for 2 h in each step as follows: ethanol: methyl salicylate (3:1), ethanol: methyl salicylate (1:1), ethanol: methyl salicylate (1:3), and followed by a 100% methyl salicylate incubation overnight107. Cleared ovaries were observed under DIC (differential interference contrast) microscope (Supplementary Fig. S1).
Ovule collection, RNA extraction, cDNA Synthesis, amplification, library construction and sequencing
Intact ovaries were dissected from fixed florets on ice using fine tweezers to avoid physical damage. Ovules were subsequently isolated by pulling apart the bifurcated stigma to separate the ovary wall from the ovaries. Fifty ovules per sample were collected. Ovule RNA was extracted using RNeasy Plant Mini Kit (QIAGEN) following the manufacturer’s protocol. The quantity and quality of RNA were checked using the Qubit 2.0 Fluorometer RNA assay (Invitrogen) and an Agilent 2100 Bioanalyzer using a RNA 6000 nano kit (Agilent Technologies), respectively. For each sample, 500 ng of total RNA was used for poly-A capture, cDNA synthesis and amplification using KAPA stranded RNA-seq Kit. Libraries were quality checked with the Qubit 2.0 Fluorometer dsDNA high sensitivity assay (Invitrogen) and Fragment Analyzer Automated CE System (Agilent Technologies), and were sequenced on an Illumina NextSeq (300 Cycles) PE150 Mid Output flow cell on which three biological replicates of two genotypes were pooled.
Tissue preparation for laser capture microdissection
Intact ovaries were dissected on ice from fixed florets using fine tweezers to avoid physical damage and were dehydrated in a graded ethanol series on ice with gentle shaking for 20 min in each step as follows: 80% ethanol, 90% ethanol, and three changes of 100% ethanol. Ovaries were stored overnight in 100% ethanol at 4 °C, and transferred the next day to a xylene series with gentle shaking for 20 min in each step as follows: ethanol: xylene (3:1), ethanol: xylene (1:1), ethanol: xylene (1:3), and three changes of 100% xylene. Xylene-cleared ovaries were transferred to a Paraplast series in an incubator (54 °C) with shaking for 9 h in each step as follows: xylene and Paraplast (1:1) mixture and seven changes of 100% Paraplast. Paraplast-infiltrated ovaries were embedded in Paraplast blocks using a tissue embedding center, and 8-µm sections were cut on a rotary microtome (Leica RM2145, Germany). Sections were floated on methanol and mounted on metal frame PET foil slides (Leica) that had been UV-irradiated (DNA transfer lamp, Fotodyne) for 30 min. Slides were heated on a slide warmer at 42 °C overnight to air-dry and stretch the Paraplast ribbons. Prior to egg identification and laser capture microdissection, PET foil slides with ovary sections were de-paraffinized in xylene for two changes of 5 min each with gentle shaking and air-dried for 1 h in a fume hood. A few ovary sections were stained with Safranin and FastGreen to check tissue quality, and the egg cell was identified under a microscope (Fig. 1). Unstained sections were used for LCM.
Laser capture microdissection and egg cell collection
A Leica LMD6000 laser microdissection system was used to capture the egg cell from tissue sections prepared as above. Before loading PCR tubes with cap into the collection device, 8 μl of RNAlater was pipetted into the lid of the cap. Microdissections were then performed by drawing a line or circle around the egg along which the laser beam cut (Supplementary Fig. S2), with the laser setting as follows: Power 45, Aperture 4, Speed 9 and Specimen balance 15. After the LCM, the egg sections fell into the lid by gravity, were preserved by the RNAlater, and stored at −80 °C until use. Around 1000 egg sections (from 500 ovules) per biological replicate (4 B-2s and 4 B-12–9) were collected.
Egg RNA extraction, cDNA Synthesis, amplification, library construction and sequencing
Egg RNA was extracted using RNeasy Plus Micro Kit (QIAGEN) following the manufacturer’s protocol with minor modifications. The quantity and quality of RNA were checked using the Qubit 2.0 Fluorometer RNA assay (Invitrogen) and an Agilent 2100 Bioanalyzer using an RNA 6000 Pico kit (Agilent Technologies), respectively. For each of the samples, 2 ng of total RNA was used for cDNA synthesis and amplification using SMART-Seq v4 Ultra Low Input RNA Kit (Clontech) with modifications. Modifications included increasing the elution buffer volume, resuspension time, constantly mixing the cDNA-bound beads as well as increasing cDNA amplification cycles to improve the cDNA recovery and yield. The concentration and profile of the amplified cDNA were checked using the Qubit 2.0 Fluorometer dsDNA high sensitivity assay (Invitrogen) and the Fragment Analyzer Automated CE System (Agilent Technologies), respectively. For each sample, 100 ng of cDNA was sheared using a Covaris E220 Evolution with 350 bp insert setting and was used for library construction with the TruSeq DNA Nano LT Kit (Illumina) following the manufacturer’s suggested protocol with modifications mainly by doing twice the Enrich DNA Fragments step. Libraries were quality checked with the Qubit 2.0 Fluorometer dsDNA high sensitivity assay (Invitrogen) and Fragment Analyzer Automated CE System (Agilent Technologies), and were sequenced in two separate runs of NextSeq (300 Cycles) PE150 Mid Output flow cell on which two biological replicates of each genotype were pooled. Further details of modified protocols can be found in Supplemental Experimental Procedures.
Data preprocessing
Both ovule and egg raw reads were cleaned by removing adapter sequences, over-represented technical sequences detected by FASTQC108, and sequences of poor-quality using Trimmomatic109. For LCM egg sequencing data, 5–7 bases from the 5′ and 3′ ends of the trimmed reads were further cut using TrimGalore110 to avoid potential sequence bias. Comprehensive rRNA removal was done using SortMeRNA111. To further identify and remove potential biological contaminants, a de novo assembly was constructed with cleaned reads from the eight LCM egg libraries using Trinity112,113. Trinity contigs were first annotated by a BLASTN114 search against the NCBI nt database (e-value cutoff of 1e−10). Using these annotations, a custom non-plant contamination database was constructed by extracting the aligned portion of the subject sequences from the contigs that hit (< 1e−10) bacterial, fungal and animal sequences stored in the NCBI nt database. The non-plant contamination database was indexed as the reference to clean each library until no significant amount of non-plant hit was seen in the final assembly. All cleaned reads were used for downstream analyses.
De novo trinity assembly
Trinity-2.8.4.simg was used for de novo transcriptome assembly. For the ovule plus egg transcriptome, cleaned egg and ovule reads (Supplementary Table S1) were combined into a single fastq file, assembled using Trinity (parameters –seqType fq –single –run_as_paired –max_memory 150G –CPU 8 –no_version_check –normalize_reads). Egg, ovule assembly was constructed using solely the cleaned egg, cleaned ovule reads respectively.
Trinotate annotation and gene ontology enrichment analyses
Trinity transcripts were annotated with Trinotate through a BLASTX114 search (e-value = 1e−5) against a comprehensive protein database comprised of the Swiss-Prot115 and UniRef90 protein databases. Putative coding regions within each Trinity transcript were predicted using TransDecoder (http://transdecoder.github.io), and the predicted coding sequences were further annotated through BLASTP (e-value = 1e−5) alignment against the comprehensive protein database mentioned above and for protein domains search using hmmer (http://hmmer.org/) and PFam116. SignalP117 was used to predict the potential signal peptides in the transcripts. All results were integrated by Trinotate, stored in an SQLite database, and then reported as a tab-delimited excel file (Supplementary Table S8). The functional enrichment analyses were done on significantly differentially expressed transcripts set against the whole expressed transcripts using Trinotate-assigned GO annotations and GOseq118, and enriched GO terms of parthenogenetic up-regulated transcripts were reported as a tab-delimited file.
BUSCO completeness analyses
The completeness of the transcriptome assembly was examined by subjecting the Trinity.fasta file to BUSCO31 analyses using the command run_busco -i Trinity.fasta -l liliopsida_odb10 -m tran.
Transcript abundance estimation and differential expression analysis
Kallisto119 software was used to quantitate the expression level for transcripts, and DESeq238 was used to identify significantly differentially expressed transcripts.
Plant transcription factor identification
The potential plant transcription factors in the trinity transcripts were identified by BLASTX search (e-value < 1e−10) against a close relative’s, Setaria italica, transcription factor database downloaded from http://planttfdb.cbi.pku.edu.cn/download.php41.
RNA in situ
Tissue for RNA in situs were prepared and sectioned according to the LCM protocol. Sections were floated on methanol and mounted on microscope slides (Fisherbrand Probe On Plus). Slides were heated on a slide warmer at 42 °C for overnight. Slides were de-paraffinized in xylene for 10 min twice, followed by a hydration series: 2 times of 100% ethanol, 95% ethanol, 70% ethanol, 50% ethanol, 30% ethanol, and 2 times of DEPC-treated water (2 min for each). Rehydrated slides were incubated in 0.2 M HCl for 10 min, and then washed in DEPC-treated water, 2 times of 2 × SSC (saline sodium citrate), then DEPC-treated water, 5 min each. Slides were then treated with a mixture of 100 mM Tris pH 8.0, 50 mM EDTA pH 8.0 and 1 μg/mL proteinase K at 37 °C for 20 min, followed by a wash in PBS (phosphate buffered saline) for 2 min. Slides were incubated in 2 mg/mL glycine in PBS for 2 min to block proteinase K, and washed in PBS for 30 s twice. Slides were fixed in 4% formaldehyde in PBS for 10 min and washed in PBS for 5 min twice, then dehydrated as follows: 2 times of DEPC-treated water, 30% ethanol, 50% ethanol, 70% ethanol, 95% ethanol and 2 times of 100% ethanol (2 min each). Slides were dried for 15 min in the fume hood.
Probe synthesis
The primers 5′-GGCGAGATCATCCTGTACCT-3′ and 5′-GTCGCAGTATCCCTTGAGCAT-3′ were designed based on the EC1.3 (egg cell-specific 1.3) Trinity transcript. The primers 5'-TTTAGCTGCTCTCAAGTACCGG-3' and 5'-CCTGGTAACCCCTCGGTAAATT-3' were designed from the PsASGR-BBML cDNA sequence (GenBank: EU559277.2). The amplicons were cloned into pGEM-T Easy Vector Systems (Promega), sequenced through Macrogen, and reverse transcribed into ssRNA probes using the MAXIscript SP6/T7 Kit (Invitrogen).
Hybridization
Hybridization buffer was made with each of the following components at a final concentration of: 50% HiDi formamide (Applied Biosystems),100 μg/ml Yeast tRNA (Invitrogen), 0.75% blocking reagents (Roche), 2.5% dextran sulfate (Millipore), 2.5 mM EDTA and DEPC-treated water to make up for the total volume. Probe (0.1 μl) was added to 10 μl of hybridization buffer and heated at 80 °C for 3 min. 190 μl hybridization buffer was added to 10 μl pre-denatured probe-hybridization buffer mixture and kept at 55 °C until use. For each slide with pretreated ovary sections, 200 μl pre-denatured probe-hybridization mixture were added, and sealed with another microscope slide on top. The slide pairs were incubated in a humidified environment at 50 °C for overnight.
Antibody reaction and detection
After incubation for hybridization, slides were first washed in 2 × SSC (Sigma-Aldrich) and 50% formamide (Sigma-Aldrich ACS reagent) for 1 h at 50 °C each, followed by another wash in TBS buffer (100 mM Tris–HCl pH 7.5, 400 mM NaCl) for 5 min. Slides were then incubated in 0.5% blocking reagents (Roche) in TBS for 1 h followed by a wash in 1% BSA (bovine serum albumin, Sigma-Aldrich), 0.3% (v/v) Triton X-100 (Sigma-Aldrich) in TBS for 30 min. Slides were further incubated in Anti-Digoxigenin-AP Fab fragments (Roche) : 1% BSA, 0.3% (v/v) Triton X-100 in TBS (1:3000) for 90 min, followed by two washes in 1% BSA, 0.3% (v/v) Triton X-100 in TBS for 30 min each. For detection, slides were first washed in detection buffer (100 mM Tris–HCl pH 9.5, 100 mM NaCl, 50 mM MgCl2) for 5 min, and then incubated in NBT/BCIP (Roche) detection buffer mixture (200 μl NBT/BCIP per 10 ml detection buffer) in dark for 12 h.
All experiments were performed in accordance with relevant institutional guidelines and regulations.
Data availability
The data that support the findings of this study are available within the paper and its supplementary materials published online.
References
Nogler, G. A. in Embryology of Angisoperms (ed B. M. Johri) 475–518 (Springer, 1984).
Bicknell, R. A. & Koltunow, A. M. Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16, S228–S245 (2004).
Ozias-Akins, P. & van Dijk, P. J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 41, 509–537 (2007).
Hojsgaard, D., Klatt, S., Baier, R., Carman, J. G. & Hörandl, E. Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit. Rev. Plant Sci. 33, 414–427 (2014).
Ozias-Akins, P., Roche, D. & Hanna, W. W. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. 95, 5127–5132 (1998).
Roche, D. et al. An apospory-specific genomic region is conserved between buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J 19, 203–208 (1999).
Akiyama, Y. et al. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 134, 1733–1741 (2004).
Akiyama, Y., Hanna, W. & Ozias-Akins, P. High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor. Appl. Genet. 111, 1042–1051. https://doi.org/10.1007/s00122-005-0020-5 (2005).
Goel, S. et al. Comparative physical mapping of the apospory-specific genomic region in two apomictic grasses: Pennisetum squamulatum and Cenchrus ciliaris. Genetics 173, 389–400 (2006).
Gualtieri, G. et al. A segment of the A pospory S pecific G enomic R egion (ASGR) is highly microsyntenic not only between the apomicts Pennisetum squamulatum and Cenchrus ciliaris, but also with a rice chromosome 11 cis-centromeric genomic region. Plant Physiol 140, 963–971 (2006).
Conner, J. A. et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 147, 1396–1411 (2008).
Conner, J. A., Mookkan, M., Huo, H. Q., Chae, K. & Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 112, 11205–11210. https://doi.org/10.1073/pnas.1505856112 (2015).
Conner, J., Podio, M. & Ozias-Akins, P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 30, 41–52. https://doi.org/10.1007/s00497-017-0298-x (2017).
Hojsgaard, D. Apomixis technology: separating the wheat from the chaff. Genes 11, 1. https://doi.org/10.3390/genes11040411 (2020).
Sharbel, T. F. et al. Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J. 58, 870–882 (2009).
Sharbel, T. F. et al. Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22, 655–671 (2010).
Okada, T. et al. Enlarging Cells Initiating Apomixis in Hieracium praealtum Transition to an Embryo Sac Program prior to Entering Mitosis. Plant Physiol. 163, 216–231. https://doi.org/10.1104/pp.113.219485 (2013).
Rabiger, D. S. et al. Generation of an integrated Hieracium genomic and transcriptomic resource enables exploration of small RNA pathways during apomixis initiation. BMC Biol. 14, 1. https://doi.org/10.1186/s12915-016-0311-0 (2016).
Galla, G. et al. Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) Accessions. Front. Plant Sci. 10, 1. https://doi.org/10.3389/fpls.2019.00654 (2019).
Ortiz, J. P. A. et al. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 20, 1. https://doi.org/10.1186/s12864-019-5881-0 (2019).
Rabiger, D. S. et al. Generation of an integrated Hieracium genomic and transcriptomic resource enables exploration of small RNA pathways during apomixis initiation. BMC Biol. 14, 86 (2016).
Galla, G. et al. Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) accessions. Front. Plant Sci. 10, 654 (2019).
Ortiz, J. P. A. et al. Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genom. 20, 487 (2019).
Anderson, S. N. et al. The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev. Cell 43, 349. https://doi.org/10.1016/j.devcel.2017.10.005 (2017).
Chen, J. Y. et al. Zygotic genome activation occurs shortly after fertilization in Maize. Plant Cell 29, 2106–2125. https://doi.org/10.1105/tpc.17.00099 (2017).
Wendrich, J. R. et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370, 810. https://doi.org/10.1126/science.aay4970 (2020).
Xu, X. et al. Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev. Cell 56, 557–568 (2021).
Vielle, J. P., Burson, B. L., Bashaw, E. C. & Hussey, M. A. Early fertilization events in the sexual and aposporous egg apparatus of Pennisetum ciliare (L.) Link. Plant J. 8, 309–316 (1995).
Sherwood, R. T., Young, B. A. & Bashaw, E. C. Facultative apomixis in buffelgrass. Crop Sci. 20, 375–379. https://doi.org/10.2135/cropsci1980.0011183X002000030022x (1980).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098. https://doi.org/10.1038/nmeth.2639 (2013).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. https://doi.org/10.1093/bioinformatics/btv351 (2015).
Donadio, S., Giussani, L. M., Kellogg, E. A., Zuolaga, F. O. & Morrone, O. A preliminary molecular phylogeny of Pennisetum and Cenchrus (Poaceae-Paniceae) based on the trnL-F, rp116 chloroplast markers. Taxon 58, 392–404 (2009).
Chemisquy, M. A., Giussani, L. M., Scataglini, M. A., Kellogg, E. A. & Morrone, O. Phylogenetic studies favour the unification of Pennisetum, Cenchrus and Odontelytrum (Poaceae): a combined nuclear, plastid and morphological analysis, and nomenclatural combinations in Cenchrus. Ann. Bot. 106, 107–130. https://doi.org/10.1093/aob/mcq090 (2010).
Kellogg, E. A. Evolution of Setaria. Vol. 19 (2017).
Sprunck, S. et al. Egg Cell-Secreted EC1 Triggers Sperm Cell Activation During Double Fertilization. Science 338, 1093–1097. https://doi.org/10.1126/science.1223944 (2012).
Kasahara, R. D., Portereiko, M. F., Sandaklie-Nikolova, L., Rabiger, D. S. & Drews, G. N. MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17, 2981–2992. https://doi.org/10.1105/tpc.105.034603 (2005).
Ohnishi, T. et al. Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol. 155, 881–891. https://doi.org/10.1104/pp.110.167502 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Schmidt, A. et al. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional hormonal and epigenetic regulation. . Plos Genet. 10, 1. https://doi.org/10.1371/journal.pgen.1004476 (2014).
Aliyu, O. M., Schranz, M. E. & Sharbel, T. F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 97, 1719–1731. https://doi.org/10.3732/ajb.1000188 (2010).
Jin, J. P., Zhang, H., Kong, L., Gao, G. & Luo, J. C. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42, D1182-D1187, doi:https://doi.org/10.1093/nar/gkt1016 (2014).
Zuo, J. R., Niu, Q. W., Frugis, G. & Chua, N. H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis . Plant J. 30, 349–359 (2002).
El Ouakfaoui, S. et al. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 74, 313–326. https://doi.org/10.1007/s11103-010-9674-8 (2010).
Abiko, M., Maeda, H., Tamura, K., Hara-Nishimura, I. & Okamoto, T. Gene expression profiles in rice gametes and zygotes: identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J. Exp. Bot. 64, 1927–1940. https://doi.org/10.1093/jxb/ert054 (2013).
Baroux, C. & Grossniklaus, U. in Maternal-to-Zygotic Transition Vol. 113 Current Topics in Developmental Biology (ed H. D. Lipshitz) 351–371 (2015).
Boutilier, K. et al. Ecotopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749 (2002).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95. https://doi.org/10.1038/s41586-018-0785-8 (2019).
Passarinho, P. et al. BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 68, 225–237 (2008).
Lowe, K. et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 28, 1998–2015. https://doi.org/10.1105/tpc.16.00124 (2016).
Laux, T., Mayer, K. F. X., Berger, J. & Jurgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96 (1996).
Gallois, J. L., Nora, F. R., Mizukami, Y. & Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18, 375–380. https://doi.org/10.1101/gad.291204 (2004).
Leibfried, A. et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. https://doi.org/10.1038/nature04270 (2005).
Tsuwamoto, R., Yokoi, S. & Takahata, Y. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol. Biol. 73, 481–492. https://doi.org/10.1007/s11103-010-9634-3 (2010).
Horstman, A. et al. AIL and HDG proteins act antagonistically to control cell proliferation. Development 142, 454–464. https://doi.org/10.1242/dev.117168 (2015).
Horstman, A. et al. The BABY BOOM Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 175, 848–857. https://doi.org/10.1104/pp.17.00232 (2017).
Mu, J. Y., Tan, H. L., Hong, S. L., Liang, Y. & Zuo, J. R. Arabidopsis Transcription Factor Genes NF-YA1, 5, 6, and 9 Play Redundant Roles in Male Gametogenesis, Embryogenesis, and Seed Development. Mol. Plant 6, 188–201. https://doi.org/10.1093/mp/sss061 (2013).
Carlier, M. F. et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 136, 1307–1322. https://doi.org/10.1083/jcb.136.6.1307 (1997).
Podio, M. et al. A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis. Journal of Experimental Botany 65, 6411–6424, doi:https://doi.org/10.1093/jxb/eru354 (2014).
Vaillant, I. & Paszkowski, J. Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10, 528–533. https://doi.org/10.1016/j.pbi.2007.06.008 (2007).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45. https://doi.org/10.1038/47412 (2000).
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. https://doi.org/10.1038/38664 (1997).
Struhl, K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. https://doi.org/10.1101/gad.12.5.599 (1998).
Ng, H. H. & Bird, A. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25, 121–126 (2000).
Zhang, Z. et al. Genome-wide Target Mapping Shows Histone Deacetylase Complex1 Regulates Cell Proliferation in Cucumber Fruit. Plant Physiol. 182, 167–184. https://doi.org/10.1104/pp.19.00532 (2020).
Greer, C. B. et al. Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 13, 1444–1455. https://doi.org/10.1016/j.celrep.2015.10.013 (2015).
Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T. & Allis, C. D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 And H4. Proc. Natl. Acad. Sci. USA 92, 1237–1241. https://doi.org/10.1073/pnas.92.4.1237 (1995).
Rice, J. C. & Allis, C. D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273. https://doi.org/10.1016/s0955-0674(00)00208-8 (2001).
Burgers, W. A., Fuks, F. & Kouzarides, T. DNA methyltransferases get connected to chromatin. Trends Genet. 18, 275–277. https://doi.org/10.1016/s0168-9525(02)02667-7 (2002).
Jackson, J. P., Lindroth, A. M., Cao, X. F. & Jacobsen, S. E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. https://doi.org/10.1038/nature731 (2002).
Naumann, K. et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24, 1418–1429. https://doi.org/10.1038/sj.emboj.7600604 (2005).
Cao, X. F. & Jacobsen, S. E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144. https://doi.org/10.1016/s0960-9822(02)00925-9 (2002).
Jullien, P. E., Susaki, D., Yelagandula, R., Higashiyama, T. & Berger, F. DNA Methylation Dynamics during Sexual Reproduction in Arabidopsis thaliana. Curr. Biol. 22, 1825–1830. https://doi.org/10.1016/j.cub.2012.07.061 (2012).
Fan, D. et al. Histone H3K9 demethylase JMJ25 epigenetically modulates anthocyanin biosynthesis in poplar. Plant J. 96, 1121–1136. https://doi.org/10.1111/tpj.14092 (2018).
Chen, X. S., Hu, Y. F. & Zhou, D. X. Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 421–426, 2011. https://doi.org/10.1016/j.bbagrm.2011.03.004 (1809).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849. https://doi.org/10.1038/nrm1761 (2005).
Jiang, D. H., Gu, X. F. & He, Y. H. Establishment of the Winter-Annual Growth Habit via FRIGIDA-Mediated Histone Methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733–1746. https://doi.org/10.1105/tpc.109.067967 (2009).
Choi, S. C. et al. Trithorax group protein oryza sativa trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol. 164, 1326–1337. https://doi.org/10.1104/pp.113.228049 (2014).
Jiang, P. F. et al. SIP1 participates in regulation of flowering time in rice by recruiting OsTrx1 to Ehd1. New Phytol. 219, 422–435. https://doi.org/10.1111/nph.15122 (2018).
Alvarez-Venegas, R. et al. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. https://doi.org/10.1016/s0960-9822(03)00243-4 (2003).
Pien, S. et al. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20, 580–588. https://doi.org/10.1105/tpc.108.058172 (2008).
Tamada, Y., Yun, J. Y., Woo, S. C. & Amasino, R. M. ARABIDOPSIS TRITHORAX-RELATED7 Is Required for Methylation of Lysine 4 of Histone H3 and for Transcriptional Activation of FLOWERING LOCUS C. Plant Cell 21, 3257–3269. https://doi.org/10.1105/tpc.109.070060 (2009).
Yun, J. Y., Tamada, Y., Kang, Y. E. & Amasino, R. M. ARABIDOPSIS TRITHORAX-RELATED3/SET DOMAIN GROUP2 is Required for the Winter-Annual Habit of Arabidopsis thaliana. Plant Cell Physiol. 53, 834–846. https://doi.org/10.1093/pcp/pcs021 (2012).
Jacob, Y. et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16, 763-U796. https://doi.org/10.1038/nsmb.1611 (2009).
Yarden, R. I., Pardo-Reoyo, S., Sgagias, M., Cowan, K. H. & Brody, L. C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30, 285–289. https://doi.org/10.1038/ng837 (2002).
Russell, P. & Nurse, P. Negative Regulation of Mitosis By Wee1+, A Gene Encoding a Protein-Kinase Homolog. Cell 49, 559–567. https://doi.org/10.1016/0092-8674(87)90458-2 (1987).
Ogita, N. et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 94, 439–453. https://doi.org/10.1111/tpj.13866 (2018).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. https://doi.org/10.1038/nature02085 (2003).
Pagnussat, G. C., Alandete-Saez, M., Bowman, J. L. & Sundaresan, V. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689 (2009).
Matzk, F. A novel approach to differentiated embryos in the absence of endosperm. Sex. Plant Reprod. 4, 88–94 (1991).
Ruegger, M. et al. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207. https://doi.org/10.1101/gad.12.2.198 (1998).
Dharmasiri, N., Dharmasiri, S. & Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. https://doi.org/10.1038/nature03543 (2005).
Kepinski, S. & Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. https://doi.org/10.1038/nature03542 (2005).
Chapman, E. J. & Estelle, M. in Annual Review of Genetics Vol. 43 Annual Review of Genetics 265–285 (2009).
Shirley, N. J. et al. Translating auxin responses into ovules, seeds and yield: Insight from Arabidopsis and the cereals. J. Integr. Plant Biol. 61, 310–336. https://doi.org/10.1111/jipb.12747 (2019).
Hepworth, S. R., Zhang, Y. L., McKim, S., Li, X. & Haughn, G. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434–1448. https://doi.org/10.1105/tpc.104.030536 (2005).
Wang, Y. et al. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 180, 937–951. https://doi.org/10.1104/pp.18.00805 (2019).
Zemlyanskaya, E. V., Omelyanchuk, N. A., Ubogoeva, E. V. & Mironova, V. V. Deciphering Auxin-Ethylene Crosstalk at a Systems Level. Int. J. Mol. Sci. 19, 1. https://doi.org/10.3390/ijms19124060 (2018).
Uranga, J. A., Pedersen, R. A. & Arechaga, J. Parthenogenetic activation of mouse oocytes using calcium ionophores and protein kinase C stimulators. Int. J. Dev. Biol. 40, 515–519 (1996).
Digonnet, C., Aldon, D., Leduc, N., Dumas, C. & Rougier, M. First evidence of a calcium transient in flowering plants at fertilization. Development 124, 2867–2874 (1997).
Antoine, A. F. et al. A calcium influx is triggered and propagates in the zygote as a wavefront during in vitro fertilization of flowering plants. Proc. Natl. Acad. Sci. USA 97, 10643–10648. https://doi.org/10.1073/pnas.180243697 (2000).
Denninger, P. et al. Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 5, 1. https://doi.org/10.1038/ncomms5645 (2014).
Means, A. R. & Dedman, J. R. Calmodulin - an intracellular calcium receptor. Nature 285, 73–77. https://doi.org/10.1038/285073a0 (1980).
Braun, A. P. & Schulman, H. The multifunctional calcium calmodulin-dependent protein-kinase - from form to function. Annu. Rev. Physiol. 57, 417–445. https://doi.org/10.1146/annurev.ph.57.030195.002221 (1995).
Farmer, A., Thibivilliers, S., Schiefelbein, R. K. H. & Libault, M. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell leve. Mol. Plant 1071, 1. https://doi.org/10.1016/j.molp.2021.01.001 (2021).
Sherwood, R. T., Berg, C. C. & Young, B. A. Inheritance of apospory in buffelgrass. Crop Sci. 34, 1490–1494 (1994).
Kerk, N. M., Ceserani, T., Tausata, S. L., Sussex, I. M. & Nelson, T. M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 132, 27–35 (2003).
Young, B., Sherwood, R. & Bashaw, E. Cleared-pistil and thick-sectioning techniques for detecting aposporous apomixis in grasses. Can. J. Bot. 57, 1668–1672 (1979).
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Krueger, F. (2012). Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. (Accessed 28 April 2016); http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
Kopylova, E., Noe, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. https://doi.org/10.1093/bioinformatics/bts611 (2012).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644-U130. https://doi.org/10.1038/nbt.1883 (2011).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. https://doi.org/10.1038/nprot.2013.084 (2013).
Camacho, C. et al. BLAST plus : architecture and applications. BMC Bioinf. 10, 1. https://doi.org/10.1186/1471-2105-10-421 (2009).
Boeckmann, B. et al. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. C.R. Biol. 328, 882–899. https://doi.org/10.1016/j.crvi.2005.06.001 (2005).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. https://doi.org/10.1093/nar/gkt1223 (2014).
discriminating signal peptides from transmembrane regions. Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0. Nat. Methods 8, 785–786. https://doi.org/10.1038/nmeth.1701 (2011).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. goseq: Gene Ontology testing for RNA-seq datasets. R Bioconductor 8, 1–25 (2012).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. https://doi.org/10.1038/nbt.3519 (2016).
Acknowledgements
We would like to thank Yinping Guo and Nuengsap Thangthong for assistance in the laboratory and Tracey Vellidis for figure creation. This research was funded through a sub-award from the CSIRO under the Capturing Heterosis for Smallholder Farmers grant from the Bill and Melinda Gates Foundation.
Author information
Authors and Affiliations
Contributions
P.O.-A. and J.C. conceptualized this project; Methodology and investigation was done by Y.K. and M.P. Formal Analysis was done by Y.K., J.C., and P.O.-A. P.O.-A. supervised the project. Y.K. wrote the original draft. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ke, Y., Podio, M., Conner, J. et al. Single-cell transcriptome profiling of buffelgrass (Cenchrus ciliaris) eggs unveils apomictic parthenogenesis signatures. Sci Rep 11, 9880 (2021). https://doi.org/10.1038/s41598-021-89170-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89170-y
- Springer Nature Limited
This article is cited by
-
Apomixis: oh, what a tangled web we have!
Planta (2023)