Abstract
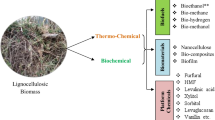
Polyhydroxybutyrate (PHB) is a bio-based, biodegradable and biocompatible plastic that has the potential to replace petroleum-based plastics. Lignocellulosic biomass is a promising feedstock for industrial fermentation to produce bioproducts such as polyhydroxybutyrate (PHB). However, the pretreatment processes of lignocellulosic biomass lead to the generation of toxic byproducts, such as furfural, 5-HMF, vanillin, and acetate, which affect microbial growth and productivity. In this study, to reduce furfural toxicity during PHB production from lignocellulosic hydrolysates, we genetically engineered Cupriavidus necator NCIMB 11599, by inserting the nicotine amide salvage pathway genes pncB and nadE to increase the NAD(P)H pool. We found that the expression of pncB was the most effective in improving tolerance to inhibitors, cell growth, PHB production and sugar consumption rate. In addition, the engineered strain harboring pncB showed higher PHB production using lignocellulosic hydrolysates than the wild-type strain. Therefore, the application of NAD salvage pathway genes improves the tolerance of Cupriavidus necator to lignocellulosic-derived inhibitors and should be used to optimize PHB production.








Similar content being viewed by others
References
Saini JK, Saini R, Tewari L (2015) Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments. 3 Biotech 5:337–353. https://doi.org/10.1007/s13205-014-0246-5
Bhatia SK, Jagtap SS, Bedekar AA et al (2021) Renewable biohydrogen production from lignocellulosic biomass using fermentation and integration of systems with other energy generation technologies. Sci Total Environ 765:144429. https://doi.org/10.1016/j.scitotenv.2020.144429
Kawaguchi H, Hasunuma T, Ogino C, Kondo A (2016) Bioprocessing of bio-based chemicals produced from lignocellulosic feedstocks. Curr Opin Biotechnol 42:30–39. https://doi.org/10.1016/j.copbio.2016.02.031
Pu Y, Hu F, Huang F et al (2013) Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol Biofuels 6:15. https://doi.org/10.1186/1754-6834-6-15
Zoghlami A, Paës G (2019) Lignocellulosic biomass: understanding recalcitrance and predicting hydrolysis. Front Chem. https://doi.org/10.3389/fchem.2019.00874
Gurav R, Bhatia SK, Choi T-R et al (2020) Utilization of different lignocellulosic hydrolysates as carbon source for electricity generation using novel Shewanella marisflavi BBL25. J Clean Prod 277:124084. https://doi.org/10.1016/j.jclepro.2020.124084
Sakai S, Tsuchida Y, Okino S et al (2007) Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol 73:2349–2353. https://doi.org/10.1128/AEM.02880-06
Bhatia SK, Jagtap SS, Bedekar AA et al (2020) Recent developments in pretreatment technologies on lignocellulosic biomass: effect of key parameters, technological improvements, and challenges. Biores Technol 300:122724. https://doi.org/10.1016/j.biortech.2019.122724
van der Pol EC, Bakker RR, Baets P, Eggink G (2014) By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio)chemicals and fuels. Appl Microbiol Biotechnol 98:9579–9593. https://doi.org/10.1007/s00253-014-6158-9
Parawira W, Tekere M (2011) Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review. Crit Rev Biotechnol 31:20–31. https://doi.org/10.3109/07388551003757816
Almeida JR, Modig T, Petersson A et al (2007) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. https://doi.org/10.1002/jctb.1676
Kim D (2018) Physico-chemical conversion of lignocellulose: inhibitor effects and detoxification strategies: a mini review. Molecules 23:309. https://doi.org/10.3390/molecules23020309
Mamman AS, Lee J-M, Kim Y-C et al (2008) Furfural: hemicellulose/xylosederived biochemical. Biofuels, Bioprod Biorefin 2:438–454. https://doi.org/10.1002/bbb.95
Larsson S, Palmqvist E, Hahn-Hägerdal B et al (1999) The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159. https://doi.org/10.1016/S0141-0229(98)00101-X
Kim S-K, Jin Y-S, Choi I-G et al (2015) Enhanced tolerance of Saccharomyces cerevisiae to multiple lignocellulose-derived inhibitors through modulation of spermidine contents. Metab Eng 29:46–55. https://doi.org/10.1016/j.ymben.2015.02.004
Jung H-R, Lee J-H, Moon Y-M et al (2019) Increased tolerance to furfural by introduction of polyhydroxybutyrate synthetic genes to Escherichia coli. J Microbiol Biotechnol 29:776–784. https://doi.org/10.4014/jmb.1901.01070
Lewis Liu Z, Moon J, Andersh BJ et al (2008) Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753. https://doi.org/10.1007/s00253-008-1702-0
Li Q, Metthew Lam LK, Xun L (2011) Biochemical characterization of ethanol-dependent reduction of furfural by alcohol dehydrogenases. Biodegradation 22:1227–1237. https://doi.org/10.1007/s10532-011-9477-x
Wang X, Miller EN, Yomano LP et al (2011) Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77:5132–5140. https://doi.org/10.1128/AEM.05008-11
Miller EN, Jarboe LR, Yomano LP et al (2009) Silencing of NADPH-dependent oxidoreductase genes ( yqhD and dkgA ) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75:4315–4323. https://doi.org/10.1128/AEM.00567-09
Wang X, Yomano LP, Lee JY et al (2013) Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc Natl Acad Sci 110:4021–4026. https://doi.org/10.1073/pnas.1217958110
Song H-S, Jeon J-M, Kim H-J et al (2017) Increase in furfural tolerance by combinatorial overexpression of NAD salvage pathway enzymes in engineered isobutanol-producing E. coli. Biores Technol 245:1430–1435. https://doi.org/10.1016/j.biortech.2017.05.197
Wang X, Miller EN, Yomano LP et al (2012) Increased furan tolerance in Escherichia coli due to a cryptic ucpA gene. Appl Environ Microbiol 78:2452–2455. https://doi.org/10.1128/AEM.07783-11
Muhammadi S, Afzal M, Hameed S (2015) Bacterial polyhydroxyalkanoates-eco-friendly next generation plastic: production, biocompatibility, biodegradation, physical properties and applications. Green Chem Lett Rev 8:56–77. https://doi.org/10.1080/17518253.2015.1109715
Lee SM, Lee H-J, Kim SH et al (2021) Screening of the strictly xylose-utilizing Bacillus sp. SM01 for polyhydroxybutyrate and its co-culture with Cupriavidus necator NCIMB 11599 for enhanced production of PHB. Int J Biol Macromol 181:410–417. https://doi.org/10.1016/j.ijbiomac.2021.03.149
Philip S, Keshavarz T, Roy I (2007) Polyhydroxyalkanoates: biodegradable polymers with a range of applications. J Chem Technol Biotechnol 82:233–247. https://doi.org/10.1002/jctb.1667
Koller M (2014) Poly (Hydroxyalkanoates) for food packaging: application and attempts towards implementation. Appl Food Biotechnol 1:3–15
Lee SM, Lee H-J, Kim SH et al (2021) Engineering of Shewanella marisflavi BBL25 for biomass-based polyhydroxybutyrate production and evaluation of its performance in electricity production. Int J Biol Macromol 183:1669–1675. https://doi.org/10.1016/j.ijbiomac.2021.05.105
Singh AK, Bilal M, Iqbal HMN et al (2021) Bioremediation of lignin derivatives and phenolics in wastewater with lignin modifying enzymes: Status, opportunities and challenges. Sci Total Environ 777:145988. https://doi.org/10.1016/j.scitotenv.2021.145988
Wang Y, Yin J, Chen G-Q (2014) Polyhydroxyalkanoates, challenges and opportunities. Curr Opin Biotechnol 30:59–65. https://doi.org/10.1016/j.copbio.2014.06.001
Park Y-L, Bhatia SK, Gurav R et al (2020) Fructose based hyper production of poly-3-hydroxybutyrate from Halomonas sp. YLGW01 and impact of carbon sources on bacteria morphologies. Int J Biol Macromol 154:929–936. https://doi.org/10.1016/j.ijbiomac.2020.03.129
Valappil SP, Peiris D, Langley GJ et al (2007) Polyhydroxyalkanoate (PHA) biosynthesis from structurally unrelated carbon sources by a newly characterized Bacillus spp. J Biotechnol 127:475–487. https://doi.org/10.1016/j.jbiotec.2006.07.015
Lee HS, Lee H-J, Kim SH et al (2021) Novel phasins from the Arctic Pseudomonas sp. B14–6 enhance the production of polyhydroxybutyrate and increase inhibitor tolerance. Int J Biol Macromol 190:722–729. https://doi.org/10.1016/j.ijbiomac.2021.08.236
Benesova P, Kucera D, Marova I, Obruca S (2017) Chicken feather hydrolysate as an inexpensive complex nitrogen source for PHA production by Cupriavidus necator on waste frying oils. Lett Appl Microbiol 65:182–188. https://doi.org/10.1111/lam.12762
Sen KY, Baidurah S (2021) Renewable biomass feedstocks for production of sustainable biodegradable polymer. Curr Opin Green Sustain Chem 27:100412. https://doi.org/10.1016/j.cogsc.2020.100412
Sirohi R, Prakash Pandey J, Kumar Gaur V et al (2020) Critical overview of biomass feedstocks as sustainable substrates for the production of polyhydroxybutyrate (PHB). Biores Technol 311:123536. https://doi.org/10.1016/j.biortech.2020.123536
Al-Battashi HS, Annamalai N, Sivakumar N et al (2019) Lignocellulosic biomass (LCB): a potential alternative biorefinery feedstock for polyhydroxyalkanoates production. Rev Environ Sci Bio/Technol 18:183–205. https://doi.org/10.1007/s11157-018-09488-4
Bhatia SK, Gurav R, Choi T-R et al (2019) Bioconversion of plant biomass hydrolysate into bioplastic (polyhydroxyalkanoates) using Ralstonia eutropha 5119. Biores Technol 271:306–315. https://doi.org/10.1016/j.biortech.2018.09.122
Phylactides M (1997) Molecular biology series 3. Tools of molecular biology: gene cloning. Br J Hosp Med 57:49–50
Park SL, Cho JY, Choi T-R et al (2021) Improvement of polyhydroxybutyrate (PHB) plate-based screening method for PHB degrading bacteria using cell-grown amorphous PHB and recovered by sodium dodecyl sulfate (SDS). Int J Biol Macromol 177:413–421. https://doi.org/10.1016/j.ijbiomac.2021.02.098
Jung H-R, Jeon J-M, Yi D-H et al (2019) Poly (3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate) terpolymer production from volatile fatty acids using engineered Ralstonia eutropha. Int J Biol Macromol 138:370–378. https://doi.org/10.1016/j.ijbiomac.2019.07.091
Liang L, Liu R, Wang G et al (2012) Regulation of NAD(H) pool and NADH/NAD+ ratio by overexpression of nicotinic acid phosphoribosyltransferase for succinic acid production in Escherichia coli NZN111. Enzyme Microb Technol 51:286–293. https://doi.org/10.1016/j.enzmictec.2012.07.011
Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ (2010) Identification and characterization of the furfural and 5-(hydroxymethyl) furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci 107:4919–4924. https://doi.org/10.1073/pnas.0913039107
Bhati R, Mallick N (2015) Poly (3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production by the diazotrophic cyanobacterium nostoc muscorum agardh: process optimization and polymer characterization. Algal Res 7:78–85. https://doi.org/10.1016/j.algal.2014.12.003
Wubbolts MG, Terpstra P, van Beilen JB et al (1990) Variation of cofactor levels in Escherichia coli. Sequence analysis and expression of the pncB gene encoding nicotinic acid phosphoribosyltransferase. J Biol Chem 265:17665–17672. https://doi.org/10.1016/S0021-9258(18)38215-2
San K-Y, Bennett GN, Berrı́os-Rivera SJ et al (2002) Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab Eng 4:182–192. https://doi.org/10.1006/mben.2001.0220
Jawed M, Pi J, Xu L et al (2016) Enhanced H2 production and redirected metabolic flux via overexpression of fhlA and pncB in klebsiella HQ-3 strain. Appl Biochem Biotechnol 178:1113–1128. https://doi.org/10.1007/s12010-015-1932-4
Berríos-Rivera S (2002) The effect of NAPRTase overexpression on the total levels of NAD, the NADH/NAD+ ratio, and the distribution of metabolites in Escherichia coli. Metab Eng 4:238–247. https://doi.org/10.1006/mben.2002.0229
Funding
This study was supported by the National Research Foundation of Korea (NRF) (NRF-2019M3E6A1103979 and NRF2022R1A2C2003138). This work was also supported by the R&D Program of MOTIE/KEIT (20015373, 20016324) and with the support of the R&D Program for Forest Science Technology [grant number 2020261C10-2022-AC02] provided by the Korea Forest Service (Korea Forestry Promotion Institute).
Author information
Authors and Affiliations
Contributions
SML and Y-HY conceived and designed the study; SML, D-HC, HJJ, BK, SHK performed the experiments and drafted the manuscript; SKB., RG., and Y-HY interpreted the experimental results; SML, J-MJ, J-JY, J-HP, J-HP, Y-GK and Y-HY revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, S.M., Cho, DH., Jung, H.J. et al. Enhanced tolerance of Cupriavidus necator NCIMB 11599 to lignocellulosic derived inhibitors by inserting NAD salvage pathway genes. Bioprocess Biosyst Eng 45, 1719–1729 (2022). https://doi.org/10.1007/s00449-022-02779-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-022-02779-9