Abstract
Main conclusion
Some salt stress response mechanisms can translate into sorghum forage yield and thus act as targets for genetic improvement.
Abstract
Sorghum is a drought-tolerant cereal that is widely grown in the vast Africa’s arid and semi-arid areas. Apart from drought, salinity is a major abiotic factor that, in addition to natural causes, has been exacerbated by increased poor anthropological activities. The importance of sorghum as a forage crop in saline areas has yet to be fully realized. Despite intraspecific variation in salt tolerance, sorghum is generally moderately salt-tolerant, and its productivity in saline soils can be remarkably limited. This is due to the difficulty of replicating optimal field saline conditions due to the great heterogeneity of salt distribution in the soil. As a promising fodder crop for saline areas, classic phenotype-based selection methods can be integrated with modern -omics in breeding programs to simultaneously address salt tolerance and production. To enable future manipulation, selection, and genetic improvement of sorghum with high yield and salt tolerance, here, we explore the potential positive correlations between the reliable indices of sorghum performance under salt stress at the phenotypic and genotypic level. We then explore the potential role of modern selection and genetic improvement programs in incorporating these linked salt tolerance and yield traits and propose a mechanism for future studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinization is widespread resulting naturally from the retention of soluble salt in the soil. Continued adoption of improper anthropogenic activities specifically farming activities has exacerbated soil salinization (Endo et al. 2011; Munns and Gilliham 2015; Sharma et al. 2016; Bui 2017). Irrigation with saline water combined with an inefficient drainage system has increased leakage rising the water tables. Raised water tables also raise salt toward the root zone which perturbs normal plant function and soil structure (Greenway and Munns 1980).
The net effects have been a disruption of ionic homeostasis to toxic levels as well as an osmotic imbalance which contribute to physiological drought which ultimately limits crop performance and productivity (Munns and Tester 2008). This poses an even more serious threat to food security and economic development in Africa and other less-developed countries (Fao 2009).
Sorghum (Sorghum bicolor L. Moench) is a key food and feed crop in Africa with the potential to improve household food security because of its excellent adaptability to drought (Taylor 2003; Wagaw et al. 2020). However, despite Africa accounting for more than a third of the global sorghum production, there is a lag in research focused on exploiting the full potential of sorghum in addressing food and fodder insecurity affecting millions of people and livestock across the globe. Other than Africa being the only continent that straddles both tropics, it is the origin of sorghum. Sorghum in Africa is processed into various highly nutritious traditional foods and fodder (Osmanzai 1992). The potential for sorghum to drive economic development in Africa and other regions is enormous.
Due to the heterogeneous nature of salinity in diverse soil, in toto studies are complex. This leaves a gap for focusing on the various crop adaptive mechanisms to salinity which will enable the selection, cultivation, and breeding of superior crops with high forage value. Despite its high drought tolerance, sorghum can withstand moderate levels of soil salinity, while high salinity stress limits its growth and productivity. Like other crops, salt stress perturbs the biochemical, morphological, physiological, and agronomical performances whose net effect is a limitation to growth and productivity (Fig. 1).
We calculated the average sorghum production for countries that produced more than 3000 metric tons between 1994 and 2021 as indexed by web platforms of Index Mundi, Statista, World Atlas, and FAO (1995). We then mapped the producers with global saline soil distribution and observed that high production regions within consistently top global sorghum producers, i.e., the US, Ethiopia, Nigeria, Mexico, India, Argentina, China, and Sudan fell within the low and moderately saline areas (Fig. 2). This suggests also that high salinity has notably negative effects on large-scale sorghum production; hence, there is an urgent need to address sorghum production in saline areas.
Salt stress effect and coping mechanisms in sorghum
So far, incredible advances have been made in sorghum selection and genetic enhancement, for instance involving a decade of research by the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) (Reddy et al. 2010; Kumar et al. 2011), while recent advances are summarized by Huang (2018). Despite the progress, efficient selection for rapid breeding of high-yielding salt-tolerant individuals remains underexplored. Wide intraspecific tolerance level and performance as well as the complexity of soil salinity are partly to blame for limited research in forage sorghum (Negrao et al. 2017). However, recent developments and breakthroughs in plant phenotyping and genotyping offer high potential for identifying and selecting salt-tolerant sorghum with high forage productivity and incorporating them with advanced breeding programs.
The principal consequence of salt stress on plants is excessive Na+ and Cl− accumulation which disrupts the ionic homeostasis and causes phytotoxicity, osmotic stress causing physiological drought, and photosynthesis disruption which causes nutrient imbalance (Gupta and Huang 2014). Like other plants, sorghum has evolved a complex but efficient coping mechanism consisting of the antioxidant enzyme machinery, ionic homeostasis, osmotic adjustment, photosynthesis rearrangement, and hormonal and transcriptional regulation (Table 1).
However, our objective is not to focus merely on response mechanisms but on how these mechanisms correlate with yield for potential selection and genetic improvement purposes. Thus, we collate and review advances and bottlenecks in sorghum research to highlight the potential interplay between the salt tolerance parameters and qualitative/quantitative values that are relevant for future high-yielding forage selection under salt stress. We propose a model for future prospective genetic improvement programs. Future directions and opportunities for sorghum breeding under salt stress are highlighted to stimulate discussion among sorghum academic and industrial communities.
Antioxidant machinery and the salt overly sensitive pathway (SOS)
Salt stress causes cytotoxic accumulation of reactive oxygen species (ROS) and other free radicals (Apel and Hirt 2004; Munns and Tester 2008; Ashraf 2009). However, tolerant plants have developed an intricate and systematic antioxidant system at the genetic and physiological levels to scavenge excess ROS. In two sorghum varieties with converse salt tolerance degrees, Costa et al (2005) observed that accessions that displayed higher superoxide dismutase to hydrogen peroxide (SOD: POD) scavenging ratio also exhibited greater biomass accumulation and height. Later, Hefny and Abdel-Kader (2009) used ROS scavenging as a selection basis for a sorghum panel growing in salinity. They observed a positive correlation between antioxidant enzyme activities and relative growth rate and biomass. Recently, Ibrahim et al. (2020) demonstrated a high positive correlation between antioxidant enzymes activity and sorghum height, fresh weight, and photosynthesis rate under salinity stress. Similarly, Yilmaz et al. (2020) and Sudhakar et al. (2016) observed a positive association between glutathione reductase, glutathione S-transferase, and ascorbate peroxidase with chlorophyll and carotene content in redbine and transgenic sorghum, respectively. Ali et al. (2020) reported a positive relationship between peroxidase content and salt tolerance index, total dry weight, root length, seedling vigor index, shoot length, total chlorophyll, and protein content in the seedlings. Also, salt-resistant sorghum varieties displayed higher photosynthetic pigments than sensitive varieties (Baiseitova et al. 2018). These observations suggest that antioxidant enzyme-mediated salt tolerance could be working in tandem with events linked to the accumulation of photosynthesis assimilates and growth in sorghum.
Antioxidant enzymes are products of elaborate transcription machinery triggered by sensed and transduced salt signals (Yang and Guo 2018). Through structural and functional analysis, Avashthi et al. (2020) observed a conservation of glutathione dehydrogenase, ascorbate peroxidase, glutathione reductase, Fe and Cu–Zn superoxide dismutase-related gene preserves which played key functions in desiccation tolerance, as well as growth of sorghum and other cereals. Mulaudzi et al. (2020) reported that salt stress induced the upregulation of SbAPX2, and SbCAT3 genes in sorghum which code for peroxidase and catalase enzymes that scavenge excessive ROS. They also observed that, under salt stress, sorghum plants overexpressing the Salt Overlay Sensitive1 (SbSOS1) gene displayed as a lower accumulation of ROS and exhibited a faster growth rate. The plant salt overly sensitive (SOS) is an important pathway that responds to salt stress by excluding Na+ ions from the cell (Rolly et al. 2020). Based on these observations, we suggest that these genes alongside SOS1 can be targeted to develop highly antioxidative potential and salt-induced desiccation-tolerant sorghum genotypes for forage production. Table 2 summarizes correlations between various antioxidant enzyme activities and yields calculated from previous studies.
Photosystem II photochemistry and photosynthesis
Chlorophyll-a fluorescence has been increasingly used as a determiner of plant health under stressful conditions, due to its high sensitivity to environmental stress including salt stress. The chlorophyll a fluorescence reflects the plant's photosystem II (PSII) which in turn can tell the plant photochemical quenching (Force et al. 2003). Netondo et al. (2004) observed that a decline in the photochemical efficiency of PSII and photochemical quenching coefficient in sorghum shrinked the leaf area, reduced stomatal conductance, and transpiration rate. In a subsequent comparative study, more salt-sensitive sorghum genotype exhibited a significant decline in PSII and the actual PSII efficiency under elevated salt stress compared to tolerant genotypes which negatively affected leaf expansion, CO2 intake, and transpiration (Sui et al. 2015). A decline in PSII efficiency under salt stress was also followed by a reduction in the relative water content (RWC), CO2, and stomatal closure in sorghum (Lawlor 2002). These observations indicate that the ability of sorghum to maintain high photochemical quenching and PSII efficiency could be associated with NADPH and ATP consumption reflecting a higher photosynthetic electron transport. In a later study, Zhang et al. (2018) observed that the obstructive consequences of salt stress on the PSII and eventually growth rate of sorghum in medium salinity (8 dS/m) were greater than that under slightly lower salt stress (6 dS/m). This suggest that slight changes in salinity level may have pronounced effects on the sorghum performance; thus, this should be considered when choosing planting area.
Sorghum being a C4 plant, carbon is fixed initially in the mesophyll cells via the phosphoenolpyruvate carboxylase (PEPC) pathway. Recently, the novel genes within the PEPC family have been described to play integral roles in plants photosynthetic resilience to salinity (Aldous et al. 2014). Through a cloning experiment in sorghum under salt stress, Echevarría et al. (2001) recorded an increase in the Ca2+-independent phosphoenolpyruvate carboxylase kinase (PEPC-k) gene activity. Later, García-Mauriño et al. (2003) observed that salt-induced activity of the PEPC-k gene not only played a positive role in the mitigation of ion toxicity but also contributed to carbon fixation efficiency in sorghum leaves in dark conditions. These observations indicate that the phosphorylation of the target protein of the PEPC-k gene could play an important role in sorghum’s key metabolic process that improve carbon assimilation and photosynthetic efficiency under salt stress. Through a transcriptome study, Sui et al. (2015) unraveled the genes that may influence photosynthesis response to salt stress in sorghum. Among these genes, Sb02g002830 and Sb09g021810 were upregulated, and they play an integral role to stabilize the construction of the oxygen-evolving complex and ATP synthase enzyme. They were also associated with increased levels of CO2, pyruvate, and NADPH, which in turn enhanced CO2 assimilation under salt stress conditions. Guo et al. (2018) reported that the NADP + -malate dehydrogenase genes which encode important enzymes involved in carbon fixation were overexpressed in salt-tolerant sweet sorghum genotypes in response to salt stress, and subsequently exhibited an increase in the chlorophyll concentration, PSI oxidoreductive function, and PSII photochemical efficiency. This suggests that a simultaneous expression of the genes could trigger an elevation of NADPH and pyruvate levels which in turn can enhance CO2 assimilation in sorghum under salt stress.
However, we notice that despite remarkable progress in photosynthesis research, there is limited information on the influence of respiration on sorghum forage yield and salt tolerance. Studies in other plants have shown that the day–night temperature changes significantly affect other vital processes like flowering (Prasad and Djanaguiraman 2011). Therefore, there is a need for modeling sorghum response to fluctuating temperature. The focus should be given to photorespiration under salt stress and its effect on yield and tolerance. Table 3 highlights research gaps on the role of respiration to sorghum yield under salt stress as compared to photosynthesis.
Osmotic adjustment may defy the ‘cost on growth, a return on photosynthesis’ hypothesis
The role of osmotic adjustment (OA) to dehydration including salt-induced physiological drought is well documented on the plant stress website (https://plantstress.com/). However, the role of OA in improving yield under dehydration in plants has raised an intensive debate in the scientific community with skepticisms notably from Munns (1988). They suggested that for the compatible solutes that boost OA to improve productivity, they must be diverted from vital cellular processes. Their argument was based on the hypothesis that growth may be hindered by water stress which precedes photosynthesis, hence producing OA-active solutes (Munns and Weir 1981). This was concurrent with an earlier dilemma postulated by Blum et al. (1980) that OA could inhibit growth while protecting the photosynthetic machinery. Furthermore, in sorghum growing under salt stress, Turner and Jones (1980) had indicated that OA not only maintained cell integrity but also improved the RWC and CO2 influx. They suggested that suppressed growth in favor of photosynthesis actively generated osmotically active compatible solutes contributing to OA. Out of this phenomenon, the term ‘cost on growth and return on photosynthesis’ was coined. Considering the old-time of these studies, our view here is that whether this cost-return concept is valid or not, consideration should be given to the net balance between sorghum yields under salt stress with OA. Thus, we focus on subsequent studies that attempted to address the positive role of OA on sorghum yield for potential forage selection. Interestingly, in a follow-up review 4 decades later, Turner acknowledged that OA could have a positive impact on yield (Turner 2018). On the other side, Blum observed a positive correlation between OA and yields of twelve crops (Blum 2017).
Salt-stressed sorghum was reported to maintain OA by actively transporting compatible and incompatible solutes in the vacuole (Lacerda et al. 2003; Girma and Krieg 1992). Recently, researchers have reported improved production traits under salt stress with high OA-active solute accumulation. For example, Curt et al. (1995) observed that at the physiological maturity phase, sorghum exhibited the highest stem yield and sugar content compared to the flowering stage, while Santamaria et al. (1990) observed that sorghum hybrids with high OA expressed better panicle exertion, higher total dry matter, higher root length density, and greater water use efficiency in late-maturing sorghum. These two studies suggested that the OA effect on sorghum yield may rely on the physiological phase during harvest. Gill et al. (2001) observed that sorghum seedlings with high OA-active sucrose content also exhibited faster recovery and growth under salt stress. de Oliveira et al. (2020) documented that salinity induced a greater release in spermine which were positively correlated with growth of salt-tolerant sorghum genotype. Earlier, Chai et al. (2010) had shown that the exogenous application of spermine improved OA and growth of sorghum seedlings under salt stress.
Since salt stress causes physiological drought, we also highlight studies that addressed OA in sorghum under desiccation and water-deficit stress. Ludlow et al. (1990) found that increasing OA corresponded with enhanced yield of sorghum exposed to water-deficit post-anthesis. Tangpremsri et al. (1995) observed that sorghum lines with high OA also had better leaf retention, grain number per unit area, yield, and total dry matter. Sinclair and Muchow (2001) simulated the response of sorghum to dehydration and the effect on yield. They observed that OA could be beneficial for sorghum survival under desiccation but simultaneously improved soil moisture capture leading to greater yield. These studies present a new argument that other than enhancing salt tolerance, OA can enhance yield.
Phenotypic responses to salt stress are linked with downstream transcriptional machinery. Through a transcriptome study, Sui et al. (2015) profiled genes that may play vital roles in sugar biosynthesis during salt stress. From their transcriptome data deposited to the NCBI, we searched for and identified abundant and upregulated genes that were previously reported to code for sugars which may play vital roles in OA and nutritive yield (Table 4).
Other than sugar, proteins act as important compatible osmolytes. Ndimba et al. (2010) and Ngara et al. (2018) conducted a proteomic study of proteins responsive to salt stress in different sorghum varieties. After studying their proteome deposited at the NCBI, we observed that many upregulated proteins fell within the glycoside hydrolase superfamily which constitutes important enzymes for hydrolysis of inactive glycosides. Glycoside hydrolysis releases free sugars such as fructose, sucrose, lactose, maltose, and galactose which play important role in OA as well as plant nutritive value. We randomly selected one of the proteins, alpha-galactosidase which hydrolyses alpha galactoside releasing galactose (an important OA sugar and a constituent of lactose) and designed its probable hydrolysis pathway (Fig. 3). This suggests that salt stress may activate the hydrolysis of inactive sugar compounds to improve OA, water uptake, and sugar yield.
The other important OA-active osmolyte is proline. Transgenic lines of sorghum overexpressing pyrroline-5-carboxylate synthetase (P5CSF129A), a gene encoding key enzymes for proline biosynthesis, exhibited an increase in the photosynthetic rate, chlorophyll contents, stomatal conductance, and carbon dioxide concentration under salt stress (Surrender et al. 2015). Taken together, these observations suggest that sugar and proline-mediated OA could occur simultaneously with plant growth, photosynthesis, and maintaining high nutritive yield value in sorghum under salt stress. Also, considering the complexity in identifying a singular plant trait responsible for yield improvement under salt stress, these observations remarkably hint that OA may sustain sorghum nutritive yield under salt-induced water stress.
Ionic homeostasis
Due to its high-water solubility, NaCl is the most abundant and significant contributor to soil salinity whose bioaccumulation results in phytotoxicity. Thus, Na+ and Cl− exclusion is major adaptive mechanisms of plant tolerance to salt stress (Ali et al. 2012; Amtmann and Sanders 1999; Apse et al. 1999; Munns and Tester 2008). The important role of HKT family genes in Na+ exclusion from glycophytic monocots has been well described (Yao et al. 2010; Horie et al. 2011; Assaha et al. 2017; Hamamoto et al. 2015; Kronzucker and Britto 2011; Maathuis et al. 2014; Munns 2002; Volkov 2015). Also, the role of K+/Na+ homeostasis in improving crop yield has been documented (Zorb et al. 2018; Hanin et al. 2016). Azooz et al. (2004) observed that K+/Na+ ratios were higher in the most salt-tolerant sorghum cultivar than in sensitive ones, and in the youngest than in the oldest leaf. They also observed that cultivars with a higher K+/Na+ ratio also exhibited larger leaf area, dry mass, and relative water content. The high-affinity potassium transporter HKT gene family has been reported to play important role in plant K uptake under salinity stress. Wang et al. (2014) characterized SbHKT1;4, which is a member of the HKT gene family from sorghum. He observed that under Na+ stress, SbHKT1;4 expressions were highly upregulated in a salt-tolerant sorghum accession and was correlated with a more balanced K+/Na+ ratio and enhanced shoot and root biomass. Besides, SbHKT1;4 was found to play an important role in maintaining the optimal K+/Na+ balance under Na+ stress (Wang et al. 2014). They found that the Arabidopsis thaliana plant overexpressing SbHKT1;4 maintained excellent growth and yield. While Munns et al. (2012) revealed that the presence of TmHKT1;5-A, another HKT member remarkably limited leaf Na+ ion concentration and increased grain yield by 25%. Other than the HKT family, sodium proton antiporter-like protein (NHXLP) is a plasma membrane-bound protein associated with Na+ exclusion and helps to maintain ion homeostasis under saline conditions. Transgenic peanut overexpressing the sorghum SbNHXLP genes displayed higher biomass and pod yield when compared with wild types of plants under salt stress (Kandula et al. 2019); while it conferred salt tolerance and improved fruit yield in tomatoes (Kumari et al. 2016). This highlights the potential of SbNHXLP as a target candidate gene to impart salt stress tolerance and improve yield in sorghum. Also, the co-expression of the HKT and NHXLP-related genes may initiate key K+/Na+ homeostatic events that are also linked with biomass addition in sorghum under salt stress.
Other than K and Na, Ca plays an important role in plant salt tolerance and production (White and Broadley 2003). Mulaudzi et al. (2020) observed that Ca2+ accumulation coincided with reduced H2O2 and Na+ to K+ ratio, hence counteracting their adverse effects on seed germination and growth.
Root system architecture (RSA)
Roots are vital organs for water and nutrient uptake which in turn determine plant growth, productivity, and stress response. The root–system architecture (RSA) is the structural and dimensional organization of the root which plays vital role in adaptation to stress (Parra-Londono et al. 2018). In sorghum, large genetic diversity levels in the root architecture associated with water deficit were observed in recombinant lines (Mace et al. 2012). Later, Chen et al. (2020) provided a theoretical platform for designing more efficient RSA to achieve greater yield and tolerance to water deficit. They presented that deep root of sorghum improved the dry matter of leaves, panicles, stems, and leaf sheaths. Deeper roots were also linked with increased soluble carbohydrates, proteins, and hormones which improved tolerance to desiccation. This offers breeders’ opportunities to design sorghum with a customized root system architecture that are not only better adapted to salt-induced physiological drought but also achieve high forage yield. Also, the identification of novel quantitative trait loci with the traits of interest will be a fundamental research platform in dissecting the large genetic variabilities of root system attributes of sorghum under salt stress.
Molecular markers as potential selection tools
Using molecular markers as selection tools is projected to improve breeding efficiency (Xie and Xu 1998; Salgotra and Stewart 2020; Nadeem et al. 2018; Collard and Mackill 2008; Fu et al. 2017). However, for sorghum, there is still much to be done in this direction. For effective sorghum breeding, advanced methods in molecular marker-assisted selection should be exploited. So far, SSR (Shehzad et al. 2009), SNP (Luo et al. 2016), RAPD (Akhare et al. 2008), and ISSR (Basahi 2015) markers have been developed for sorghum. From these markers, SNPs have been the most intensively studied in sorghum as far as salt tolerance and yield are concerned. For instance, Bekele et al. (2013) developed and tested a robust SNP platform that allowed screening for genome-wide and trait-linked polymorphisms in sorghum. Luo et al. (2016) presented the SNP database based on the assembled and annotated genome sequences of sorghum and recently published sorghum re-sequencing genomic data. The SNPs have subsequently been used to dissect genetic diversity in sorghum (Menamo et al. 2021; Karla et al. 2021; Zeleke et al. 2021; Afolayan et al. 2019), a fundamental step in selecting desirable parents for conventional and genomic-assisted breeding.
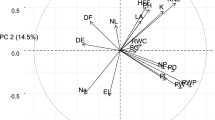
With the rapid development of genome-wide association studies (GWAS), many sorghum accessions have been collected for association mapping. Recently, Ruperao et al. (2021) and Cuevas and Prom (2020), through association analysis, identified SNPs that were significantly associated with important agronomic traits. Figure 4 shows some significant associations between SNPs and agronomic traits we compiled from previous mapping studies. We observe that chromosome number 9 appears to carry most traits hence acting as a potential target for further genetic improvement.
Wang et al. (2020) through a recombinant inbred line study mapped QTL clusters controlling plant height, total biomass, and fresh weight. Besides, they observed that some SNPs were significantly associated with the salt tolerance index as an indicator for the growth response of each accession to salt stress. These locations might serve as target sites for marker-assisted selection in improving the salt tolerance of forage sorghum. Besides, Yamazaki et al. (2020) conducted a genome-wide association study for salt tolerance and identified two SNPs that were significantly associated with biomass production in sorghum. They also observed that the genetic factors that affected biomass production under salt stress were varied remarkably from those without salt stress. These results offer candidate genetic resources and SNP markers for breeding salt-tolerant sorghum with high yield under salt stress. Upadhyaya et al. (2013) through linkage disequilibrium identified SNP loci linked to biomass and height in sorghum under salinity. They also identified a disequilibrium block that carried a gene homologous to the Arabidopsis flowering time gene, LUMINIDEPENDENS. This gene promotes plant growth at flowering maturity (Lee et al. 1994). Thus, these newly mapped SNP markers will facilitate the identification of beneficial traits in sorghum under salt stress. In another study, Ruperao et al. (2021) found a significant association between SNPs and starch biosynthesis genes located in five QTLs. Also, among the associations, some SNPs were associated with genes Sobic.002G022500, Sobic.003G173400, and Sobic.004G350800 which belong to the NAC-domain family of genes linked to flowering, PSII protein complex for photosynthesis, lignin biosynthesis, and folate metabolism (Adeyanju et al. 2021; Tian et al. 2016). These results suggest that genomic signatures of salt stress tolerance may be useful for sorghum improvement, enhancing germplasm identification and marker-assisted selections using SNPs.
Apart from their abundance in all genomes with elevated levels of polymorphism compared to other molecular markers, SSR markers with moderate density are more informative than SNPs for assessing genetic relatedness in a population (Yang et al. 2011). Billot et al. (2013) presented a platform for designing a robust reference SSR kit for sorghum. They then presented a diversity survey for thousands of sorghum accessions using the SSRs which provided an entry to global sorghum germplasm collections. Later, Zhan et al. (2019) evaluated a panel of sorghum using an SSR marker for salt tolerance. However, all these studies have focused on genetic diversity remaining entirely silent on the potential simultaneous associations with salt tolerance and yield in a breeding perspective. Therefore, to facilitate genotyping toward a refined functional understanding and yield of sorghum under salt stress, a core integrated reference containing all the markers will be mandatory.
Incorporation of sorghum into modern genetic breeding strategies
With the availability of sorghum genome, genomic selection (GS) described by Luan et al. (2009), Nakaya and Isobe (2012) and Xu et al. (2020) can be exploited to breed sorghum varieties with excellent salt tolerance and yield for forage use cost-effectively without QTL related to target traits as well as phenotypes. Also, by exploiting the advantages of both linkage analysis and linkage disequilibrium progress in sorghum, the Nested Association Mapping (NAM) first described by Yu et al. (2008) can be applied to sorghum to detect loci of yield and salt tolerance with minimum false positives commonly observed in GWAS. To the best of our knowledge, only two true NAM sorghum populations have been developed. One by Bouchet et al. (2017), while Perumal (2021) has recently registered the sorghum NAM population in RTx430 background with KS-RTx430NAM. With these developments, recombinant inbred lines should be exploited to identify QTL markers containing salt tolerance and yield-related traits. Another important genetic breeding strategy is targeting induced local lesions in genomes (TILLING) as first described by Kurowska et al. (2011). High-throughput TILLING in sorghum will allow rapid and low-cost discovery of new alleles related to salt tolerance and yield. So far, TILLING has been successfully performed in Arabidopsis (Horst et al. 2007), wheat (Slade et al. 2005), and maize (Till et al. 2004). In sorghum, Xin et al. (2008), Nida et al. (2016), and Addo-Quaye et al. (2018) used mutagenic sorghum as a reference to develop two mutant populations which may serve as vital resources for forward and reverse sorghum genetic studies for forage breeding. Due to its high conserved nature, crop breeders can exploit meiosis to create sorghum lines with high forage yield and salt tolerance combinations through engineering. Dhaka et al. (2020) presented in-depth profile candidates for engineering male fertility in sorghum. However, this appears to be the only study related to reproductive engineering in sorghum. Studies have been in part hindered by complexities in crossovers per chromosome per meiosis as well as centromere flanking. Therefore, to overcome this, sorghum breeders should generate a large population to recover the desired recombinants. Figure 5 shows our proposed selection and genetic improvement model. GWAS and NAM can be exploited to select high-yielding sorghum and genetically improved and bred using meiotic recombination. Also, selection of the more exploratory RSA through NAM or GWAS can offer candidates for genetic improvement through meiotic recombination.
Proposed mechanism and future management
So far, the knowledge gained concerning sorghum simultaneous tolerance and yield is far from being translated into real breeding practices. We propose using the genome-based approach for designing sorghum as a forage crop combining the above-reviewed salt tolerance values aiming at high yield. To accelerate the breeding of superior-performing sorghum varieties with high yield potential and salt stress tolerance to quality forage standards, improved genetic and speed breeding concepts should be considered. Also, the role of growth regulators and seed priming should be considered.
From our synthesis, we hypothesize that salt stress induces signals prompting the upregulation of SbCAT3, SbSOD1, SbSOS1, and SbHKT1; SbCAT3 and SbSOD1 codes for CAT and SOD enzymes which scavenge excess ROS. SbHKT1, SbNHLP, and SbSOS1 initiate downstream events that lead to Na exclusion restoring the K/Na homeostasis. This simultaneous activity activates or works in tandem with P5CSF129A genes leading to osmotic adjustment and the promotion of root growth. Osmotic adjustment improves water uptake increasing the RWC which activates the PEPC-k gene that drives the photosynthetic machinery improving the accumulation of photosynthetic products such as starch, crude protein, and other nutrients which trigger further growth and biomass increase. Breeding programs should optimize toward efficient identification and manipulating qualitative/quantitative loci carrying both yield and salt tolerance-related parameters (Fig. 6).
Author contribution statement
EA: data analysis and writing; DA, AH, LK, AO, CG, MEG: revision of first drafts and final version; AN: conceptualization, supervision and revision. All authors have read and approved the final document.
References
Addo-Quaye C, Tuinstra M, Carraro N, Weil C, Dilkes BP (2018) Whole-genome sequence accuracy is improved by replication in a population of mutagenized sorghum. G3 (bethesda) 8(3):1079–1094. https://doi.org/10.1534/g3.117.300301
Adeyanju AO, Sattler SE, Rich PJ, Rivera-Burgos LA, Xu X, Ejeta G (2021) Sorghum Brown midrib19 (Bmr19) gene links lignin biosynthesis to folate metabolism. Genes 12(5):660. https://doi.org/10.3390/genes12050660
Afolayan G, Deshpande S, Aladele S, Kolawole A, Angarawai I, Nwosu D, Danquah E (2019) Genetic diversity assessment of sorghum (Sorghum bicolor (L.) Moench) accessions using single nucleotide polymorphism markers. Plant Genet Resour C 17(5):412–420
Akhare A, Sakhare SB, Kulwal P, Dhumale DB, Kharkar A (2008) RAPD profile studies in Sorghum for identification of hybrids and their parents. Int J Integr Biol 3:18–24
Aldous SH, Weise SE, Sharkey TD, Waldera-Lupa DM, Stühler K, Mallmann J, Groth G, Gowik U, Westhoff P, Arsova B (2014) Evolution of the phosphoenolpyruvate carboxylase protein kinase family in C3 and C4 flaveria spp. Plant Physiol 165(3):1076–1091. https://doi.org/10.1104/pp.114.240283
Ali Z, Park HC, Ali A, Oh DH, Aman R, Kropornicka A, Hong H, Choi W, Chung WS, Kim WY, Bressan RA, Bohnert HJ, Lee SY, Yun DJ (2012) TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K(+) specificity in the presence of NaCl. Plant Physiol 158(3):1463–1474. https://doi.org/10.1104/pp.111.193110
Ali AYA, Ibrahim MEH, Zhou G, Nimir NEA, Jiao X, Zhu G, Elsiddig AMI, Suliman MSE, Elradi SBM, Yue W (2020) Exogenous jasmonic acid and humic acid increased salinity tolerance of sorghum. Agron J 112:871–884. https://doi.org/10.1002/agj2.20072
Ali AYA, Ibrahim MEH, Zhou G, Nimir NEA, Elsiddig AMI, Jiao X, Zhu G, Salih EGI, Suliman MSE, Elradi SBM (2021) Gibberellic acid and nitrogen efficiently protect early seedlings growth stage from salt stress damage in Sorghum. Sci Rep 11(1):6672. https://doi.org/10.1038/s41598-021-84713-9
Amtmann A, Sanders D (1999) Mechanism of Na+ uptake by plant cells. Adv Bot Res 29:75–112
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285(5431):1256–1258. https://doi.org/10.1126/science.285.5431.1256
Ashraf M (2009) Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol Adv 27(1):84–93. https://doi.org/10.1016/j.biotechadv.2008.09.003
Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8:509. https://doi.org/10.3389/fphys.2017.00509
Avashthi H, Pathak RK, Gaur VS, Singh S, Gupta VK, Ramteke PW, Kumar A (2020) Comparative analysis of ROS-scavenging gene families in finger millet, rice, sorghum, and foxtail millet revealed potential targets for antioxidant activity and drought tolerance improvement. Netw Model Anal Health Inform Bioinform 9:33. https://doi.org/10.1007/s13721-020-00240-z
Azooz M, Shaddad MA, Abdel LA (2004) The accumulation and compartmentation of proline in relation to salt tolerance of three sorghum cultivars. Indian J Plant Physiol 9:1–8
Baiseitova G, Sarsenbayev B, Kirshibayev E, Kamunur M (2018) Influence of salinity (NaCl) on the photosynthetic pigments content of some sweet sorghum varieties: All-Russia scientific-practical conference on the prospects of development and challenges of modern botany BIO web of conferences 11, p 00003
Basahi M (2015) ISSR-based analysis of genetic diversity among sorghum landraces growing in some parts of Saudi Arabia and Yemen. Compte Rendus Biol 338(11):723–727. https://doi.org/10.1016/j.crvi.2015.09.003
Bekele WA, Wieckhorst S, Friedt W, Snowdon RJ (2013) High-throughput genomics in sorghum: from whole-genome resequencing to a SNP screening array. Plant Biotechnol J 11(9):1112–1125. https://doi.org/10.1111/pbi.12106
Billot C, Ramu P, Bouchet S, Chantereau J, Deu M, Gardes L, Noyer JL, Rami JF, Rivallan R, Li Y, Lu P, Wang T, Folkertsma RT, Arnaud E, Upadhyaya HD, Glaszmann JC, Hash CT (2013) Massive sorghum collection genotyped with SSR markers to enhance use of global genetic resources. PLoS ONE 8(4):e59714. https://doi.org/10.1371/journal.pone.0059714
Blum A (2017) Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ 40(1):4–10. https://doi.org/10.1111/pce.12800
Blum A, Sinmena B, Ziv O (1980) An evaluation of seed and seedling drought tolerance screening tests in wheat. Euphytica 29P:727–736. https://doi.org/10.1007/BF00023219
Bouchet S, Olatoye MO, Marla SR, Perumal R, Tesso T, Yu J, Tuinstra M, Morris GP (2017) Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 206(2):573–585. https://doi.org/10.1534/genetics.116.198499
Bui EN (2017) Causes of soil salinization, sodification, and alkalinization. Environ Sci. https://doi.org/10.1093/acrefore/9780199389414.013.264
Chai YY, Jiang CD, Shi L, Shi T, Gu W (2010) Effects of exogenous spermine on sweet sorghum during germination under salinity. Biol Plant 54:145–148. https://doi.org/10.1007/s10535-010-0023-1
Chen X, Wu Q, Gao Y, Zhang J, Wang Y, Zhang R, Zhou Y, Xiao M, Xu W, Huang R (2020) The role of deep roots in sorghum yield production under drought conditions. Agronomy 10(4):611. https://doi.org/10.3390/agronomy10040611
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363(1491):557–572. https://doi.org/10.1098/rstb.2007.2170
Costa P, Azevedo N, André B, Marlos P, José T, Gomes-Filho E (2005) Antioxidant-enzymatic system of two sorghum genotypes differing in salt tolerance. Braz J Plant Physiol 17:4. https://doi.org/10.1590/S1677-04202005000400003
Cuevas HE, Prom LK (2020) Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan Sorghum Core collection. BMC Genom 21(1):88. https://doi.org/10.1186/s12864-020-6489-0
Curt MD, Fernandez J, Martinez M (1995) Productivity and water use efficiency of sweet sorghum (Sorghum bicolor (L.) Moench) cv. “Keller” in relation to water regime. Biomass Bioenergy 8(6):401–409. https://doi.org/10.1016/0961-9534(95)00036-4
de Oliveira DF, Lopes LS, Gomes-Filho E (2020) Metabolic changes associated with differential salt tolerance in sorghum genotypes. Planta 252(3):34. https://doi.org/10.1007/s00425-020-03437-8
Dhaka N, Krishnan K, Kandpal M, Ira Vashisht I, Pal M, Sharma MK, Sharma R (2020) Transcriptional trajectories of anther development provide candidates for engineering male fertility in sorghum. Sci Rep 10:897. https://doi.org/10.1038/s41598-020-57717-0
Echevarría C, Garcia-Mauriño S, Alvarez R, Soler A, Vidal J (2001) Salt stress increases the Ca2+-independent phosphoenolpyruvate carboxylase kinase activity in Sorghum leaves. Planta 214(2):283–287. https://doi.org/10.1007/s004250100616
Endo T, Yamamoto S, Larrinaga-M FJ, Honna H (2011) Status and causes of soil salinization of irrigated agricultural lands in southern Baja California. Mexico Appl Environ Soil Sci. https://doi.org/10.1155/2011/873625
FAO (1995) Sorghum and millets in human nutrition. Food and Agriculture Organization of the United Nations, Rome
FAO (2009) High-level expert forum—how to feed the world in 2050, economic and social development. Food and Agriculture Organization of the United Nations, Rome
Force L, Critchley C, van Rensen JJ (2003) New fluorescence parameters for monitoring photosynthesis in plants. Photosynth Res 78:17. https://doi.org/10.1023/A:1026012116709
Fu YB, Yang MH, Zeng F, Biligetu B (2017) Searching for an accurate marker-based prediction of an individual quantitative trait in molecular plant breeding. Front Plant Sci 8:1182. https://doi.org/10.3389/fpls.2017.01182
Fu L, Ding Z, Sun X, Zhang J (2019) Physiological and transcriptomic analysis reveals distorted ion homeostasis and responses in the freshwater plant Spirodela polyrhiza L. under salt stress. Genes (basel) 10(10):743. https://doi.org/10.3390/genes10100743
García-Mauriño S, Monreal J, Alvarez R, Vidal J, Echevarría C (2003) Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 216:648–655. https://doi.org/10.1007/s00425-002-0893-3
Gill K, Sharma P, Singh A, Bhullar P (2001) Effect of various abiotic stresses on the growth, soluble sugars and water relations of Sorghum seedlings grown in light and darkness. Bulg J Plant Physiol 27(1–2):72–84
Girma S, Krieg DR (1992) Osmotic adjustment in sorghum: I. Mechanisms of diurnal osmotic potential changes. Plant Physiol. https://doi.org/10.1104/pp.99.2.577
Greenway H, Munns R (1980) Mechanisms of salt tolerance in non-halophytes. Annu Rev Plant Biol 31:149–190. https://doi.org/10.1146/annurev.pp.31.060180.001053
Guo Y, Song Y, Zheng H, Zhang Y, Guo J, Sui N (2018) NADP-Malate dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J Agric Food Chem 66(24):5992–6002. https://doi.org/10.1021/acs.jafc.8b02159
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genom. https://doi.org/10.1155/2014/701596
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32:113–120. https://doi.org/10.1016/j.copbio.2014.11.025
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci 7:1787. https://doi.org/10.3389/fpls.2016.01787
Hefny M, Abdel-Kader DZ (2009) Antioxidant-enzyme system as selection criteria for salt tolerance in forage sorghum genotypes (Sorghum bicolor L. Moench). In: Ashraf M, Ozturk M, Athar H (eds) Salinity and water stress. Tasks for vegetation sciences, vol 44. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-9065-3_3
Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI (2011) K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol 156(3):1493–1507. https://doi.org/10.1104/pp.110.168047
Horst I, Welham T, Kelly S, Kaneko T, Sato S, Tabata S, Parniske M, Wang TL (2007) TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol 144(2):806–820. https://doi.org/10.1104/pp.107.097063
Huang R (2018) Research progress on plant tolerance to soil salinity and alkalinity in sorghum. J Integr Agric 17(4):739–746. https://doi.org/10.1016/S2095-3119(17)61728-3
Ibrahim ME, Adam A, Zhou G, Aboagla E, Zhu G, Nimir N, Ahmed I (2020) Biochar application affects forage sorghum under salinity stress. Chil J Agric Res 80:317–325. https://doi.org/10.4067/S0718-58392020000300317
Kandula V, Pudutha A, Kumari H, Kumar S, Kishor P, Anupalli R (2019) Overexpression of sorghum plasma membrane-bound Na+/H+ antiporter-like protein (SbNHXLP) enhances salt tolerance in transgenic groundnut (Arachis hypogaea L.). Plant Cell Tissue Organ Cult. https://doi.org/10.1007/s11240-019-01628-0
Karla J, Maria P, Claudia G, Magalhães V, Pimentel J, Schaffert L, Pinto L, de Souza M, Bernardino V, Silva K, Borém K, de Menezes A (2021) Genetic diversity and heterotic grouping of sorghum lines using SNP markers. Scientia Agricola. https://doi.org/10.1590/1678-992X-2020-0039
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189(1):54–81. https://doi.org/10.1111/j.1469-8137.2010.03540.x
Kumar A, Reddy B, Sharma H, Hash H, Rao P, Ramaiah B, Reddy P (2011) Recent advances in sorghum genetic enhancement research at ICRISAT. Am J Plant Sci 2:589–600. https://doi.org/10.4236/ajps.2011.24070
Kumari PH, Kumar SA, Sivan P, Katam R, Suravajhala P, Rao KS, Varshney RK, Kishor PBK (2016) Overexpression of a plasma membrane bound Na+/H+ antiporter-like protein (SbNHXLP) confers salt tolerance and improves fruit yield in tomato by maintaining ion homeostasis. Front Plant Sci 7:2027. https://doi.org/10.3389/fpls.2016.02027
Kurowska M, Daszkowska-Golec A, Gruszka D, Marzec M, Szurman M, Szarejko I, Maluszynski M (2011) TILLING: a shortcut in functional genomics. J Appl Genet 52(4):371–390. https://doi.org/10.1007/s13353-011-0061-1
Lacerda CDF, Cambraia J, Olivia MA, Ruiz HA, Prisco JT (2003) Solute accumulation and distribution during shoot and leaf development in two sorghum genotypes under salt stress. Environ Exp Bot 49(2):107–120. https://doi.org/10.1016/S0098-8472(02)00064-3
Lawlor DW (2002) Limitation to photosynthesis in water-stressed leaves: stomata vs. metabolism and the role of ATP. Ann Bot 89(7):871–885. https://doi.org/10.1093/aob/mcf110
Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6(1):75–83. https://doi.org/10.1105/tpc.6.1.75
Lin S, Zeng S, Biao A, Yang X, Yang T, Zheng G, Mao G, Wang Y (2021) Integrative analysis of transcriptome and metabolome reveals salt stress orchestrating the accumulation of specialized metabolites in Lycium barbarum L. Fruit Int J Mol Sci 22(9):94414. https://doi.org/10.3390/ijms22094414
Liu Q, Tian Z, Guo Y (2021) The maize hexokinase gene ZmHXK7 confers salt resistance in transgenic Arabidopsis plants. https://doi.org/10.1101/2021.04.26.441367
Luan T, Woolliams JA, Lien S, Kent M, Svendsen M, Meuwissen TH (2009) The accuracy of genomic selection in Norwegian red cattle assessed by cross-validation. Genetics 183:1119–1126. https://doi.org/10.1534/genetics.109.107391
Ludlow MM, Santamaria JM, Fukai S (1990) Contribution of osmotic adjustment to grain yield in Sorghum bicolor (L.) Moench under water-limited conditions. II. Water stress after anthesis. Crop Pasture Sci 41:67–78. https://doi.org/10.1071/AR9900067
Luo H, Zhao W, Wang Y, Yan Xia Y, Wu X, Zhang L, Tang B, Zhu J, Fang L, Du Z, Bekele WA, Tai S, Jordan DR, Godwin ID, Snowdon RJ, Mace ES, Luo J, Jing HC (2016) SorGSD: a sorghum genome SNP database. Biotechnol Biofuels. https://doi.org/10.1186/s13068-015-0415-8
Maathuis FJ, Ahmad I, Patishtan J (2014) Regulation of Na (+) fluxes in plants. Front Plant Sci 5:467. https://doi.org/10.3389/fpls.2014.00467
Mace ES, Singh V, Van Oosterom EJ, Hammer GL, Hunt CH, Jordan DR (2012) QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor Appl Genet 124(1):97–109. https://doi.org/10.1007/s00122-011-1690-9
Menamo T, Kassahun B, Borrell AK, Jordan DR, Tao Y, Hunt C, Mace E (2021) Genetic diversity of Ethiopian sorghum reveals signatures of climatic adaptation. Theor Appl Genet 134:731–742. https://doi.org/10.1007/s00122-020-03727-5
Morari F, Meggio F, Lunardon A, Scudiero E, Forestan C, Farinati S, Varotto S (2015) Time course of biochemical, physiological, and molecular responses to field-mimicked conditions of drought, salinity, and recovery in two maize lines. Front Plant Sci 12(6):314. https://doi.org/10.3389/fpls.2015.00314
Mulaudzi T, Hendricks K, Mabiya T, Muthevhuli M, Ajayi RF, Mayedwa N, Gehring C, Iwuoha E (2020) Calcium improves germination and growth of Sorghum bicolor seedlings under salt stress. Plants (basel) 9(6):730. https://doi.org/10.3390/plants9060730
Munns R (1988) Why measure osmotic adjustment. Funct Plant Biol 15:717–726. https://doi.org/10.1071/pp9880717
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25(2):239–250. https://doi.org/10.1046/j.0016-8025.2001.00808.x
Munns R, Gilliham M (2015) Salinity tolerance of crops—what is the cost? New Phytol 208(3):668–673. https://doi.org/10.1111/nph.13519
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Munns R, Weir R (1981) Contribution of sugars to osmotic adjustment in elongating and expanded zones of wheat leaves during moderate water deficits at two light levels. Aust J Plant Physiol 8(1):93–105. https://doi.org/10.1071/PP9810093
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30(4):360–364. https://doi.org/10.1038/nbt.2120
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Faheem SH (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32:261–285. https://doi.org/10.1080/13102818.2017.1400401
Nakaya A, Isobe SN (2012) Will genomic selection be a practical method for plant breeding? Ann Bot 110(6):1303–1316. https://doi.org/10.1093/aob/mcs109
Ndimba B, Ludivine T, Ngara R (2010) Sorghum 2-dimensional proteome profiles and analysis of HSP70 expression under salinity stress. Nat Sci 44:768–775
Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119(1):1–11. https://doi.org/10.1093/aob/mcw191
Netondo GW, Onyango JC, Beck J (2004) Gas exchange and chlorophyll fluorescence of sorghum under salt stress. Crop Sci 44(3):806–811. https://doi.org/10.2135/cropsci2004.8060
Ngara R, Ramulifho E, Movahedi M, Shargie NG, Brown AP, Chivasa S (2018) Identifying differentially expressed proteins in sorghum cell cultures exposed to osmotic stress. Sci Rep 8:8671. https://doi.org/10.1038/s41598-018-27003-1
Nida H, Blum S, Zielinski D, Srivastava DA, Elbaum R, Xin Z, Erlich Y, Fridman E, Shental N (2016) Highly efficient de novo mutant identification in a Sorghum bicolor TILLING population using the ComSeq approach. Plant J 86(4):349–359. https://doi.org/10.1111/tpj.13161
Osmanzai M (1992) Sorghum response to water deficit. In: ‘SADC/ICRISAT Southern Africa Programs: annual report 1992’, ICRISAT, Bulawayo, Zimbabwe (1992), pp 8
Parra-Londono S, Kavka M, Samans B, Snowdon R, Wieckhorst S, Uptmoor R (2018) Sorghum root-system classification in contrasting P environments reveals three main rooting types and root-architecture-related marker–trait associations. Ann Bot 121(2):267–280. https://doi.org/10.1093/aob/mcx157
Peng J, Liu J, Zhang L, Luo J, Dong H, Ma Y, Zhao X, Chen B, Sui N, Zhou Z, Meng Y (2016) Effects of soil salinity on sucrose metabolism in cotton leaves. PLoS ONE 11(5):e0156241. https://doi.org/10.1371/journal.pone.0156241
Perumal R, Tesfaye T, Geoffrey PM, Krishna J, Christopher R, Scott R, Yu J, Vara P, Tuinstra R (2021) Registration of the sorghum nested association mapping (NAM) population in RTx430 background. J Plant Regist 15(2):395–402. https://doi.org/10.1002/plr2.20110
Prasad PVV, Djanaguiraman M (2011) High night temperature decreases leaf photosynthesis and pollen function in grain sorghum. Funct Plant Biol 38:993–1003. https://doi.org/10.1071/FP11035
Reddy B, Are A, Reddy S (2010) Recent advances in sorghum improvement research at ICRISAT. Nat Sci 44:499–506
Rolly NK, Imran QM, Lee IJ, Yun BW (2020) Salinity stress-mediated suppression of expression of salt overly sensitive signaling pathway genes suggests negative regulation by AtbZIP62 transcription factor in Arabidopsis thaliana. Int J Mol Sci 21(5):1726. https://doi.org/10.3390/ijms21051726
Ruperao P, Thirunavukkarasu N, Gandham P, Selvanayagam S, Govindaraj M, Nebie B, Manyasa E, Gupta R, Das RR, Odeny DA, Gandhi H, Edwards D, Deshpande SP, Rathore A (2021) Sorghum pan-genome explores the functional utility for genomic-assisted breeding to accelerate the genetic gain. Front Plant Sci 12:666342. https://doi.org/10.3389/fpls.2021.666342
Salgotra RK, Stewart CN Jr (2020) Functional markers for precision plant breeding. Int J Mol Sci 21(13):4792. https://doi.org/10.3390/ijms21134792
Santamaria JM, Ludlow MM, Fukai S (1990) Contribution of osmotic adjustment to grain yield in Sorghum bicolor (L.) Moench under water-limited conditions. I. Water stress before anthesis. Crop Pasture Sci 41:51–65. https://doi.org/10.1071/AR9900067
Sellami S, Le Hir R, Thorpe MR, Vilaine F, Wolff N, Brini F, Dinant S (2019) Salinity effects on sugar homeostasis and vascular anatomy in the stem of the Arabidopsis thaliana inflorescence. Int J Mol Sci 20(13):3167. https://doi.org/10.3390/ijms20133167
Sharma A, Rana C, Singh C, Katoch V (2016) Soil salinity causes, effects, and management in cucurbits. In: Pessarakli M (ed) Handbook of cucurbits: growth, cultural practices, and physiology. CRC Press, Taylor and Francis Group, pp 419–434
Shehzad T, Okuizumi H, Kawase M, Okuno K (2009) Development of SSR-based sorghum (Sorghum bicolor (L.) Moench) diversity research set of germplasm and its evaluation by morphological traits. Genet Resour Crop Evol 56:809–827. https://doi.org/10.1007/s10722-008-9403-1
Sinclair T, Muchow R (2001) System analysis of plant traits to increase grain yield on limited water supplies. Agron J 93(3):263–270. https://doi.org/10.2134/agronj2001.932263x
Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol 23:75–81. https://doi.org/10.1038/nbt1043
Sudhakar RP, Srinivas RD, Sivasakthi K, Bhatnagar-Mathur P, Vadez V, Sharma KK (2016) Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front Plant Sci 7:529. https://doi.org/10.3389/fpls.2016.00529
Sui N, Yang Z, Liu M, Wang B (2015) Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genom 16:534. https://doi.org/10.1186/s12864-015-1760-5
Surender RP, Jogeswar G, Rasineni GK, Maheswari M, Reddy AR, Varshney RK, Kishor PBK (2015) Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol Biochem 94:104–113. https://doi.org/10.1016/j.plaphy.2015.05.014
Tangpremsri T, Fukai S, Fischer KS (1995) Growth and yield of sorghum lines extracted from a population for differences in osmotic adjustment. Crop Pasture Sci 46:61–74. https://doi.org/10.1071/AR9950061
Taylor J (2003) Overview: importance of sorghum in Africa. In: Belton PS, Taylor JRN (eds) Proceedings of the workshop on the proteins of sorghum and millets: enhancing nutritional and functional properties for Africa. Pretoria, South Africa, Department of Food Science. http://www.afripro.org.uk/papers/paper01Taylor.pdf. Accessed 17 Nov 2021
Tian T, You Q, Zhang L, Yi X, Yan H, Xu W, Su Z (2016) Sorghum FDB: sorghum functional genomics database with multidimensional network analysis. Database (oxford). 2016:baw099. https://doi.org/10.1093/database/baw099
Tian L, Xie Z, Lu C, Hao X, Wu S, Huang Y, Li D, Chen L (2019) The trehalose-6-phosphate synthase TPS5 negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep 38(8):869–882. https://doi.org/10.1007/s00299-019-02408-y
Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR, Greene EA, Comai L, Henikoff S (2004) Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol 4:12. https://doi.org/10.1186/1471-2229-4-12c
Turner NC (2018) Turgor maintenance by osmotic adjustment: 40 years of progress. J Exp Bot 69(13):3223–3233. https://doi.org/10.1093/jxb/ery181
Turner NC, Jones HG (1980) Turgor maintenance by osmotic adjustment: a review and evaluation. In: Turner NC, Kramer PJ (eds) Adaptation of plants to water and high temperature stress. Wiley, New York, pp 87–103
Upadhyaya HD, Wang YH, Gowda CL, Sharma S (2013) Association mapping of maturity and plant height using SNP markers with the sorghum mini core collection. Theor Appl Genet 126(8):2003–2015. https://doi.org/10.1007/s00122-013-2113-x
Volkov V (2015) Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front Plant Sci 6:873. https://doi.org/10.3389/fpls.2015.00873
Wagaw K, Bantte K, Tadesse T (2020) Gene action, combining ability and heterotic performance of Ethiopian Sorghum (Sorghum bicolor (L.) Moench) lines under moisture stress areas in Ethiopia. Afr J Plant Sci 14(9):372–394. https://doi.org/10.5897/AJPS2019.1813
Wang TT, Ren ZJ, Liu ZQ, Feng X, Guo RQ, Li BG, Li LG, Jing HC (2014) SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+ /K+ balance under Na+ stress. J Integr Plant Biol 56(3):315–332. https://doi.org/10.1111/jipb.12144
Wang H, Wang R, Liu B, Yang Y, Qin L, Chen E, Zhang H, Guan Y (2020) QTL analysis of salt tolerance in Sorghum bicolor during whole-plant growth stages. Plant Breed 139(3):455–465. https://doi.org/10.1111/pbr.12805
Wang A, Ma C, Ma H, Qiu Z, Wen X (2021) Physiological and proteomic responses of pitaya to PEG-induced drought stress. Agriculture 11:632. https://doi.org/10.3390/agriculture11070632
Weimberg R, Lerner HR, Poljakoff-Mayber A (1982) A relationship between potassium and proline accumulation in salt-stressed Sorghum bicolor. Physiol Plant 55:5–10. https://doi.org/10.1111/j.1399-3054.1982.tb00276.x
White P, Broadley MR (2003) Calcium in plants. Ann Bot 92:487–511. https://doi.org/10.1093/aob/mcg164
Xie C, Xu S (1998) Efficiency of multistage marker-assisted selection in the improvement of multiple quantitative traits. Heredity (edinb) 4:489–498. https://doi.org/10.1046/j.1365-2540.1998.00308.x
Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8:103. https://doi.org/10.1186/1471-2229-8-103
Xu Y, Liu X, Fu J, Wang H, Wang J, Huang C, Boddupalli P, Michael O, Wang G, Zhang A (2020) Enhancing genetic gain through genomic selection: From livestock to plants. Plant Commun 1(1):2590–3462. https://doi.org/10.1016/j.xplc.2019.100005
Yamazaki K, Ishimori M, Kajiya-Kanegae H, Takanashi H, Fujimoto M, Yoneda J, Yano K, Koshiba T, Tanaka R, Iwata H, Tokunaga T, Tsutsumi N, Fujiwara T (2020) Effect of salt tolerance on biomass production in a large population of sorghum accessions. Breed Sci 70(2):167–175. https://doi.org/10.1270/jsbbs.19009
Yang Y, Guo Y (2018) Unraveling salt stress signaling in plants. J Integr Plant Biol 60(9):796–804. https://doi.org/10.1111/jipb.12689
Yang X, Xu Y, Shah T, Li H, Han Z, Li J, Yan J (2011) Comparison of SSRs and SNPs in assessment of genetic relatedness in maize. Genetica 139(8):1045–1054. https://doi.org/10.1007/s10709-011
Yang Z, Li JL, Liu LN, Xie Q, Sui N (2020) Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet sorghum. Front Plant Sci 10:1722. https://doi.org/10.3389/fpls.2019.01722
Yao X, Horie T, Xue S, Leung HY, Katsuhara M, Brodsky DE, Wu Y, Schroeder JI (2010) Differential sodium and potassium transport selectivities of the rice OsHKT2;1 and OsHKT2;2 transporters in plant cells. Plant Physiol 152(1):341–355. https://doi.org/10.1104/pp.109.145722
Yilmaz S, Temizgü R, Yürürdurmaz C, Kaplan M (2020) Oxidant and antioxidant enzyme response of redbine sweet sorghum under NaCl salinity stress. Bioagro 32(1):31–38
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178(1):539–551. https://doi.org/10.1534/genetics.107.074245
Zeleke W, Dong H, Paterson AP, Worku W, Bantte K (2021) Genetic diversity, population structure, and selection signature in Ethiopian sorghum [Sorghum bicolor L. (Moench)] germplasm. G Genes Genomes Genet. https://doi.org/10.1093/g3journal/jkab087
Zhan Q, Shu C, Li X, Zhan M, Li J (2019) Screening of SSR primers and evaluation of salt tolerance in 20 sweet sorghum varieties for silage. In: International Grassland Congress Sydney, Australia from September 15 through September 19, 2013. Proceedings Editors: David L. Michalk, Geoffrey D. Millar, Warwick B. Badgery, and Kim M. Broadfoot. Publisher: New South Wales Department of Primary Industry, Kite St., Orange New South Wales, Australia
Zhang HH, Xu N, Wu X, Wang J, Ma S, Li X, Sun G (2018) Effects of four types of sodium salt stress on plant growth and photosynthetic apparatus in sorghum leaves. J Plant Interact 13(1):506–513. https://doi.org/10.1080/17429145.2018.1526978
Zhao W, Faust F, Schubert S (2020) Potassium is a potential toxicant for Arabidopsis thaliana under saline conditions. J Plant Nutr Soil Sci 3:455–467. https://doi.org/10.1002/jpln.201900491
Zorb C, Geilfus CM, Dietz KJ (2018) Salinity and crop yield. Plant Biol (stuttg) 21(Suppl 1):31–38. https://doi.org/10.1111/plb.12884
Acknowledgements
We thank the OCP Phosboucraa Foundation for supporting this project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amombo, E., Ashilenje, D., Hirich, A. et al. Exploring the correlation between salt tolerance and yield: research advances and perspectives for salt-tolerant forage sorghum selection and genetic improvement. Planta 255, 71 (2022). https://doi.org/10.1007/s00425-022-03847-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03847-w