Abstract
Rapid chromosome movement during prophase of the first meiotic division has been observed in many organisms. It is generally concomitant with formation of the “meiotic chromosome bouquet,” a special chromosome configuration in which one or both chromosome ends attach to the nuclear envelope and become concentrated within a limited area. The precise function of the chromosomal bouquet is still not fully understood. Chromosome mobility is implicated in homologous chromosome pairing, synaptonemal complex formation, recombination, and resolution of chromosome entanglements. The basic mechanistic module through which forces are exerted on chromosomes is widely conserved; however, phenotypic differences have been reported among various model organisms once movement is abrogated. Movements are transmitted to the chromosome ends by the nuclear membrane-bridging SUN/KASH complex and are dependent on cytoskeletal filaments and motor proteins located in the cytoplasm. Here we review the recent findings on chromosome mobility during meiosis in an animal model system: the Caenorhabditis elegans nematode.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Rapid chromosome movement is widely conserved
Sexual reproducing organisms must reduce their DNA content by half prior to gamete formation to preserve their genome size in subsequent generations. To achieve this, a diploid cell undergoes two rounds of consecutive chromosome segregation during meiosis, thus giving rise to four haploid products that can, in principle, develop into gametes. During the first meiotic division, homologous chromosomes are separated from each other; in the second, sister chromatids are partitioned. To enable the faithful segregation of parental homologous chromosomes to the opposite poles in anaphase I, a stable connection must be established between them. In most organisms, such as yeasts, ciliates, flies, mammals, or Caenorhabditis elegans, such stable connections between homologous chromosomes are formed by crossovers, which result from double-strand break (DSB) repair by homologous recombination, involving the use of the homolog as a repair template, and subsequent cohesion. In most species, homologous chromosomes, unlike sister chromatids, are not associated during pre-meiotic growth; this is also the case for C. elegans (Dernburg et al. 1998; Lorenz et al. 2003). Consequently, homologous chromosomes must implement a search process to locate each other, identify each other as “self,” form pairs, and stabilize chromosome pairs by establishing a proteinaceous structure, the synaptonemal complex (SC) between them. All of these events take place during extended meiotic prophase I to form the obligatory crossovers prior to anaphase I. At the same time, unwanted interactions, such as entanglements or non-homologous interactions, must be prevented or resolved.
In many organisms, chromosome end-led movements have been studied in prophase of meiosis I by combining genetics with in vivo imaging of various chromatin or chromosomal markers, such as tagged telomere proteins, Hoechst dyes, LacO/LacR-inserted repressor-binding sites, tagged SC proteins or chromosome associated proteins (Conrad et al. 2008; Ding et al. 2004; Koszul et al. 2008; Morimoto et al. 2012; Parvinen and Soderstrom 1976; Scherthan et al. 2007; Sheehan and Pawlowski 2009). Characterization of both wild-type movement and impaired movement in mutants led to proposed roles for chromosome movement in the homology search process, in synapsis, in minimizing undesirable chromosomal interactions, and even in actively promoting recombination (Koszul and Kleckner 2009; Wanat et al. 2008). C. elegans is one of the most intensively studied model organisms to date. Many of the requirements and functions of meiotic chromosome mobility are understood in this 1-mm-long roundworm. Here we attempt to summarize recent insights into the properties, biological significance, regulation, and mechanism of chromosome movement during meiotic prophase in this well-established meiotic model system.
Why use C. elegans as a model to study chromosome mobility?
In the two gonad tubes of C. elegans hermaphrodites (or the single one in males), complete meiotic prophase I is both temporally and spatially organized (Fig. 1). In the adult worm, meiocytes progress through the meiotic cell cycle as they move along the gonad tube from the distal tip to the spermatheca. This means that nuclei at any given disto-proximal position in the syncytial gonad have spent the same time in meiosis (Crittenden et al. 1994; Hirsh et al. 1976); therefore, the gonad represents a highly synchronous meiotic time course (Fig. 2). The gonads of an adult hermaphrodite contain half the total number of nuclei of the entire animal (Hirsh et al. 1976); as a result, sufficient meiocytes are available for biochemical analysis. In addition, because the worm is transparent, the gonad can be visualized in whole mounted animals. Furthermore, it can easily be extruded from the adult hermaphrodite (Figs. 1 and 2). These properties make C. elegans a highly suitable model for both high-resolution immunofluorescence analysis of fixed tissues and live imaging of meiocytes progressing through the different stages of meiotic prophase. The availability of several GFP- and mCherry-tagged transgenes has substantially contributed to our improved understanding of meiotic chromosome motion in C. elegans. Elegant methods for single-copy transgene insertion (Frokjaer-Jensen et al. 2008) and targeted genomic deletions (Friedland et al. 2013; Frokjaer-Jensen et al. 2010; Wood et al. 2011) have enabled the efficient generation of transgenic animals as powerful tools for analyzing meiotic chromosome motion.
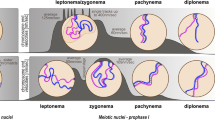
Dissected gonad of a hermaphrodite. A sun-1::gfp transgene highlights the nuclear envelope (co-stained with DAPI). The mitotic zone represents a stem cell niche. Upon meiotic entry, meiocytes pass through the progressive stages of prophase I. They move along the gonad tube with an approximate speed of one cell row per hour. SUN-1 localization changes in leptotene/zygotene, where it becomes concentrated to chromosome end attachments at the nuclear envelope. Autosomal aggregates redistribute in early pachytene; at this stage, only the X chromosome aggregate is seen. In the late pachytene, SUN-1 is evenly distributed throughout the nuclear envelope
Meiosis in C. elegans
After pre-meiotic DNA replication and passage through the meiotic entry zone, meiocytes with unpaired homologous chromosomes enter the leptotene/zygotene stage, located at approximately 20 cell rows from the distal tip cell (see Fig. 2) (Crittenden et al. 2006; Dernburg et al. 1998). Leptotene and zygotene cannot be readily distinguished by cytology. For this reason, the leptotene/zygotene stage is referred to as the transition zone in worms. Chromatin in nuclei within the transition zone is concentrated to one side and thus appears crescent-shaped (Hirsh et al. 1976). It is at this stage that chromosome pairing is achieved (Dernburg et al. 1998). Concomitantly, the SC progressively stabilizes the association between individual homologous chromosome pairs at a distance of roughly 100 nm apart along the length of the chromosomal axes (Smolikov et al. 2008). SPO-11-generated DSBs are first introduced within the transition zone (Alpi et al. 2003; Colaiacovo et al. 2003; Mets and Meyer 2009). However, unlike in other organisms, pairing and SC formation in C. elegans are independent of both meiotic DSBs and recombination (Dernburg et al. 1998). In this organism, homolog recognition can be genetically separated from synapsis (Colaiacovo et al. 2003); however, DSB repair must take place in the context of the SC to form the obligate crossovers between homologous chromosome pairs (Alpi et al. 2003; Colaiacovo et al. 2003; MacQueen et al. 2002). So-called homology recognition regions or pairing centers are responsible for DSB-independent pairing in C. elegans (Herman and Kari 1989; Herman et al. 1982; McKim et al. 1988; Rose et al. 1984; Rosenbluth and Baillie 1981; Villeneuve 1994). These pairing centers are localized to a single subtelomeric region of each chromosome (McKim et al. 1988). C. elegans expresses four C2H2 zinc finger proteins [HIM-8 and ZIM-1, ZIM-2, ZIM-3, referred to as ZIMs (i.e., zinc finger in meiosis)] that bind specifically to the highly repetitive sequences of the pairing center in one or two of the six chromosomes (ZIM-1, ZIM-2, and ZIM-3 bind to the autosomes and HIM-8 binds to the X chromosome) (Phillips and Dernburg 2006; Phillips et al. 2009, 2005; Sanford and Perry 2001). Pairing center repeats are sufficient to trigger ZIM loading. Furthermore, ZIMs are required for aligning homologs and synapsis of the specific chromosomes with which they are associated (Phillips and Dernburg 2006; Phillips et al. 2009, 2005).
Meiocytes move along the gonad with an approximate speed of one cell row per hour (Crittenden et al. 2006). Pairing and synapsis between homologous chromosomes is completed within the 7–12 cell rows of the transition zone, and the cells then enter early pachytene stage. In the early pachytene, chromatin starts to be redistributed but remains loosely clustered toward one pole. Later in pachytene, chromatin is found distributed throughout the entire nuclear periphery. Most DSBs are repaired in early pachytene, and sites of the maturing crossover become visible in late pachytene (see Fig. 3) (Alpi et al. 2003; Bhalla et al. 2008; Colaiacovo et al. 2003; Jantsch et al. 2004; Yokoo et al. 2012). In C. elegans, recombination using the homologous chromosome as a repair template depends on SC formation (Alpi et al. 2003; Colaiacovo et al. 2003; MacQueen et al. 2002).
Representative nuclei at pre-meiotic and meiotic (leptotene/zygotene, early pachytene, and late pachytene) stages. Images of SUN-1::GFP, SUN-1 phosphoserine-12 (S12), and HIM-8 (marks the X chromosome pairing center) labeling, and DAPI staining. The following structures and markers were hand-drawn: chromosome axes, synaptonemal complex, tubulin, and dynein. RAD-51, MSH-5, ZHP-3, and COSA-1 indicate the precursors of crossover recombination. In leptotene/zygotene, SUN-1 is concentrated at chromosome ends, where it becomes phosphorylated at serine-12 (S12). SUN-1 aggregation is mirrored on the cytoplasmic side of the membrane by the aggregation of SUN-1’s KASH partner ZYG-12 and components of the dynein motor. Chromosome axes load the lateral element SC components upon meiotic entry. In leptotene/zygotene, chromosomes vigorously move and install the central elements of the SC. At the same time, DSBs are induced and the RAD-51 strand invasion protein, and other markers of maturing crossovers allow us to follow ongoing recombination. COSA-1 labels the six crossover sites in late pachytene. In early pachytene, synapsis is complete and SUN-1 which is concentrated around autosomal chromosome end attachments is dephosphorylated and probably relocalizes; only X chromosome end attachment persists and is dissolved in the late pachytene. Pairing center regions at chromosome end attachments at the nuclear envelope recruit pairing center proteins (HIM-8 marks the X chromosome). These act as recruitment sites for PLK-2. SUN-1 serine-12 is a substrate for PLK-2. PLK-2 colocalizes with SUN-1 aggregates at chromosome ends. PLK-2 also decorates the developing SC
The diplotene stage begins with the disassembly of the SC (Nabeshima et al. 2005), and consequently, chiasmata, the cytological manifestations of crossovers, become apparent during this process. During diakinesis, meiocytes leave the syncytium and cellularize. Following further chromosome condensation, six DAPI-positive structures become visible, corresponding to each of the six bivalents. These pairs of homologs are usually held together by only one chiasma per chromosome pair (Villeneuve 1994), reflecting the presence of robust crossover interference in this organism.
Core components of the C. elegans meiotic chromosome movement machinery
Pairing centers and pairing center-binding proteins drive and regulate the highly conserved meiotic prophase chromosome movements that occur in C. elegans (Fig. 3). HIM-8 is expressed and localized as foci to X chromosome pairing centers throughout the gonad (Phillips et al. 2005). Autosomal pairing centers behave differently; ZIM-1, ZIM-2 and ZIM-3 foci are only found at the autosomal pairing centers during leptotene/zygotene (and to a reduced extent in early pachytene) stage. Loading of ZIMs onto autosomal pairing centers depends on the chk-2 kinase (Phillips and Dernburg 2006; Phillips et al. 2005), a key master regulator of C. elegans meiosis (MacQueen and Villeneuve 2001). The pairing center bearing chromosome ends are found at the nuclear envelope while loading pairing center proteins to the appropriate chromosome region, (Phillips et al. 2009). Subsequent polo-like kinase 2 (PLK-2) recruitment to these sites is dependent on the presence of pairing center proteins. PLK-2 kinase activity induces dramatic changes at the nuclear envelope (Harper et al. 2011; Labella et al. 2011): The inner and outer nuclear envelope proteins SUN-1 and ZYG-12 form distinct aggregates at pairing center localization sites (Penkner et al. 2009; Sato et al. 2009). SUN-1 and ZYG-12 form a functional SUN/KASH domain protein–protein interaction module (Fig. 4) (Fridkin et al. 2004; Malone et al. 2003; Minn et al. 2009; Penkner et al. 2007), which bridges the nuclear membranes and connects the cytoskeleton to chromatin [for review, see Burke (2012)]. For simplicity, we term the multiprotein complexes at the nuclear envelope, consisting of pairing centers, ZIMs, PLK-2, and the SUN-1/ZYG-12 nuclear envelope bridging complex, the “movement-mediating complex” (MMC). In the absence of SUN/KASH interaction, ZYG-12 retention at the nuclear envelope is defective (Malone et al. 2003; Penkner et al. 2007), despite the assembly of SUN-1 aggregates following the loading of pairing center proteins to the one chromosome end at the nuclear envelope (Penkner et al. 2009). Therefore, the trigger for aggregation is most likely to be transmitted from the nucleus to the cytoplasm. MMC formation is independent of DSBs, recombination, pairing, and synapsis, but requires chk-2 (Penkner et al. 2009; Sato et al. 2009).
Schematic diagram of the SUN-1/KASH bridge spanning the nuclear membranes. The SUN-1/KASH bridge spans the outer and inner nuclear membranes (ONM and INM). ZYG-12 connects to the movement apparatus (comprising dynein and cytoplasmic microtubules). Chromosomes connect to the SUN-1/KASH bridge via the pairing centers, which are bound by pairing center proteins (ZIMs) and PLK-2
MMCs persist on all autosomal pairing centers until the end of leptotene/zygotene (Fig. 3). In early pachytene, SUN-1 and ZYG-12 start to redistribute throughout the nuclear envelope, and the autosomal ZIM foci diminish (Penkner et al. 2009; Sato et al. 2009). Only the MMC formed around the X chromosome pairing center persists throughout early pachytene. In middle and late pachytene stages, most nuclei lack SUN-1/ZYG-12 aggregates, despite the presence of a HIM-8 focus at the nuclear envelope (Penkner et al. 2009; Phillips et al. 2005; Sato et al. 2009). PLK-2 diffuses from MMCs to synapsed chromosome axes throughout prophase and is found on the synapsed chromosomes during pachytene stage (Harper et al. 2011; Labella et al. 2011).
Features of chromosome mobility in wild-type C. elegans
Several fluorescent markers have been used to study chromosome movement in vivo. GFP-tagged transgenes of SUN-1 (Penkner et al. 2009) and ZYG-12 (Malone et al. 2003; Sato et al. 2009) mark all chromosome ends (the sex chromosome and autosomes). GFP-tagged HIM-8 can specifically visualize the single X chromosome pairing center and was used to assess pre-meiotic movement (Wynne et al. 2012). In addition, delayed pre-meiotic replication of the X chromosome enables specific X chromosome labeling for live imaging (Jaramillo-Lambert et al. 2007). Hoechst dye or mCherry-tagged histone H2B (McNally et al. 2006) has been employed to assess the mobility of the entire chromatin mass.
It is clear that MMCs display highly mobile behavior (see Fig. 5). During leptotene/zygotene, an average of four to six MMCs can be observed moving within a nucleus, but the number can fluctuate between 1 and 12 (in C. elegans, 2n = 12) (Baudrimont et al. 2010; Penkner et al. 2009; Sato et al. 2009; Wynne et al. 2012). In contrast to the highly mobile chromosome ends, other chromosome regions are relatively static. A high degree of chromosome elasticity enables the pairing center to explore a larger area of the nucleus than the distal ends of the chromosomes (Wynne et al. 2012). In addition, pairing center chromosome ends have been observed to move independently. Over time, MMCs fuse into local clusters; from there they either continue to move together or “bud out” of those clusters and resume moving individually. Thus, chromosomes interact with both homologous and heterologous partners during the period of homology search and SC formation. Most interactions exist for around 1 min, a smaller fraction for up to 3 min, and an even smaller fraction for more than 3 min (Baudrimont et al. 2010).
Chromosome ends display highly dynamic behavior. All chromosome ends are highlighted by SUN-1–GFP aggregates; these form around chromosome ends adjacent to the chromosome pairing center regions. The X chromosome pairing center is visualized by a him-8::mCherry transgene. Six time points of a time-lapse imaging series are shown. Bottom right, displacement tracks of SUN-1 and HIM-8 signals
High-resolution 4D in vivo analysis of MMC mobility suggests that two types of movement alternate. When analyzing the motion of a single-MMC, processive chromosome movements, in the sense that an MMC moves continuously in the same direction for several seconds, are superimposed on undirected movements, in which an MMC oscillates close to its origin. MMCs only rarely engage in processive movements; thus, fast trajectories only contribute to 15 % of displacement tracks. Nevertheless, this processive movement accounts for most surface area exploration by a single MMC. These chromosome-end trajectories have an average length of 0.5 μm (with an average speed of 0.19 μm/s) but can be as long as 2 μm (Wynne et al. 2012).
It is important to note that limitations to the time resolution of chromosome movement (minimum recording intervals used in the most technically advance study were 0.4 s in a single plane and 2 s in Z stacks) currently impedes the accurate measurement of MMC mobility (Wynne et al. 2012). It also complicates the determination of background movements in the complex tissue of the gonad. Recordings of euthanized animals suggest that the lower speed ranges (which are especially overrepresented in non-processive movement) may represent background motion (Baudrimont et al. 2010). Residual muscle activity and gonad pumping in anesthetized animals may even increase background movement during live imaging. Therefore, we would like to refrain from making a distinction between processive and non-processive motions and refer to “rapid prophase movements” (RPMs) to specifically describe movements above the background level of diffusion and Brownian motion.
Chromosome end movements during leptotene/zygotene are mainly restricted to one hemisphere of the nucleus (Baudrimont et al. 2010; Labrador et al. 2013; Penkner et al. 2009). After completion of pairing and synapsis, meiocytes enter early pachytene and chromatin clustering is loosened but not completely abrogated (Fig. 3). RPMs of the single remaining MMC (induced by the pairing center of the X chromosome) are reduced and the explored area of the nuclear envelope is smaller than during leptotene/zygotene. Nevertheless, the X chromosome remains mobile and continues to undergo rapid prophase motion (RPM) in early pachytene (Wynne et al. 2012). Although it is obvious that chromosomes continue to pair and synapse while they progress through leptotene/zygotene, the characteristics of chromosome movement remain essentially the same with respect to the number of MMCs, the distance traveled on the nuclear envelope, and the rate of MMC fusion and splitting events (Baudrimont et al. 2010; Wynne et al. 2012), indicating that paired/synapsed chromosome pairs continue to move throughout this stage.
How are rapid chromosome movements generated?
In the gonad syncytium of C. elegans, nuclei are embedded in a dense meshwork of microtubule bundles (Minn et al. 2009). Components of the dynein motor complex are enriched at sites of SUN-1/ZYG-12 aggregation (Sato et al. 2009), and ZYG-12 is required for dynein localization to the nuclear envelope (Malone et al. 2003; Sato et al. 2009). Furthermore, an analysis of RPM characteristics found that they fitted well with known parameters for motor protein-driven movement. Consistent with this, actin was found to be dispensable for RPMs (Wynne et al. 2012).
RPMs are absent following dynein knockdown or tubulin depolymerization, despite correct MMC assembly (Labrador et al. 2013; Wynne et al. 2012). Since dynein localizes to MMCs that are not engaged in RPM, it is apparent that dynein localization to MMCs is insufficient to engage them in RPMs (Labrador et al. 2013; Wynne et al. 2012). Dynein motor function requires ATP. Consequently, ATP depletion by cytochrome C oxidase inhibition caused by the administration of low doses of sodium azide severely abrogates RPMs. Furthermore, experiments using a specific allele of the mitochondria-localized SPD-3 protein showed that mitochondria are likely to produce the energy required for RPMs, but not for MMC formation or dynein complex localization to MMCs (Labrador et al. 2013).
Taken together, these observations suggest that RPMs are the result of dynein–ATP-driven MMC mobilization along cytoplasmic microtubules.
Are RPMs required for chromosome pairing?
Under conditions that prevent the formation of functional MMCs (such as mutations in sun-1, chk-2, or plk-2 or ZIM knockdown), both homologous pairing and RPMs are absent (Harper et al. 2011; Labella et al. 2011; MacQueen and Villeneuve 2001; Penkner et al. 2007; Phillips and Dernburg 2006; Phillips et al. 2005). When MMCs are intact but RPMs are impaired for other reasons (e.g., disruption of the SUN/KASH bridge, tubulin depolymerization, ATP depletion, spd-3 mutation, or dynein knockdown), chromosome pairing is severely reduced or absent and SCs start to be formed between non-homologs. This is accompanied by a lack of chromosome end fusion (and splitting) events (Baudrimont et al. 2010; Labrador et al. 2013; Wynne et al. 2012). Taken together, this strongly suggests that RPMs are in fact required for homologous pairing and not only for preventing non-homologous synapsis, since spd-3 or sun-1 mutants combined with a synapsis mutant do not result in a significant improvement in autosome pairing; only X chromosome homologous pairing is somewhat improved (Labrador et al. 2013; Penkner et al. 2007).
How do RPMs contribute to pairing? RPMs of individual MMCs are not directed toward each other (Wynne et al. 2012). Movement, the number of MMCs, and pairing center collision events do not change during leptotene/zygotene, despite ongoing pairing and synapsis (Baudrimont et al. 2010). Furthermore, during leptotene/zygotene, paired HIM-8 foci disassociate and there are no differences in movement properties between paired and unpaired pairing centers (Wynne et al. 2012). This suggests that RPMs do not bring chromosome ends closer together, but rather that when chromosome ends meet randomly, RPMs induce collisions and “shuffle” chromosome ends (Baudrimont et al. 2010) until their interaction is stabilized along the entire chromosome length by the SC.
When RPMs are absent (e.g., by ATP or dynein depletion), chromosomes exhibit reduced pairing in regions both proximal and distal to the pairing center regions (Labrador et al. 2013). In this background, MMCs can still explore a larger nuclear surface area, as shown by comparing HIM-8 mobility in pre-meiotic and meiotic nuclei. This RPM-independent mobility depends largely on the CHK-2 kinase. It is speculated that CHK-2-dependent movement enables the residual pairing observed in RPM-deficient conditions (Wynne et al. 2012). Alternatively, or perhaps additionally, chromosome clustering to one hemisphere of the nucleus upon meiotic entry may contribute to homologous pairing. Such clustering is independent of RPMs but dependent on MMC formation and nuclear bridging by the SUN/KASH complex (Labrador et al. 2013; Penkner et al. 2007).
How is chromosome movement controlled?
Chromosome movement does not occur in meiocytes throughout prophase I. For autosomes, RPMs are likely to be restricted to leptotene/zygotene; in contrast, the X chromosome undergoes rapid chromosome movements until the end of early pachytene (Wynne et al. 2012). This movement is not controlled by the position of the nucleus within the gonad. For instance, when synapsis is abrogated, the nuclei arrest in leptotene (or leptotene/zygotene) and throughout the gonad all chromosomes display RPM for an extended period. Therefore, the signal controlling RPMs must emanate from within the nucleus. Ongoing pairing and synapsis initiation, which happens during leptotene/zygotene, does not alter MMC mobility or persistence (Baudrimont et al. 2010; Wynne et al. 2012). Nevertheless, several observations suggest that unsynapsed chromosome axes lead to continued stimulation of chromosome movement of all chromosomes within a nucleus. First, synchronous termination of autosome movement is concomitant with SC coverage of all chromosome axes in the wild-type gonad (Baudrimont et al. 2010). Second, when synapsis is defective but the movement apparatus is functional, movement persists (Baudrimont et al. 2010; Wynne et al. 2012). Third, in mutants in which SC central elements are prematurely installed onto chromosome axes (and even onto non-homologs) movement ceases prematurely. HAL-2, a recently discovered C. elegans nuclear protein, was found to prevent premature SC central element localization to unpaired chromosomes. HAL-2 depletion leads to a failure in MMC formation, leading to an absence of pairing and most likely movement, inferred from the absence of chromatin clustering in the TZ. Co-depletion of SC components and HAL-2 restores MMC formation and chromosome pairing (Zhang et al. 2012). The mechanism how hal-2 establishes a response of unsynapsed chromosomes to the regulation of the movement machinery has not been elucidated yet. Fourth, mutation in htp-1, one of the four HORMA-domain (HORMAD) Hop1 orthologs that binds to the meiotic chromosome axis, appears to abrogate the prolonged chromatin movement in SC mutants (Martinez-Perez and Villeneuve 2005), provided that chromatin clustering reflects ongoing movement. Furthermore, the HTP-3 and HIM-3 HORMAD proteins are probably required to construct MMCs in the first place; this is suggested by an absence of ZIMs and SUN-1 aggregates concentrating at chromosome ends in htp-3 or him-3 mutants (Baudrimont et al. 2010; Labella et al. 2011; Penkner et al. 2009).
Since RPMs require MMCs, their formation and disintegration provide an elegant way to switch chromosome movement on and off. Moreover, no mutant has been identified in which the assembled MMCs are stable but fail to move when the cytosolic movement apparatus is intact. Therefore, it is reasonable to speculate that synapsis completion switches off the signal that leads to MMC formation or persistence and thereby terminates movement. One factor required for MMC stability is SUN-1 phosphorylation at several residues in its nuclear amino-terminus. These phosphorylation events are dependent on CHK-2 and PLK-2 and are initiated upon meiotic entry (Harper et al. 2011; Labella et al. 2011; Penkner et al. 2009). SUN-1 serine-12 phosphorylation is restricted to SUN-1 molecules located within MMCs (Fig. 3) (Penkner et al. 2009). When all putative target phosphorylation sites are replaced with non-phosphorylatable amino acids, smaller, less distinct MMCs are observed; however, these can still undergo RPM and chromosome pairing follows wild-type kinetics, albeit with a decreased rate of SC polymerization. MMCs disassemble prematurely in these SUN-1 phosphorylation-deficient mutants when synapsis is compromised (Penkner et al. 2009; Woglar et al. 2013). Therefore, SUN-1 phosphorylations are required to sustain PLK-2 recruitment to MMCs under a challenged condition and thus they are able to prolong the time window of chromosome movement. By analogy, PLK-2 deletion is somewhat compensated for by the PLK-1 homolog, and MMCs form upon meiotic entry but fail to be sustained in synapsis-deficient mutants (Harper et al. 2011; Labella et al. 2011). PLK-2 recruitment to chromosome ends and SUN-1 phosphorylation are therefore interdependent. Since PLKs have an essential role in chromosome movement [(Harper et al. 2011; Labella et al. 2011); Monique Zetka, personal communication], SUN-1 phosphorylation and PLK-2 localization to the ends of unsynapsed chromosomes could represent a positive feedback loop to sustain chromosome end-led mobility for as long as necessary to complete synapsis.
Chromosome mobility during early pachytene
RPMs of all chromosomes are required for chromosome pairing and homolog synapsis during leptotene/zygotene (Baudrimont et al. 2010; Penkner et al. 2007; Penkner et al. 2009; Sato et al. 2009; Wynne et al. 2012). Although RPMs seem to persist in early pachytene (Wynne et al. 2012), to date we have information only on X chromosome mobility during early pachytene. At this stage, the pairing is complete and SC installation has progressed to a large extent (Dernburg et al. 1998; Tang et al. 2010). Recombinational repair of spo-11-induced meiotic DSBs that can result in crossover formation takes place during this meiotic stage (Fig. 3). Most RAD-51 foci start to appear in the transition zone, peak in early to mid-pachytene, and then disappear before late pachytene (Alpi et al. 2003; Colaiacovo et al. 2003); sites of future crossovers are marked by ZHP-3 and MSH-5 proteins during pachytene (Fig. 3) (Bhalla et al. 2008; Jantsch et al. 2004; Yokoo et al. 2012). During early pachytene, the period of chromosome mobility is prolonged when repair or recombination are prevented (Woglar et al. 2013), similar to the extension of leptotene/zygotene mobility observed when synapsis is abrogated (Baudrimont et al. 2010; MacQueen et al. 2002; Wynne et al. 2012).
In other organisms in which movement plays a minor role in the actual chromosome pairing process, ectopic recombination and chromosome entanglements have been observed when movement is limited, suggesting that movement can overcome non-homologous (i.e., weaker) chromosomal interactions (Conrad et al. 2008; Davis and Smith 2006; Goldman and Lichten 2000; Golubovskaya et al. 2002; Koszul et al. 2008; Niwa et al. 2000; Schlecht et al. 2004).
Furthermore, the movement apparatus plays a role in crossover interference in budding yeast (Chua and Roeder 1997). It was previously speculated that chromosome oscillations could account for negative crossover interference (Hulten 2011). Also, the enlargement of chromosomal territories caused by the pull on chromosomes (Wynne et al. 2012) may aid axis elongation and SC extension in early pachytene stage. This would improve the efficiency of crossover interference, since complete SC localization to chromosome axes is required for the execution of crossover interference (Hayashi et al. 2010; Hillers and Villeneuve 2003; Sym and Roeder 1994).
To date, there is no experimental evidence in C. elegans to directly link the residual early pachytene movement with any of these tasks, since movement is required for the preceding homologous chromosome pairing. However, the easy-to-access C. elegans gonad offers the possibility of transient movement inhibition in meiotic nuclei in which chromosome pairing and synapse have already taken place. This would facilitate the use of the worm as an animal model to study all aspects of chromosome mobility.
Synopsis
The power of C. elegans genetics has enabled the generation of particularly informative alleles of genes required to drive the movement machinery of chromosomes in meiotic prophase I. Furthermore, drug inhibition of cytoskeletal forces and the establishment of live imaging in whole mounted animals, together with the insight provided by studying these key mutants, have enabled the construction of a basic framework to understand the major role of prophase chromosome movement in an important animal model system (also see Fig. 3).
-
Following meiotic entry, one end of each chromosome is tethered to the nuclear envelope, where cytoskeletal forces are transmitted to chromosomes via a SUN/KASH bridge spanning the nuclear membranes.
-
Chromosome tethering to the nuclear envelope is accompanied by the binding of pairing center-binding zinc finger proteins to pairing centers/homology recognition regions on one end of each chromosome.
-
The pairing centers/homology recognition regions recruit PLK(s) that couple chromosomes to the movement apparatus.
-
Chromosome movement depends on microtubules, the dynein motor complex, and ATP supplied by mitochondria.
-
Rapid chromosome movements are essential for homolog searching through random chromosome encounters.
-
Reduced chromosome end-led motion has an equally negative effect on pairing for chromosome regions distant from pairing centers.
-
Rapid chromosome movements prevent synapsis between non-homologs.
-
Although frequently seen, chromosome end-led movements correlate with the presence of clustered chromatin in the transition zone; however, this is not obligatory.
-
Meiotic chromosome axis proteins and PLK-2 and CHK-2 kinases control movement.
-
It is possible that chromosome movement has a positive effect on the processivity of SC polymerization. Delayed synapsis correlates with reduced plk-2 activity.
-
A surveillance mechanism couples events leading to the establishment of the obligate crossover to chromosome end mobilization in prophase I. In this mechanism, reversible phospho-modifications of SUN-1 establish a positive feedback loop for recruiting PLK-2 to chromosome ends.
Abbreviations
- DSB:
-
DNA double-strand break
- MMC:
-
Movement-mediating complex
- RPM:
-
Rapid prophase motion
- SC:
-
Synaptonemal complex
- ZIM:
-
Zinc finger in meiosis
References
Alpi A, Pasierbek P, Gartner A, Loidl J (2003) Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112:6–16
Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P et al (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6:e1001219
Bhalla N, Wynne DJ, Jantsch V, Dernburg AF (2008) ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet 4:e1000235
Burke B (2012) It takes KASH to hitch to the SUN. Cell 149:961–963
Chua PR, Roeder GS (1997) Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev 11:1786–1800
Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, La Volpe A, Villeneuve AM (2003) Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell 5:463–474
Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME (2008) Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133:1175–1187
Crittenden SL, Troemel ER, Evans TC, Kimble J (1994) GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120:2901–2911
Crittenden SL, Leonhard KA, Byrd DT, Kimble J (2006) Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell 17:3051–3061
Davis L, Smith GR (2006) The meiotic bouquet promotes homolog interactions and restricts ectopic recombination in Schizosaccharomyces pombe. Genetics 174:167–177
Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94:387–398
Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y (2004) Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6:329–341
Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y (2004) Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci U S A 101:6987–6992
Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10:741–743
Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40:1375–1383
Frokjaer-Jensen C, Davis MW, Hollopeter G, Taylor J, Harris TW, Nix P, Lofgren R, Prestgard-Duke M, Bastiani M, Moerman DG et al (2010) Targeted gene deletions in C. elegans using transposon excision. Nat Methods 7:451–453
Goldman AS, Lichten M (2000) Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A 97:9537–9542
Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ (2002) The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162:1979–1993
Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF (2011) Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev Cell 21:934–947
Hayashi M, Mlynarczyk-Evans S, Villeneuve AM (2010) The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 186:45–58
Herman RK, Kari CK (1989) Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics 121:723–737
Herman RK, Kari CK, Hartman PS (1982) Dominant X-chromosome nondisjunction mutants of Caenorhabditis elegans. Genetics 102:379–400
Hillers KJ, Villeneuve AM (2003) Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol: CB 13:1641–1647
Hirsh D, Oppenheim D, Klass M (1976) Development of the reproductive system of Caenorhabditis elegans. Dev Biol 49:200–219
Hulten MA (2011) On the origin of crossover interference: a chromosome oscillatory movement (COM) model. Mol Cytogenet 4:10
Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J (2004) Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol 24:7998–8006
Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J (2007) Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol 308:206–221
Koszul R, Kleckner N (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19:716–724
Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133:1188–1201
Labella S, Woglar A, Jantsch V, Zetka M (2011) Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell 21:948–958
Labrador L, Barroso C, Lightfoot J, Muller-Reichert T, Flibotte S, Taylor J, Moerman DG, Villeneuve AM, Martinez-Perez E (2013) Chromosome movements promoted by the mitochondrial protein SPD-3 are required for homology search during Caenorhabditis elegans meiosis. PLoS Genet 9:e1003497
Lorenz A, Fuchs J, Burger R, Loidl J (2003) Chromosome pairing does not contribute to nuclear architecture in vegetative yeast cells. Eukaryot Cell 2:856–866
MacQueen AJ, Villeneuve AM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15:1674–1687
MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM (2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16:2428–2442
Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115:825–836
Martinez-Perez E, Villeneuve AM (2005) HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19:2727–2743
McKim KS, Howell AM, Rose AM (1988) The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120:987–1001
McNally K, Audhya A, Oegema K, McNally FJ (2006) Katanin controls mitotic and meiotic spindle length. J Cell Biol 175:881–891
Mets DG, Meyer BJ (2009) Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139:73–86
Minn IL, Rolls MM, Hanna-Rose W, Malone CJ (2009) SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell 20:4586–4595
Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198(2):165–172. doi:10.1083/jcb.201204085
Nabeshima K, Villeneuve AM, Colaiácovo MP (2005) Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol 168(5):683–689
Niwa O, Shimanuki M, Miki F (2000) Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J 19:3831–3840
Parvinen M, Soderstrom KO (1976) Chromosome rotation and formation of synapsis. Nature 260:534–535
Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V (2007) The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 12:873–885
Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y et al (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139:920–933
Phillips CM, Dernburg AF (2006) A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell 11:817–829
Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF (2005) HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123:1051–1063
Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF (2009) Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol 11:934–942
Rose AM, Baillie DL, Curran J (1984) Meiotic pairing behavior of two free duplications of linkage group I in Caenorhabditis elegans. Mol Gen Genet 195:52–56
Rosenbluth RE, Baillie DL (1981) The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99:415–428
Sanford C, Perry MD (2001) Asymmetrically distributed oligonucleotide repeats in the Caenorhabditis elegans genome sequence that map to regions important for meiotic chromosome segregation. Nucleic Acids Res 29:2920–2926
Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139:907–919
Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB (2007) Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104:16934–16939
Schlecht HB, Lichten M, Goldman AS (2004) Compartmentalization of the yeast meiotic nucleus revealed by analysis of ectopic recombination. Genetics 168:1189–1203
Sheehan MJ, Pawlowski WP (2009) Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci U S A 106:20989–20994
Smolikov S, Schild-Prufert K, Colaiacovo MP (2008) CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet 4:e1000088
Sym M, Roeder GS (1994) Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79:283–292
Tang L, Machacek T, Mamnun YM, Penkner A, Gloggnitzer J, Wegrostek C, Konrat R, Jantsch MF, Loidl J, Jantsch V (2010) Mutations in Caenorhabditis elegans him-19 show meiotic defects that worsen with age. Mol Biol Cell 21:885–896
Villeneuve AM (1994) A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136:887–902
Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E (2008) Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet 4:e1000188
Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V (2013) Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet 9:e1003335
Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X et al (2011) Targeted genome editing across species using ZFNs and TALENs. Science 333:307
Wynne DJ, Rog O, Carlton PM, Dernburg AF (2012) Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196:47–64
Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM (2012) COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149:75–87
Zhang W, Miley N, Zastrow MS, MacQueen AJ, Sato A, Nabeshima K, Martinez-Perez E, Mlynarczyk-Evans S, Carlton PM, Villeneuve AM (2012) HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet 8:e1002880
Acknowledgments
We would like to thank Anahita Daryabeigi, Monique Zetka and Enrique Martinez-Perez for sharing unpublished data. We are indebted to Monique Zetka and Dimitra Paouneskou for critically reading this manuscript. This work was funded by FWF grants P-23638-B12 and SFB-F34.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Woglar, A., Jantsch, V. Chromosome movement in meiosis I prophase of Caenorhabditis elegans . Chromosoma 123, 15–24 (2014). https://doi.org/10.1007/s00412-013-0436-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-013-0436-7