Abstract
Background
It is now nearly a century since it was first discovered that crossovers between homologous parental chromosomes, originating at the Prophase stage of Meiosis I, are not randomly placed. In fact, the number and distribution of crossovers are strictly regulated with crossovers/chiasmata formed in optimal positions along the length of individual chromosomes, facilitating regular chromosome segregation at the first meiotic division. In spite of much research addressing this question, the underlying mechanism(s) for the phenomenon called crossover/chiasma interference is/are still unknown; and this constitutes an outstanding biological enigma.
Results
The C hromosome O scillatory M ovement (COM) model for crossover/chiasma interference implies that, during Prophase of Meiosis I, oscillatory movements of the telomeres (attached to the nuclear membrane) and the kinetochores (within the centromeres) create waves along the length of chromosome pairs (bivalents) so that crossing-over and chiasma formation is facilitated by the proximity of parental homologs induced at the nodal regions of the waves thus created. This model adequately explains the salient features of crossover/chiasma interference, where (1) there is normally at least one crossover/chiasma per bivalent, (2) the number is correlated to bivalent length, (3) the positions are dependent on the number per bivalent, (4) interference distances are on average longer over the centromere than along chromosome arms, and (5) there are significant changes in carriers of structural chromosome rearrangements.
Conclusions
The crossover/chiasma frequency distribution in humans and mice with normal karyotypes as well as in carriers of structural chromosome rearrangements are those expected on the COM model. Further studies are underway to analyze mechanical/mathematical aspects of this model for the origin of crossover/chiasma interference, using string replicas of the homologous chromosomes at the Prophase stage of Meiosis I. The parameters to vary in this type of experiment will include: (1) the mitotic karyotype, i.e. ranked length and centromere index of the chromosomes involved, (2) the specific bivalent/multivalent length and flexibility, dependent on the way this structure is positioned within the nucleus and the size of the respective meiocyte nuclei, (3) the frequency characteristics of the oscillatory movements at respectively the telomeres and the kinetochores.
Similar content being viewed by others
Background
Positive crossover interference, also termed genetic or chiasma interference, i.e. the non-random placement of crossovers along the length of individual chromosomes with a reduced probability of occurrence of one crossover in the vicinity of another, is a universal feature in the outstanding majority of eukaryotic organisms. The patterns of crossovers/chiasmata on individual chromosome pairs, as governed by interference, are of crucial importance for regular segregation of the homologous parental chromosomes at the meiosis I division [review in [1–3]] as schematically illustrated in Figure 1a.
Schematic illustration of the meiotic process. (a) Homologous chromosome synapsis and crossing over/chiasma formation at the Pachytene stage of Prophase 1 and the derivative bivalents at the following Metaphase I. (b) Progression through Metaphase I to Anaphase I leading to the halving of the chromosome number, Metaphase II and Anaphase II where the chromatids separate (similar to mitotic Anaphase) and Telophase II comprising the four haploid daughter cell nuclei. Reproduced from [3.]
Completion of reciprocal recombination/crossing-over between parental half chromosomes (chromatids) together with chromatid cohesion, leads to the formation of chiasmata, i.e. physical connections that hold parental homologs (bivalents) together. The positional control by interference seemingly creates bivalents of optimal mechanical stability, promoting regular segregation at the subsequent Meiosis I Anaphase. This first meiotic, reductional, division leads to the chromosome number of the two daughter nuclei being halved, with the second meiotic division giving rise to haploid gametes, as illustrated in Figure 1b.
Crossover interference was first described nearly a century ago by Sturtevant and Muller in Drosophila melanogaster [4, 5], in fact only a decade after the chromosome theory of Bovery and Sutton had been established [review in [6, 7]]. Muller in his paper published in 1916 [5] wrote that "In a sense then, the occurrence of one crossing-over interferes with the coincident occurrence of another crossing-over in the same pair of chromosomes, and I have accordingly termed this phenomenon "interference"."
In the interim this phenomenon, that each homologous chromosome pair will receive at least one crossover/chiasma (the so-called obligate chiasma) has also been called 'crossover assurance'. On the other hand, the reduced probability of occurrence of one crossover in the vicinity of another has been termed 'crossover homeostasis'. Some authors have suggested that the underlying mechanism for these two phenomena is different, while others have argued that both are likely to originate from the same cause.
Positive crossover interference governs the patterns of inheritance of blocks of genes, the linkage groups. It is therefore of outstanding importance to get to grips with the underlying mechanism(s), not only for theoretical, genetic, reasons but also to facilitate the design of breeding experiments in plants and domestic animals as well as the development of personalized medicine and drug treatment. It goes without saying that numerous investigations have been undertaken to understand its origin.
The identification of crossover/chiasma interference has been based on (1) genetic recombination maps, more recently created primarily by tracing DNA markers along the length of individual chromosomes between parents and offspring, (2) chiasma maps illustrating the positioning of crossovers/chiasmata by light microscopy at the Diakinesis/Metaphase I stages of meiocytes, and (3) Late Recombination Nodules/MLH1 maps showing the positions of crossovers/chiasma formation at the earlier Pachytene stage of Meiosis I Prophase, using electron/immuno-fluorescence microscopy. At this stage of Meiosis I homologs are normally held together by a meiosis-specific proteinaceous structure, the so-called Synaptonemal Complex (SC) illustrated in Figure 2.
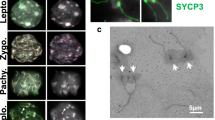
The Synaptonemal Complex. The Synaptonemal Complex (SC) is a meiosis-specific supra-molecular protein assembly that supports synapsis of homologs, crossover/chiasma formation and reciprocal recombination between sister chromatids at the Pachytene stage of Meiosis I. The chromatids of each homolog are held together by the Lateral Element (LE) consisting of cohesin proteins, formed already at the earlier Leptotene stage and then called the Axial Element [150]. The LE holds the two chromatids of each homolog tightly together until the onset of Anaphase I (see Fig 1). The central Element (CE) of the SC, made up of additional meiosis-specific proteins that hold the homologs together in a Velcro type of fashion, is required for maturation of early recombination events into crossovers/chiasmata [153]. (a) Electron-microscopy picture of the SC from a human male showing the Lateral Elements (LE) holding the two chromatids of each homolog together, the Central Element (CE), and the surrounding chromatin loops. Courtesy of N. Saadallah. (b) Schematic illustration showing the Lateral Elements (green), the Central Element, consisting of transverse fibrils (red), and the surrounding Chromatin (blue). (c) Electron-microscopy picture of the same bivalent as in (a) focused in such a way that the Late Recombination Nodules, corresponding to the crossovers/chiasmata are highlighted (arrows). The telomeres at each end, forming so-called attachment plaques, are associated with the nuclear membrane. Courtesy of N. Saadallah. Revised from [3].
Remarkably, the basic underlying mechanism(s) for positive crossover/chiasma interference is/are still not understood, and this constitutes an outstanding biological enigma. A number of different models have been proposed, reviewed in [8–15]. In this paper I present a model for positive crossover/chiasma interference, based on the relative mechanical impact of oscillatory movements of homologous chromosome pairs during the Prophase stage of Meiosis I, induced respectively at the telomeres via the nuclear membrane and the centromeres via the kinetochores. In so doing I presume that both crossover assurance and crossover homeostasis are caused by the same basic mechanism.
I suggest that crossing-over and chiasma formation is facilitated by the proximity of parental homologs at the nodal regions of the waves thus created. I further propose that this model may adequately explain the salient features of crossover/chiasma patterns and interference, i.e. (1) there is normally at least one, obligate, crossover/chiasma per bivalent, (2) the number is correlated to bivalent length, (3) the positions are dependent on number, (4) the interference distances are on average longer over the centromere than along chromosome arms, and (5) there are significant changes in carriers of structural chromosome rearrangements.
Mathematical aspects of this and previously published interference models will be presented separately (Clocksin et al. in preparation).
Results and Discussion
The only way in which it is possible to visualize crossover distribution along the length of all the individual chromosomes simultaneously is by cytogenetic analysis of meiocytes. Cytogenetic methods thus provide a means to determine directly the patterns of recombination both across the whole genome and at the chromosomal level, information that cannot readily be obtained in any other way [reviewed in [16–18]]. I will therefore here focus attention on the results illustrated by this type of investigation. As my special interest concerns the crossover picture of human chromosomes in relation to that in the mouse, my analysis will be biased to this effect. With reference to the COM model the parameters to consider include: (1) the mitotic karyotype, i.e. ranked length and centromere index of the chromosomes involved, (2) the specific bivalent/multivalent length and condensation/flexibility, dependent on the way this structure is positioned within the nucleus and the size of the respective meiocyte nuclei, (3) the frequency characteristics of the oscillatory movements at respectively the telomeres and the kinetochores.
I will in the following be looking at the cytogenetic information relevant to the understanding of the origin of crossover/chiasma interference separately as regards (1) Chiasmata at the Diakinesis/Metaphase I stage of Meiosis, (2) MLH1 foci at the Pachytene stage of Meiosis I Prophase, (3) Crossover patterns in mammals other than humans and mice, (4) Crossover patterns in other eukaryotes, (5) Telomere and kinetochore movements during Meiosis I.
1. Chiasmata at the Diakinesis/Metaphase I stage of Meiosis
Most information on the frequency and distribution of chiasmata along the lengths of individual human chromosomes has been obtained by microscopy analysis of spermatocytes at the Diakinesis/Metaphase I stage in testicular samples from adult males (Figure 3a, b). By comparison there is little corresponding information on the chiasma frequency distribution in the human female. One of the main reasons for this discrepancy is likely to be the access to the material for study.
The Diakinesis/Metaphase I stage in oocytes takes place just before ovulation, usually with only a single oocyte in division at any one time. Also, the morphology of the chromosome pairs and the identification of the chiasmata at this oocyte stage (Figure 3c) are not as clear as that in spermatocytes (Figure 3a, b). This difference has precluded detailed information from being obtained on the chiasma frequency and distribution in human oocytes in comparison to spermatocytes. On the other hand, information on chiasma frequency and distribution in female mice has been obtained following short time in vitro culture of oocytes (Figure 4; see e.g. [19, 20]). There is now also quite a lot of information on the crossover patterns in human and mouse spermatocytes and oocytes obtained by immuno-fluorescence analysis of MLH1 foci at the Pachytene stage of Meiosis I, as described in more detail in the following section.
The chiasma pattern in normal mouse chromosomes. Mouse spermatocytes (top) and oocytes (bottom) at the Metaphase I stage from mice with normal karyotypes after block staining (a, c) followed by C-banding (b, d). The XY bivalent of the spermatocyte is arrowed. Note examples of mono-chiasmate (I) and di-chiasmate (II) bivalents. Reproduced from [20].
1.1 The chiasma patterns in males with a normal karyotype/spermatogenesis
The first detailed analysis of chiasmata in spermatocytes, obtained by testicular biopsies from human males with normal mitotic karyotypes and normal spermatogenesis, was performed in the 1970s [21–26]. In summary these studies demonstrate: (1) the occurrence of a so-called obligate chiasma, i.e. the fact that normally each chromosome pair (bivalent) undergoes at least one crossover, (2) a positive correlation between bivalent length and number of chiasmata, (3) the distribution of chiasmata being dependent on their numbers with a single chiasma often localized in the middle of the respective bivalents, in contrast to the situation in bivalents with higher number of chiasmata, showing a tendency for additional chiasmata to become placed nearer to the telomeres, (4) the interference distances being increasingly shorter with increasing number of chiasmata, (5) the interference distance on average being longer over the centromere in comparison to that along the lengths of individual chromosome arms, and (6) the pattern of interference being significantly changed in carriers of structural chromosome rearrangements. Further studies during the next few decades have substantiated these observations, and also demonstrated the existence of inter-individual variation in chiasma frequency and distribution between normal human males [27–31]. Similar observations have been made in mice [see e.g. [19, 20, 32, 33]].
Measurements of chiasmata along the lengths of individual chromosomes (Figure 5) have allowed Ch iasma I nterference M aps (CHIMs) to be produced for each individual human and mouse chromosome, examples of which are shown in Figure 6, 7. Looking at these CHIMs it would appear that the frequency distribution of chiasmata is dependent on some specific features, located at the nuclear membrane as well as at the centromeres. One straight forward interpretation for these patterns is that this reflects oscillatory chromosome movements, taking place at the time of chiasma formation during Meiosis I Prophase. I would thus suggest that chiasmata are preferentially formed at the nodal regions of any such waves, created at the telomeres attached to the nuclear membrane and the kinetochores within the centromeres. In order to accommodate the interference distance spanning the centromere being longer than that within chromosome arms, I presume that the nuclear envelope/telomeric oscillatory movements are counteracted by those created at the centromeres/kinetochores, operating in both directions. I also envisage that once established the accumulation of crossover proteins (such as the MLH1 and MLH3) may clamp homologs, causing adjacent chromosome segments to splay, thus preventing additional adjacent crossovers to be formed at any nodal regions created by subsequent waves.
Measurements of chiasma positions. Spermatocytes are photographed, following consecutive triple staining with Quinacrine Mustard, Orcein (top right) and C-staining (bottom right) and drawings then made from the projected pictures at approximately 7000 times enlargement. Measurements are made of the chiasma positions in relation to the centromeres (left). Revised from [25].
Chiasma Interference Map (CHIM) of human chromosome 9. The data are based on direct measurements (see Fig 5) in 366 spermatocytes from 10 normal human males. The × axis represents the chromosome and the vertical black line the centromere position. The figure illustrates the frequency of chiasmata along the length of the chromosome arms and the interference distances in each spermatocyte separately. Interference loops, which involve nearly the whole chromosome are black, the near telomere plus interstitial ones are green and blue respectively, dependent on whether the near terminal chiasma is located at 9p or 9q, while those which involve interstitial chiasmata only are red. The barcode diagram shows the chiasma-derived 1 cM genetic map calculated from the centromere towards the telomeres. Note the large pericentromeric gap. The near telomere gaps are artifacts due to any chiasmata within the width of the chromosome being recorded as strictly terminal. Revised from [31].
Chiasma interference maps (CHIMs) of mouse chromosomes. Chiama interference maps (CHIMs) for spermatocyte (upper) and oocyte (lower) chromosome rank sizes with the centromeric heterochromatin situated to the left. Single chiasmata are represented by vertical red bars, crossing the axis with distal clusters projected outside the axis. Multiple chiasmata within each bivalent are joined by loops, illustrating the chiasma interference patterns. Loops joining extreme proximal and interstitial chiasmata are shown in green, and those joining extreme distal and interstitial ones in blue. Loops joining extreme proximal and extreme distal chiasmata are black and those joining two interstitially located chiasmata are red. Note the wider distribution of interstitial chiasmata in comparison to the corresponding, near terminal ones in multiple chiasma bivalents. Revised from [20].
It is in this context essential to note that it is not the centromere per se that inhibits chiasmata to be formed near its vicinity. This is obvious from the patterns seen in acrocentric chromosomes (Figure 3, 4, 7, 8; see e.g. [34, 35]). The small human acrocentrics (21 and 22) usually harbor only a single interstitially or near terminally positioned chiasma, while the longer ones (13-15) have one, two or three chiasmata. Singles are positioned interstitially/medially, or near terminally. In double- and triple chiasma bivalents, however, the proximal chiasma is located adjacent to the centromere and the distal nearer to the telomere. This pattern is very similar to that in wild type/normal laboratory mice (Figure 4, 7; [19, 20, 33]). In accordance with the COM model I have interpreted this standardized pattern of chiasma frequency distribution in human and mouse acrocentrics to be the result of the conjoined oscillatory action of the telomeres and the kinetochores (via the heterochromatc short arm) being abutted to the nuclear membrane.
The three types of chiasma formation in acrocentric chromosomes. The drawing illustrates the crossover patterns/chiasma formation and reciprocal recombination between homologous chromatids in acrocentric chromosomes, which as regards singles and doubles are basically the same in mice and humans. Note that the different chromatids from the two homologs are randomly selected, i.e. there is no chromatid interference. A single chiasma is most often positioned either medially or more distal towards the telomere (top). Two chiasmata are located more near to the centrome and telomere (middle). Three chiasmata are located, respectively, adjacent the centromere, medially and adjacent to the telomere. Reproduced from [155].
On a more general note it is also important to recognize that chiasmata are in fact already locked into their original positions at the transition between the Pachytene and Diplotene stage of Meiosis I. Thus, the original suggestion in 1929 by Darlington [36] as recently reiterated by e.g. de Boer et al. [37] that the frequent occurrence of near-terminal positions of chiasmata is due to their movement from their original interstitial positions (so-called chiasma terminalization) is a misconception [27, 30, 38–41].
Looking at the bivalent in 2D it would seem necessary for chiasma terminalization to take place before homologs are able to separate (Figure 9). However, as soon as the telomeres are disconnected from the nuclear membrane in the transition between the Diplotene and Diakinesis stages, the chromosomes condense and at the same time they are transformed into 3D structures, where each interference/inter-chiasma segment is located perpendicular to the next. Chiasmata are thus bound to remain in their original positions as laid down at the Pachytene stage of Meiosis Prophase I. One mechanically favorable result of this 3D arrangement (similar to that in an ordinary metallic chain) is that any kinetochore-induced chromosome movement towards the opposite spindle poles at Anaphase I of Meiosis induces a separation of chromatids in adjacent interference loops.
Chiasma positions at the Diplotene stage as seen in 2D. It would from this picture in 2D appear that chiasmata would have to move towards the ends of the chromosome (terminalize) in order for homologs to be able to separate at the following Anaphase I stage. In reality, however, any such movement is prohibited by the inter-chiasma loops being orientated perpendicular to each other. Reproduced from [30].
It is also essential to appreciate that the large-scale preferential/optimal crossover placement/chiasma formation along the length of each individual bivalent as dictated by crossover/chiasma interference is not related to G-banding/chromomeres or any DNA specification but primarily dependent on chromosome morphology and bivalent length per se [42–46]. On the other hand, it is now well known that, at the DNA level, certain DNA sequences within these chromosome segments constitute so-called 'crossover hotspots'. Mammalian crossover hotspots, corresponding to initial DNA breaks are around 1-2 Kb long DNA segments that are separated by larger intervals with very low frequencies [47–52].
Only a small proportion (around 1 in 500 in the human male) of the specific DNA motif (recognized by the PRDM9 protein) within these 1-2 Kb long crossover hotspot are, however, selected for the final crossover and chiasma formation; and I am here discussing a model aimed at explaining the classical type of crossover/chiasma interference, involving many Mb of DNA. Thus, I am not addressing the mechanism(s) underlying any interference involved in the interaction between homologs, taking place as part and parcel of the complex molecular pathway leading up to final crossover/chiasma formation and reciprocal recombination. For a detailed analysis of these factors in relation to previous models of crossover interference readers are referred to the recent presentations in [12–15].
1.2 The chiasma patterns in carriers of structural chromosome rearrangements
Analysis of chiasma interference has also been performed in human male carriers of structural chromosome rearrangements. Most attention has focused on reciprocal translocations, where in the majority of spermatocytes at the Diakinesis/Metaphase I stage a quadrivalent configuration has been seen. The chiasma frequency distribution has been studied in a relatively large number of such human male carriers (see e.g. [23, 26, 53–75]).
In the reciprocal translocation carriers where the chiasma frequency distribution has been analyzed in detail, the most striking deviation from the situation in human males with normal karyotypes is a significant increase in the frequency of chiasmata localized within the interstitial segment, i.e. the chromosome segment positioned in between the breakpoint and the centromere (Figure 10; see also Figure 8 in [65]). This is true even when the interstitial segment is very short. In stark contrast to the normal situation in non-acrocentric chromosomes there is then a tendency for chiasmata to occupy positions near to/adjacent to the centromere, as well as a substantial reduction in the crossover/interference distance over the centromere. A similar tendency for an increased frequency of chiasmata within the interstitial segment has been seen in reciprocal translocations in mice [76, 77].
The chiasma pattern in a human male reciprocal 1;22 translocation carrier. Pictures of translocation quadrivalents at the Diakinesis/Metaphase I stage in spermatocytes from a carrier of a reciprocal 1; 22 translocation with the corresponding drawings showing the interpretation of the chiasma patterns (top). Note the high frequency and different distribution of chiasmata within the interstitial segment between the centromere and the breakpoint. The translocation carrier shows a raised chiasma frequency and altered chiasma distribution in chromosome 1, particularly in the region adjacent to the breakpoint (grey staples) in comparison to six controls with normal karyotypes (bottom). Reproduced from [64].
Under the COM model I would suggest that the explanation for this deviant pattern of chiasma frequency distribution in the quadrivalent in comparison to the normal is the change in the mechanics of the waves induced by the oscillatory movements of the telomeres/the nuclear membrane in relation to those of the kinetochores/centromeres. First of all, the quadrivalent has to accommodate waves originating from four different places along the nuclear membrane travelling to its centre. Second, the situation is further complicated by the quadrivalent having two rather than one duplex kinetochore and the potential associated alteration in effect on the nodal regions of the chromosomal waves caused by their oscillation.
The chiasma patterns seen in spermatocytes from human Robertsonian translocations are of special interest, demonstrating quite clearly the influence of karyotype and chromosome morphology. The chromosome arms of the trivalents in the common 13; 14 and 14; 21 translocations (formed by the two normal together with the translocation chromosome) show the same pattern as that in the normal situation of the respective acrocentrics (Figure 11; [26, 54]). This apparently normal chiasma pattern within individual chromosome arms in trivalents of the human heterologous Robertsonian translocations is also seen in the corresponding mouse Robertsonian fusions [78, 79]. This pattern is that expected on the COM model, as these trivalents are likely to be positioned in the same way as their corresponding normal bivalents within the meiotic cell nuclei, with the normal telomere and centromere movements thus retained.
Chiasma pattern in a carrier of a 13; 14 Robertsonian translocation. Trivalents containing either two chiasmata on one arm together with two chiasmata on the other (arrow, top) or two chiasmata on both arms (arrow, bottom) and two additional examples (cut outs, middle). Note the dicentric nature of this metacentric derivative 13; 14 Robertsonian translocation, as illustrated by the drawings (middle). These chiasma patterns of the trivalents in this carrier are similar to those of the individual chromosomes 13 and 14 in human males with normal karyotypes. Revised from [26] and [155].
In stark contrast, the univalent in the unique case of a human 14; 14 metacentric Robertsonian translocation invariably forms a ring with a single distal chiasma, different to the rings of the normal chromosome 14 bivalent, having two chiasmata, one at each end (Figure 12, cc Figure 7, 8). The derivative 14; 14 chromosome is dicentric, where the proximal telomeres have been lost. According to the COM model the single distal/near telomeric chiasma in the Diakinesis/Metahase I univalent is likely to be due to the effect of the oscillatory movements induced by the dual kinetochores counteracting those originating at the dual distal telomeres, both attached to the nuclear membrane.
Chiasma pattern in a carrier of a 14; 14 Robertsonian translocation. Spermatocyte at the Diakinesis/Metaphase I from a human male carrier of a 14; 14 Robertsonian translocation, stained with C-banding (left) and orcein (right) where the univalent 14; 14 is arrowed. Note the parallel centromeres, showing that synapsis has occurred between chromatids from the two different homologs, and the occurrence of a single chiasma very near the telomere. Reproduced from [23].
Yet again, in stark contrast, the chiasma patterns in the mouse homologous Robertsonian translocations seemingly correspond to that expected on the basis of the oscillatory movements, similar to those in the middle-sized human metacentric/submetacentric chromosomes with either two or three chiasmata, forming rings or figures of eight (cc Figure 1 in [33] and Figure 3a here).
Finally, as regards structural heterozygotes, inversion carriers present an especially complex case, associated with the particular problems encountered in the pairing and synapsis of parental homologs, which is a pre-requisite for their interaction in the process of crossing over/chiasma formation [review in [80]]. In inversion heterozygotes involving a short chromosome segment there is a possibility of its elimination from synapsis by looping out, leading to a corresponding reduction in homologous crossing-over in this particular chromosome segment [60]. From a mechanical point of view the situation in carriers of larger interstitial inversions is even more complicated, both as regards initial homologous synapsis, so called non-allelic homologous synapsis and synaptic adjustment, identified by detailed EM analysis [80–82]. Further studies on the patterns of crossover/chiasma formation are required before any firm conclusion can be drawn as regards the interpretation of their origin in relation to the COM model.
Intriguingly, in carriers of a double inversion of chromosome 1 in mice, a reduction of chiasmata has been seen in single heterozygotes but an increase in the double heterozygotes, the latter associated with a reduced strength of interference [83]. In order to provide an adequate explanation in particular for the apparent decrease in strength of interference in the double hetrozygote it would be helpful to obtain additional information on the relation between synapsis [84] and crossovers by way of MLH1 analysis at the Pachytene stage (see section 2). The same holds true as regards a double heterochromatic insertion in the middle of the mouse chromosome 1 [85]. Both such heterozygotes and homozygotes show an increase in chiasma frequency with the normally medial chiasma replaced by one proximal together with one distal/pro-terminal. It may seem likely that this is somehow related to the well known prevention of chiasma formation within heterochromatic segments. Perhaps the expected looping out of the two heterochromatic blocks includes the interstitial euchromatic section (see e.g. [86]), thereby preventing crossovers within this segment?
1.3 The chiasma patterns in human males with non-obstructive azoospermia
Chiasma analysis at the Diakinesis/Metaphase I stage in a number of studies on spermatocytes from testicular biopsy samples of men suffering from reduced fertility associated with non-obstructive oligo-azoospermia has shown that some have disturbances in chiasma formation [87–90]. In a first comprehensive study of 50 men with this condition [87] the majority (n = 41) was found to have a normal progression of spermatogenesis and a normal, or nearly normal, chiasma pattern. Among the remaining 9/50 cases, 7/50 showed spermatogenic arrest already at the Pachytene stage of Meiosis I, and no information on crossover/chiasma formation could at the time be obtained. In two exceptional cases the majority of parental homologs in spermatocytes reaching the Diakineses/Metaphase I were unpaired. Some of these spermatocytes did, however, show the occasional apparently normally paired bivalents, illustrating the notion of positional control of chiasma formation. Even in this aberrant situation a single chiasma in a large bivalent occupied a medial/central position (Figure 13). The same has more recently been seen in some oligo-azoospermic men, where the crossover pattern has been studied by MLH1 focus analysis of spermatocytes at the Pachytene stage of Meiosis Prophase I, as described in the following section.
Positional control of chiasma formation. Spermatocytes at the Diakinesis/Metaphase I stage from a normally fertile human male with normal mitotic karyotype (top) in comparison to that in a male with non-obstructive azoospermia (bottom). Note the extremely low number of chiasmata in the spermatocyte from the azoospermic male; some chromosomes lack chiasmata altogether (I) while two relatively large bivalents show a single chiasma a medial position (arrowed, left). There are also a number of univalents (arrow heads) as well as two bivalents with two chiasmata (arrowed, right). Reproduced from [156].
2. MLH1 foci at the Pachytene Stage of Meiosis I Prophase
As would be expected from the correspondence between the positions of MLH1 foci analyzed at the Pachytene stage of Meiosis I and chiasmata at the later Diplotene/Diakinesis/Metaphase I stage in Ocadaic Acid stimulated spermatocytes of mice and men ([91] and Khazanehdari and Hultén (unpubl. obs.)) the deduced crossover patterns are largely congruent. One advantage of the MLH1 approach concerns the number of spermatocytes that can be readily analyzed, dependent on the much longer duration of the Pachytene stage of Meiosis I in relation to that at the short Diakinesis/Metaphase I stage. Another advantage is that the MLH1 analysis can be performed on equally large populations of human oocytes at the Pachytene stage of Meiosis I obtained from fetal ovarian biopsies. The possibility of obtaining information on the crossover patterns in both human males and females has in particular allowed a detailed comparison to be made as regards any sex difference in crossover/chiasma interference distances. Figure 14 shows the typical pattern of MLH1 foci in a spermatocyte in comparison to that in an oocyte.
MLH1 foci along the Synaptonemal Complexes (SCs) in human males and females. The spermatocyte (top) and oocyte (bottom) have been stained using antibodies against SCP3 (red), MLH1 (yellow) and, in the spermatocyte, the kinetochore (blue). Homologs 21, 13 and 18 have been identified in the oocyte, using Fluorescence in situ hybridisation (FISH) with centromere-specific probes. There are obvious differences between the spermatocyte and oocyte: the SCs are much longer in the oocyte; there are more MLH1 foci in the oocyte; MLH1 foci tend to be positioned closer to the telomeres in the spermatocyte (arrow heads). Reproduced from [86] and revised from [94].
2.1 MLH1 focus analysis in human males with normal mitotic karyotypes
Analysis of MLH1 foci at the Pachytene stage of Meiosis I in spermatocytes from a range of human males with normal mitotic karyotypes and normal spermatogenesis has by and large produced a very similar picture of crossover interference to that obtained by chiasma analysis at the Diakinesis/Metaphase I stage (cc Figure 3a, b and 14; [42, 92–102]). This is also the case in mouse spermatocytes [103, 104].
The mathematical model on crossover interference by Falque et al. [105], based on the MLH1 data by Froenicke et al. [104], will be discussed in conjunction with our own mathematical version of the COM model to be presented separately (Clocksin et al. in preparation).
2.2 MLH1 focus analysis of carriers of structural chromosome rearrangements
MLH1 foci have also recently been investigated in a number of human male carriers of reciprocal translocations with normal spermatogenesis [104–107]. This work provides valuable new information on synaptic problems with respect to crossover frequency distribution. There is, on the other hand, no substantial new information as regards crossover interference. The same holds true for Robertsonian translocations and inversions in mice [[96, 110], review in [111]].
2.3 MLH1 focus analysis in men with non-obstructive azoospermia
By comparison to reports describing the chiasma patterns at the Diakinesis/Metaphase I stage in men with non-obstructive oligo-azoospermia discussed above there are a relatively large number of human males, where MLH1 focus analysis of spermatocytes at the Pachytene stage of Meiosis Prophase I has been used to highlight crossover frequency distribution [see e.g. [95, 98, 100, 108, 109, 112–116]]). Again, the picture revealed is very similar with the majority having some reduction in deducted crossover frequency. Figure 3c of Gonsalves et al. [95] demonstrates the positional effect with the medial/central position of a single MLH1 focus in a large chromosome (cc the position of the chiasma in Figure 13 here).
2.4 MLH1 focus analysis of human oocytes at the Pachytene stage
As illustrated in Figure 14, the numbers of crossovers estimated from MLH1 foci, is higher in oocytes than in spermatocytes. Initial MLH1 analysis [92, 94] has suggested an average of approximately 70 crossovers per oocyte with a larger inter-cell variability (range 40-100) in comparison to around 50 in spermatocytes (range 41-59). Subsequent studies have confirmed the occurrence of a large variation in crossover frequency between individual oocytes within subjects and also indicated a higher inter-individual variability [117, 118]. The higher rate of recombination in oocytes is most probably related to the considerable difference in chromosome length at the Pachytene stage of Meiosis Prophase I [42, 44]. Thus, the human female genome has a longer physical platform for establishment of crossing-over/chiasmata/reciprocal recombination than the male (Table two in [94]). Both the larger variation in crossover frequency between individual oocytes and the higher inter-individual variation is likely to be due to larger differences in oocyte nuclear size in comparison to that in spermatocytes. It should be added, on the other hand, that there is, by measurement of the meiosis-specific chromosome pairing structures, the Synaptonemal Complexes (SCs), evidence to suggest that the strength of interference is similar in both sexes in terms of actual physical distance between crossovers/chiasmata. Thus, the rate of recombination per unit length of SC is relatively constant in the two sexes, when the influence of the "obligate chiasma" is discounted [44].
Not only do the two sexes show a significant variation in recombination frequency, but they also display some differences in distribution [94, 117, 118]. In spermatocytes, the MLH1 foci/chiasmata are often located very close to the ends of the chromosomal axes. In oocytes, MLH1 foci are located more interstitially (away from chromosome ends) and only very rarely positioned so near to telomeric segments as in spermatocytes (Figure 14). On the COM model I have interpreted this sex difference in crossover numbers as well as positioning to be related to the effect of the telomeric and kinetochore oscillatory movements of the longer and thinner, and therefore more flexible, female chromosome pairs.
3. MLH1 Foci Patterns in Mammals other than Humans and Mice
Additional information on the crossover frequency distribution has more recently been obtained on normal spermatocytes (and in a few cases also oocytes) in a number of different mammalian species, i.e. by analysis of MLH1 foci in domestic animals [review in [119]]as well as cat [120], common shrew [121], dog [122], American mink [123], Rhesus [124] and silver fox [125]. By and large these studies reiterate the notion that the patterns of distribution of crossovers along the length of individual bivalents are very similar, to a large extent being dependent on chromosome morphology, as reflected by bivalent length and centromere/kinetochore position. It is noteworthy, however, that unusually short intra-arm interference distances have been identified in cat spermatocytes [120]. The reason for this exceptional behavior is not known, and requires further study. In the context of optimal mechanical stability, facilitating regular segregation of parental homologs at the Anaphase I stage, I would presume these dual crossovers would function in the same way as a single chiasma.
4. Crossover Patterns in other Eukaryotes
Investigation of crossover patterns in a wide range of eukaryotes indicates that crossover/chiasma interference is a characteristic feature in most. One extreme example of interference is seen in the nematode Caenorhabditis elegans, where all bivalents irrespective of their size have a single chiasma localized distally. The multiple sites of recombination initiation are then resolved into a single crossover, with the diffuse (holocentric) kinetic activity that extends along the length of the mitotic chromosomes being reduced to the single telomeric end of each meiotic chromosome, via direct insertion of the microtubules into the chromatin [review in [126, 127]].
The only two known exceptions to the general rule of positive crossover interference in eukaryotes concern the fission yeast, Saccharomyces pombe together with the fungus, Aspergillus nidulans. In both of these organisms crossovers are randomly distributed along the length of individual bivalents, and both lack the meiosis-specific chromosome pairing structure, the so-called Synaptonemal Complex [[128, 129], see also [130]]. Much attention has been paid to the underlying reason for this random distribution of crossovers in fission yeast, involving the clustering of telomeres in a restricted area of the nuclear membrane (bouquet) and the movement of the nucleus back and forth in the cell by a so-called horsetail formation [129–134].
5. Meiotic Telomere and Kinetochore Movements
Interest has recently focused on oscillatory movements of the telomeres during Prophase of Meiosis I, when homologous parental chromosomes align and pair intimately (synapse) to allow crossing-over between non-sister chromatids to take place [see e.g. [135–141]]. No conclusion has, however, been reached as regards their exact role(s) with respect to the patterns of crossover/chiasma frequency and distribution. Most recently it has been suggested that these movements may eliminate unwanted inter-chromosomal associations or entanglements that have arisen as part and parcel of the homolog pairing process [141].
Much less attention has been paid to any corresponding movements of the kinetochores at the Prophase stage of Meiosis I. Thus, information on meiotic kinetochore movements per se is currently restricted to that obtained at the later Metaphase I to Anaphase I transition [142–145].
With reference to the COM model I would be specially interested in further investigation of telomere and kinetochore movements at the Pachytene stage of Meiosis I in organisms with large chromosomes (such as maize, locusts and grasshoppers, mice and humans) using approaches similar to the ones already performed on human chromosomes at the mitotic Metaphase stage [see e.g. [146]]. It would also be helpful to get information on the behavior of kinesin proteins [review in [147]] and other potentially relevant proteins such as Sgo1 suggested to act at sister kinetochores to promote their bi-orientation in Saccharomyces cerevisiae [148], klp3A where Drosophila mutants show abnormal crossover distribution [149], TEL1 proposed to be involved in the regulation of interference [150] and the MCAK protein, recently found to be associated with chiasmata at the prometaphase stage in mouse oocytes [151].
Conclusions and Perspectives
I have here described a model for the origin of the meiotic crossover patterns shared between most eukaryotic organisms. I have suggested that the patterns seen, with special reference to the non random distribution and the crossover/chiasma interference is related to the oscillatory movements of the telomeres attached to the nuclear membrane and the kinetochores within the centromeres. Thus, I have presumed that these oscillatory movements, taking place at the Prophase stage of Meiosis I, lead to waves of physical interaction between the homologous chromosomes, with the highest chance of final crossovers/chiasma formation/reciprocal recombination being restricted to the chromosome segments corresponding to the nodal regions of the waves thus created.
One advantage of this type of purely mechanical/physical model for the origin of crossover interference is that it may now be tested in mechanical/mathematical experiments using any string replica of the homologous chromosomes at the Prophase stage of Meiosis I. The parameters to vary in this type of experiment would include: (1) the mitotic karyotype, i.e. ranked length and centromere index of the chromosomes involved, (2) the specific bivalent/multivalent length and flexibility, dependent on the way this structure is positioned within the nucleus and the size of the respective meiocyte nuclei, (3) the frequency characteristics of the oscillatory movements at respectively the telomeres and the kinetochores.
Should it turn out that the oscillatory movements that I have postulated do not adequately explain the crossover frequency distributions observed, then it will be essential to explore in particular what other characteristics of the centromeres/kinetochores that may underlie the increased interference distance over the centromere and the variation induced by structural chromosome rearrangements in comparison to the normal karyotype. I would be especially interested in obtaining further information on the potential impact of the differential mass of the centromere/kinetochore and the 3D spatial orientation of the chromosomes within the meiocyte nuclei, which likely will influence the progression of the waves I have hypothesized regulate the patterns of crossover/chiasma frequency and distribution along the length of individual chromosome pairs. I envisage that it might in fact be possible to modify and possibly simplify the COM model, based on the results of such mechanical/mathematical analysis. Perhaps the specific characteristics of the centromere may mean that it is not necessary to imply any oscillatory movements induced by the kintechores, and the patterns seen could be explained by waves induced by the telomeres alone?
Either way, I do nourish a hope that we will within the next few years have reached a full understanding of the origin of the phenomenon of crossover interference, so that we may celebrate the centenary since its first discovery, by Sturtevant [4] and Muller [5].
Conflicts of interests
The author declares that she has no competing interests.
References
Page SL, Hawley RS: Chromosome choreography: the meiotic ballet. Science (New York, NY) 2003, 301: 785–789. 10.1126/science.1086605
Marston AL, Amon A: Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 2004, 5: 983–997. 10.1038/nrm1526
Hultén MA: Meiosis. In Encyclopedia of Life Sciences (eLS). Chichester: John Wiley & Sons, Ltd; Published: On Line 15 Sep 2010;
Sturtevant AH: A Third Group of Linked Genes in Drosophila ampelophila. Science (New York, NY) 1913, 37: 990–992. 10.1126/science.37.965.990
Muller HJ: The Mechanism of Crossing-Over. The American Naturalist 1916, 50: 193–221. 10.1086/279534
Crow EW, Crow JF: 100 years ago: Walter Sutton and the chromosome theory of heredity. Genetics 2002, 160: 1–4.
Satzinger H: Theodor and Marcella Boveri: chromosomes and cytoplasm in heredity and development. Nature reviews 2008, 9: 231–238.
Colombo PC, Jones GH: Chiasma interference is blind to centromeres. Heredity 1997,79(Pt 2):214–227.
van Veen JE, Hawley RS: Meiosis: when even two is a crowd. Curr Biol 2003, 13: R831–833. 10.1016/j.cub.2003.10.014
Hillers KJ: Crossover interference. Curr Biol 2004, 14: R1036–1037. 10.1016/j.cub.2004.11.038
Jones GH, Franklin FC: Meiotic crossing-over: obligation and interference. Cell 2006, 126: 246–248. 10.1016/j.cell.2006.07.010
Mezard C, Vignard J, Drouaud J, Mercier R: The road to crossovers: plants have their say. Trends Genet 2007, 23: 91–99. 10.1016/j.tig.2006.12.007
Martinez-Perez E, Colaiacovo MP: Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev 2009, 19: 105–112. 10.1016/j.gde.2009.02.005
Berchowitz LE, Copenhaver GP: Genetic interference: don't stand so close to me. Curr Genomics 2010, 11: 91–102. 10.2174/138920210790886835
Youds JL, Boulton SJ: The choice in meiosis - defining the factors that influence crossover or non-crossover formation. Journal of cell science 2011, 124: 501–513. 10.1242/jcs.074427
Tease C, Hultén MA: Meiosis. In Nature Encyclopedia of the Human Genome. Volume 2. Edited by: Cooper DN. London: Nature Publishing Group; 2003:865–873.
Hultén MA, Tease C: Genetic mapping: comparison of direct and indirect approaches. In Nature Encyclopedia of the Human Genome. Volume 2. Edited by: Cooper DN. London: Nature Publishing Group; 2003:876–881.
Hultén MA, Tease C: Genetic maps: direct meiotic analysis. In Nature Encyclopedia of the Human Genome. Volume 2. Edited by: Cooper DN. London: Nature Publishing Group; 2003:882–887.
Hultén MA, Tease C, Lawrie NM: Chiasma-based genetic map of the mouse × chromosome. Chromosoma 1995, 104: 223–227. 10.1007/BF00352187
Lawrie NM, Tease C, Hultén MA: Chiasma frequency, distribution and interference maps of mouse autosomes. Chromosoma 1995, 104: 308–314. 10.1007/BF00352262
Ferguson-Smith MA: Human chromosomes in meiosis. In Human Genetics, Proceedings of the Firth International Congress of Human Genertics, Paris, 6–11 September, 1971 Edited by: de Grouchy J. 195–211.
Ferguson-Smith MA: Meiosis in the human male. In Chromosomes Today, Proceedings of the Leiden Chromosome Conference July 15–17, 1974. Edited by: Pearson PL, Lewis KR. published 1976 by John Wiley & Sons, New York, Israel Universities Press, Jerusalem; 33–41.
Hultén M, Lindsten J: Cytogenetic aspects of human male meiosis. Advances in human genetics 1973, 4: 327–387.
McDermott A: The frequency and distribution of chiasmata in man. Annals of human genetics 1973, 37: 13–20. 10.1111/j.1469-1809.1973.tb01810.x
Hultén M: Chiasma distribution at diakinesis in the normal human male. Hereditas 1974, 76: 55–78.
Hultén M, Lindsten J: The behaviour of structural aberrations at male meiosis, Information from man. In Human Population Cytogenetics. Edinburgh: University of Edinburgh Press; 1970:24–61. Pfizer Medical Monograph
Laurie DA, Hultén M, Jones GH: Chiasma frequency and distribution in a sample of human males: chromosomes 1, 2, and 9. Cytogenetics and cell genetics 1981, 31: 153–166. 10.1159/000131641
Laurie DA, Hultén MA: Further studies on bivalent chiasma frequency in human males with normal karyotypes. Annals of human genetics 1985, 49: 189–201. 10.1111/j.1469-1809.1985.tb01693.x
Laurie DA, Hultén MA: Further studies on chiasma distribution and interference in the human male. Annals of human genetics 1985, 49: 203–214. 10.1111/j.1469-1809.1985.tb01694.x
Hultén MA: The topology of meiotic chiasmata prevents terminalization. Annals of human genetics 1990, 54: 307–314. 10.1111/j.1469-1809.1990.tb00386.x
Povey S, Smith M, Haines J, Kwiatkowski D, Fountain J, Bale A, Abbott C, Jackson I, Lawrie M, Hultén M: Report and abstracts of the First International Workshop on Chromosome 9, Held at Girton College Cambridge, UK, 22–24 March, 1992. Annals of human genetics 1992, 56: 167–182. 10.1111/j.1469-1809.1992.tb01145.x
Gorlov IP, Borodin PM: [Chiasmata distribution in the normal karyotype of mice]. Genetika 1991, 27: 247–251.
Dumas D, Britton-Davidian J: Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and robertsonian populations of the house mouse. Genetics 2002, 162: 1355–1366.
Palmer RW, Hultén MA: Chiasma derived genetic maps and recombination fractions: chromosome 13 with reference to the proposed 13q14 retinoblastoma locus. Journal of medical genetics 1982, 19: 125–129. 10.1136/jmg.19.2.125
Saadallah N, Hultén M: Chiasma distribution, genetic lengths, and recombination fractions: a comparison between chromosomes 15 and 16. Journal of medical genetics 1983, 20: 290–299. 10.1136/jmg.20.4.290
Darlington C: Chromosome behaviour and structural hybridity in the Tradescantiae II. Journal of Genetics 1929, 21: 207–286. 10.1007/BF02984208
de Boer E, Lhuissier FG, Heyting C: Cytological analysis of interference in mouse meiosis. Methods in molecular biology, Clifton, NJ 2009, 558: 355–382.
Peacock WJ: Replication, recombination, and chiasmata in Goniaea australasiae (Orthoptera:Acrididae). Genetics 1970, 65: 593–617.
Jones GH: The analysis of exchanges in tritium-labelled meiotic chromosomes. Chromosoma 1971, 34: 367–382. 10.1007/BF00326312
Tease C: Cytological detection of crossing-over in BUdR substituted meiotic chromosomes using the fluorescent plus Giemsa technique. Nature 1978, 272: 823–824. 10.1038/272823a0
Hultén MA, Palmer RW, Laurie DA: Chiasma derived genetic maps and recombination fractions: chromosome 1. Annals of human genetics 1982, 46: 167–175. 10.1111/j.1469-1809.1982.tb00707.x
Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ: Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science (New York, NY) 2002, 296: 2222–2225. 10.1126/science.1071220
Kleckner N, Storlazzi A, Zickler D: Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet 2003, 19: 623–628. 10.1016/j.tig.2003.09.004
Tease C, Hultén MA: Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenetic and genome research 2004, 107: 208–215. 10.1159/000080599
Petkov PM, Broman KW, Szatkiewicz JP, Paigen K: Crossover interference underlies sex differences in recombination rates. Trends Genet 2007, 23: 539–542. 10.1016/j.tig.2007.08.015
Belonogova NM, Borodin PM: Frequency of meiotic recombination in G and R chromosome bands of the common shrew (Sorex araneus). Dokl Biol Sci 2010, 433: 268–270. 10.1134/S0012496610040095
Jeffreys AJ, Kauppi L, Neumann R: Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature genetics 2001, 29: 217–222. 10.1038/ng1001-217
Guillon H, de Massy B: An initiation site for meiotic crossing-over and gene conversion in the mouse. Nature genetics 2002, 32: 296–299. 10.1038/ng990
Myers S, Bottolo L, Freeman C, McVean G, Donnelly P: A fine-scale map of recombination rates and hotspots across the human genome. Science (New York, NY) 2005, 310: 321–324. 10.1126/science.1117196
McVean G, Myers S: PRDM9 marks the spot. Nature genetics 2010, 42: 821–822. 10.1038/ng1010-821
Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B: PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science, New York, NY 2010, 327: 836–840. 10.1126/science.1183439
Parvanov ED, Petkov PM, Paigen K: Prdm9 controls activation of mammalian recombination hotspots. Science, New York, NY 2010, 327: 835. 10.1126/science.1181495
Chandley AC, Seuanez H, Fletcher JM: Meiotic behaviour of five human translocation carriers. Cytogenetics and cell genetics 1976, 17: 98–111. 10.1159/000130694
Chandley AC, Hargreave TB, Fletcher JM: Translocation 21q22q in an infertile human male. Journal of medical genetics 1982, 19: 366–369. 10.1136/jmg.19.5.366
Chandley AC, Speed RM, McBeath S, Hargreave TB: A human 9;20 reciprocal translocation associated with male infertility analyzed at prophase and metaphase I of meiosis. Cytogenetics and cell genetics 1986, 41: 145–153. 10.1159/000132219
Egozcue J, Marina S, Templado C: Meiotic behaviour of two human reciprocal translocations. Journal of medical genetics 1981, 18: 362–365. 10.1136/jmg.18.5.362
Micic MD, Micic SR: Meiotic findings in human reciprocal 1;3 translocation. Human genetics 1981, 57: 442–443. 10.1007/BF00281703
Gonzales J, Lesourd S, Dutrillaux B: Mitotic and meiotic analysis of a reciprocal translocation t(Y;3) in an azoospermic male. Human genetics 1981, 57: 111–114. 10.1007/BF00271182
Faed MJ, Lamont MA, Baxby K: Cytogenetic and histological studies of testicular biopsies from subfertile men with chromosome anomaly. Journal of medical genetics 1982, 19: 49–56. 10.1136/jmg.19.1.49
Goldman AS, Hultén MA: Chromosome in situ suppression hybridisation in human male meiosis. Journal of medical genetics 1992, 29: 98–102. 10.1136/jmg.29.2.98
Laurent C, Chandley AC, Dutrillaux B, Speed RM: The use of surface spreading in the pachytene analysis of a human t (Y;17) reciprocal translocation. Cytogenetics and cell genetics 1982, 33: 312–318. 10.1159/000131777
Petit P, Unglik A, Fryns JP: Translocation 46, X, t(Y;14)(q122;q111) in a case of sterility in the male. Annales de genetique 1982, 25: 63–64.
Zuffardi O, Tiepolo L: Frequencies and types of chromosome abnormalities associated with human male infertility. In Genetic Control, Gametic Production and Function. Edited by: PG C, Rubin BL. London: Academic Press; 1982:261–273.
Palmer RW, Hultén MA: Chiasma-derived genetic lengths and recombination fractions: a reciprocal translocation 46, XY, t(1;22) (q32;q13). Annals of human genetics 1983, 47: 299–310. 10.1111/j.1469-1809.1983.tb01000.x
Laurie DA, Palmer RW, Hultén MA: Studies on chiasma frequency and distribution in two fertile men carrying reciprocal translocations; one with a t(9;10) karyotype and one with a t(Y;10) karyotype. Human genetics 1984, 68: 235–247. 10.1007/BF00418394
Micic M, Micic S: Meiotic studies in two infertile males with autosomal translocations. Human genetics 1984, 65: 308–310. 10.1007/BF00286524
Gabriel-Robez O, Ratomponirina C, Dutrillaux B, Carre-Pigeon F, Rumpler Y: Meiotic association between the XY chromosomes and the autosomal quadrivalent of a reciprocal translocation in two infertile men, 46, XY, t(19;22) and 46, XY, t(17;21). Cytogenetics and cell genetics 1986, 43: 154–160. 10.1159/000132314
Templado C, Navarro J, Requena R, Benet J, Ballesta F, Egozcue J: Meiotic and sperm chromosome studies in a reciprocal translocation t(1;2)(q32;q36). Human genetics 1990, 84: 159–162. 10.1007/BF00208932
Micic M, Nikolis J, Micic S: Clinical and meiotic studies in an infertile man with Y;13 translocation. Human reproduction (Oxford, England) 1992, 7: 1118–1120.
Goldman AS, Hultén MA: Analysis of chiasma frequency and first meiotic segregation in a human male reciprocal translocation heterozygote, t(1;11)(p36.3;q13.1), using fluorescence in situ hybridisation. Cytogenetics and cell genetics 1993, 63: 16–23. 10.1159/000133493
Goldman AS, Hultén MA: Meiotic analysis by FISH of a human male 46, XY, t(15;20)(q11.2;q11.2) translocation heterozygote: quadrivalent configuration, orientation and first meiotic segregation. Chromosoma 1993, 102: 102–111. 10.1007/BF00356027
Armstrong SJ, Hultén MA: Meiotic segregation analysis by FISH investigations in sperm and spermatocytes of translocation heterozygotes. Eur J Hum Genet 1998, 6: 430–431. 10.1038/sj.ejhg.5200209
Armstrong SJ, Goldman AS, Speed RM, Hultén MA: Meiotic studies of a human male carrier of the common translocation, t(11;22), suggests postzygotic selection rather than preferential 3:1 MI segregation as the cause of liveborn offspring with an unbalanced translocation. American journal of human genetics 2000, 67: 601–609. 10.1086/303052
Oliver-Bonet M, Navarro J, Codina-Pascual M, Abad C, Guitart M, Egozcue J, Benet J: From spermatocytes to sperm: meiotic behaviour of human male reciprocal translocations. Human reproduction (Oxford, England) 2004, 19: 2515–2522. 10.1093/humrep/deh492
Sarrate Z, Blanco J, Egozcue S, Vidal F, Egozcue J: Identification of meiotic anomalies with multiplex fluorescence in situ hybridization: Preliminary results. Fertility and sterility 2004, 82: 712–717. 10.1016/j.fertnstert.2004.02.122
Tease C: Chiasma distributions and chromosome segregation in male and female translocation heterozygous mice analysed using FISH. Chromosoma 1998, 107: 549–558. 10.1007/s004120050341
de Boer P: Proximal chiasma localization within an interstitial chromosome segment, a likely correlate of adjacent-2 segregation of translocation causing multivalents in the mouse. Environmental health perspectives 1979, 31: 137–140. 10.2307/3429151
Wallace BM, Searle JB, Everett CA: Male meiosis and gametogenesis in wild house mice (Mus musculus domesticus) from a chromosomal hybrid zone; a comparison between "simple" Robertsonian heterozygotes and homozygotes. Cytogenetics and cell genetics 1992, 61: 211–220. 10.1159/000133410
Bidau CJ, Gimenez MD, Palmer CL, Searle JB: The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity 2001, 87: 305–313. 10.1046/j.1365-2540.2001.00877.x
Torgasheva AA, Borodin PM: Synapsis and recombination in inversion heterozygotes. Biochemical Society transactions 2010, 38: 1676–1680. 10.1042/BST0381676
Batanian J, Hultén MA: Electron microscopic investigations of synaptonemal complexes in an infertile human male carrier of a pericentric inversion inv(1)(p32q42). Regular loop formation but defective synapsis including a possible interchromosomal effect. Human genetics 1987, 76: 81–89. 10.1007/BF00283055
Winsor EJ, Palmer CG, Ellis PM, Hunter JL, Ferguson-Smith MA: Meiotic analysis of a pericentric inversion, inv(7) (p22q32), in the father of a child with a duplication-deletion of chromosome 7. Cytogenetics and cell genetics 1978, 20: 169–184. 10.1159/000130849
Gorlov IP, Borodin PM: Recombination in single and double heterozygotes for two partially overlapping inversions in chromosome 1 of the house mouse. Heredity 1995,75(Pt 2):113–125. 10.1038/hdy.1995.114
Borodin PM, Gorlov IP, Ladygina T: Synaptic interrelationships between the segments of the heteromorphic bivalent in double heterozygotes for paracentric inversions in chromosome 1 of the house mouse. Chromosoma 1992, 101: 374–379. 10.1007/BF00346017
Gorlov IP, Ladygina T, Borodin PM: Chiasma distribution in the first bivalent of mice carrying a double insertion of homogeneously-staining regions in homo- and heterozygous states. Heredity 1993,70(Pt 6):642–647. 10.1038/hdy.1993.92
Barlow AL, Hultén MA: Combined immunocytogenetic and molecular cytogenetic analysis of meiosis I human spermatocytes. Chromosome Res 1996, 4: 562–573. 10.1007/BF02261719
Hultén M, Eliasson R, Tillinger KG: Low chiasma count and other meiotic irregularities in two infertile 46, XY men with spermatogenic arrest. Hereditas 1970, 65: 285–290.
Templado C, Marina S, Egozcue J: Three cases of low chiasma frequency associated with infertility in man. Andrologia 1976, 8: 285–289. 10.1111/j.1439-0272.1976.tb01657.x
Chaganti RS, Jhanwar SC, Ehrenbard LT, Kourides IA, Williams JJ: Genetically determined asynapsis, spermatogenic degeneration, and infertility in men. American journal of human genetics 1980, 32: 833–848.
Micic M, Micic S, Diklic V: Low chiasma frequency as an aetiological factor in male infertility. Clinical genetics 1982, 22: 266–269. 10.1111/j.1399-0004.1982.tb01443.x
Marcon E, Moens P: MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 2003, 165: 2283–2287.
Barlow AL, Hultén MA: Crossing over analysis at pachytene in man. Eur J Hum Genet 1998, 6: 350–358. 10.1038/sj.ejhg.5200200
Lynn A, Ashley T, Hassold T: Variation in human meiotic recombination. Annual review of genomics and human genetics 2004, 5: 317–349. 10.1146/annurev.genom.4.070802.110217
Tease C, Hartshorne GM, Hultén MA: Patterns of meiotic recombination in human fetal oocytes. American journal of human genetics 2002, 70: 1469–1479. 10.1086/340734
Gonsalves J, Sun F, Schlegel PN, Turek PJ, Hopps CV, Greene C, Martin RH, Pera RA: Defective recombination in infertile men. Human molecular genetics 2004, 13: 2875–2883. 10.1093/hmg/ddh302
Koehler KE, Millie EA, Cherry JP, Schrump SE, Hassold TJ: Meiotic exchange and segregation in female mice heterozygous for paracentric inversions. Genetics 2004, 166: 1199–1214. 10.1534/genetics.166.3.1199
Sun F, Oliver-Bonet M, Liehr T, Starke H, Ko E, Rademaker A, Navarro J, Benet J, Martin RH: Human male recombination maps for individual chromosomes. American journal of human genetics 2004, 74: 521–531. 10.1086/382138
Sun F, Greene C, Turek PJ, Ko E, Rademaker A, Martin RH: Immunofluorescent synaptonemal complex analysis in azoospermic men. Cytogenetic and genome research 2005, 111: 366–370. 10.1159/000086913
Sun F, Oliver-Bonet M, Liehr T, Starke H, Turek P, Ko E, Rademaker A, Martin RH: Variation in MLH1 distribution in recombination maps for individual chromosomes from human males. Human molecular genetics 2006, 15: 2376–2391. 10.1093/hmg/ddl162
Sun F, Turek P, Greene C, Ko E, Rademaker A, Martin RH: Abnormal progression through meiosis in men with nonobstructive azoospermia. Fertility and sterility 2007, 87: 565–571. 10.1016/j.fertnstert.2006.07.1531
Lian J, Yin Y, Oliver-Bonet M, Liehr T, Ko E, Turek P, Sun F, Martin RH: Variation in crossover interference levels on individual chromosomes from human males. Human molecular genetics 2008, 17: 2583–2594. 10.1093/hmg/ddn158
Garcia-Cruz R, Casanovas A, Brieno-Enriquez M, Robles P, Roig I, Pujol A, Cabero L, Durban M, Garcia Caldes M: Cytogenetic analyses of human oocytes provide new data on non-disjunction mechanisms and the origin of trisomy 16. Human reproduction (Oxford, England) 2010, 25: 179–191. 10.1093/humrep/dep347
Anderson LK, Reeves A, Webb LM, Ashley T: Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 1999, 151: 1569–1579.
Froenicke L, Anderson LK, Wienberg J, Ashley T: Male mouse recombination maps for each autosome identified by chromosome painting. American journal of human genetics 2002, 71: 1353–1368. 10.1086/344714
Falque M, Mercier R, Mezard C, de Vienne D, Martin OC: Patterns of recombination and MLH1 foci density along mouse chromosomes: modeling effects of interference and obligate chiasma. Genetics 2007, 176: 1453–1467. 10.1534/genetics.106.070235
Oliver-Bonet M, Benet J, Sun F, Navarro J, Abad C, Liehr T, Starke H, Greene C, Ko E, Martin RH: Meiotic studies in two human reciprocal translocations and their association with spermatogenic failure. Human reproduction (Oxford, England) 2005, 20: 683–688. 10.1093/humrep/deh654
Pigozzi MI, Sciurano RB, Solari AJ: Changes in crossover distribution along a quadrivalent in a man carrier of a reciprocal translocation t(11;14). Biocell 2005, 29: 195–203.
Sun F, Oliver-Bonet M, Turek PJ, Ko E, Martin RH: Meiotic studies in an azoospermic human translocation (Y;1) carrier. Molecular human reproduction 2005, 11: 361–364. 10.1093/molehr/gah173
Ferguson KA, Chow V, Ma S: Silencing of unpaired meiotic chromosomes and altered recombination patterns in an azoospermic carrier of a t(8;13) reciprocal translocation. Human reproduction (Oxford, England) 2008, 23: 988–995. 10.1093/humrep/den013
Merico V, Pigozzi MI, Esposito A, Merani MS, Garagna S: Meiotic recombination and spermatogenic impairment in Mus musculus domesticus carrying multiple simple Robertsonian translocations. Cytogenetic and genome research 2003, 103: 321–329. 10.1159/000076820
Torgasheva AA, Zhelezova AI, Rubtsov NB, Borodin PM: Effects of sex and gene order on the recombination frequency and distribution in the chromosome 1 of the house mouse. Dokl Biol Sci 2009, 429: 559–561. 10.1134/S0012496609060222
Judis L, Chan ER, Schwartz S, Seftel A, Hassold T: Meiosis I arrest and azoospermia in an infertile male explained by failure of formation of a component of the synaptonemal complex. Fertility and sterility 2004, 81: 205–209. 10.1016/j.fertnstert.2003.05.021
Codina-Pascual M, Oliver-Bonet M, Navarro J, Campillo M, Garcia F, Egozcue S, Abad C, Egozcue J, Benet J: Synapsis and meiotic recombination analyses: MLH1 focus in the XY pair as an indicator. Human reproduction (Oxford, England) 2005, 20: 2133–2139. 10.1093/humrep/dei023
Egozcue J, Sarrate Z, Codina-Pascual M, Egozcue S, Oliver-Bonet M, Blanco J, Navarro J, Benet J, Vidal F: Meiotic abnormalities in infertile males. Cytogenetic and genome research 2005, 111: 337–342. 10.1159/000086907
Topping D, Brown P, Judis L, Schwartz S, Seftel A, Thomas A, Hassold T: Synaptic defects at meiosis I and non-obstructive azoospermia. Human reproduction (Oxford, England) 2006, 21: 3171–3177. 10.1093/humrep/del281
Leng M, Li G, Zhong L, Hou H, Yu D, Shi Q: Abnormal synapses and recombination in an azoospermic male carrier of a reciprocal translocation t(1;21). Fertility and sterility 2009, 91: 1293. e1217–1222 10.1016/j.fertnstert.2008.12.049
Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE: Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis i in human oocytes. American journal of human genetics 2005, 76: 112–127. 10.1086/427268
Cheng EY, Hunt PA, Naluai-Cecchini TA, Fligner CL, Fujimoto VY, Pasternack TL, Schwartz JM, Steinauer JE, Woodruff TJ, Cherry SM, et al.: Meiotic recombination in human oocytes. PLoS genetics 2009, 5: e1000661. 10.1371/journal.pgen.1000661
Villagomez DA, Pinton A: Chromosomal abnormalities, meiotic behavior and fertility in domestic animals. Cytogenetic and genome research 2008, 120: 69–80. 10.1159/000118742
Borodin PM, Karamysheva TV, Rubtsov NB: [Immunofluorescent analysis of meiotic recombination and interference in the domestic cat]. Tsitologiia 2008, 50: 62–66.
Borodin PM, Karamysheva TV, Belonogova NM, Torgasheva AA, Rubtsov NB, Searle JB: Recombination map of the common shrew, Sorex araneus (Eulipotyphla, Mammalia). Genetics 2008, 178: 621–632. 10.1534/genetics.107.079665
Basheva EA, Bidau CJ, Borodin PM: General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res 2008, 16: 709–719. 10.1007/s10577-008-1221-y
Borodin PM, Basheva EA, Zhelezova AI: Immunocytological analysis of meiotic recombination in the American mink (Mustela vison). Animal genetics 2009, 40: 235–238. 10.1111/j.1365-2052.2008.01808.x
Hassold T, Hansen T, Hunt P, Van de Voort C: Cytological studies of recombination in rhesus males. Cytogenetic and genome research 2009, 124: 132–138. 10.1159/000207519
Basheva EA, Torgasheva AA, Sakaeva GR, Bidau C, Borodin PM: A- and B-chromosome pairing and recombination in male meiosis of the silver fox (Vulpes vulpes L., 1758, Carnivora, Canidae). Chromosome Res 2010, 18: 689–696. 10.1007/s10577-010-9149-4
Zetka M: Homologue pairing, recombination and segregation in Caenorhabditis elegans. Genome dynamics 2009, 5: 43–55. full_text
Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ: RTEL-1 enforces meiotic crossover interference and homeostasis. Science New York, NY 2010, 327: 1254–1258. 10.1126/science.1183112
Egel-Mitani M, Olson LW, Egel R: Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 1982, 97: 179–187. 10.1111/j.1601-5223.1982.tb00761.x
Bahler J, Wyler T, Loidl J, Kohli J: Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. The Journal of cell biology 1993, 121: 241–256. 10.1083/jcb.121.2.241
Lorenz A, Estreicher A, Kohli J, Loidl J: Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 2006, 115: 330–340. 10.1007/s00412-006-0053-9
Munz P: An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 1994, 137: 701–707.
Asakawa H, Haraguchi T, Hiraoka Y: Reconstruction of the kinetochore: a prelude to meiosis. Cell division 2007, 2: 17. 10.1186/1747-1028-2-17
Cromie G, Smith GR: Meiotic Recombination in Schizosaccharomyces pombe: A Paradigm for Genetic and Molecular Analysis. Genome dynamics and stability 2008, 3: 195. full_text
Asakawa H, Hiraoka Y: Live-cell fluorescence imaging of meiotic chromosome dynamics in Schizosaccharomyces pombe. Methods in molecular biology, Clifton, NJ 2009, 558: 53–64.
Scherthan H: Telomere attachment and clustering during meiosis. Cell Mol Life Sci 2007, 64: 117–124. 10.1007/s00018-006-6463-2
Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB: Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America 2007, 104: 16934–16939. 10.1073/pnas.0704860104
Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME: Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 2008, 133: 1175–1187. 10.1016/j.cell.2008.04.047
Kosaka H, Shinohara M, Shinohara A: Csm4-dependent telomere movement on nuclear envelope promotes meiotic recombination. PLoS genetics 2008, 4: e1000196. 10.1371/journal.pgen.1000196
Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S: Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 2008, 133: 1188–1201. 10.1016/j.cell.2008.04.050
Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, Alani E: Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS genetics 2008, 4: e1000188. 10.1371/journal.pgen.1000188
Koszul R, Kleckner N: Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends in cell biology 2009, 19: 716–724. 10.1016/j.tcb.2009.09.007
Bardhan A: Complex regulation of sister kinetochore orientation in meiosis-I. Journal of biosciences 2010, 35: 485–495. 10.1007/s12038-010-0053-z
Ma W, Hou Y, Sun QY, Sun XF, Wang WH: Localization of centromere proteins and their association with chromosomes and microtubules during meiotic maturation in pig oocytes. Reproduction (Cambridge, England) 2003, 126: 731–738.
Sakuno T, Watanabe Y: Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions. Chromosome Res 2009, 17: 239–249. 10.1007/s10577-008-9013-y
Wignall SM, Villeneuve AM: Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nature cell biology 2009, 11: 839–844. 10.1038/ncb1891
Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P: Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nature cell biology 2010, 12: 319–329. 10.1038/ncb2033
Xie P: Mechanism of processive movement of monomeric and dimeric kinesin molecules. International journal of biological sciences 2010, 6: 665–674.
Kiburz BM, Amon A, Marston AL: Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Molecular biology of the cell 2008, 19: 1199–1209. 10.1091/mbc.E07-06-0584
Page SL, Hawley RS: The Drosophila meiotic mutant mei-352 is an allele of klp3A and reveals a role for a kinesin-like protein in crossover distribution. Genetics 2005, 170: 1797–1807. 10.1534/genetics.105.041194
Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ: RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic acids research 2011, 39: 1647–1655. 10.1093/nar/gkq1045
Vogt E, Sanhaji M, Klein W, Seidel T, Wordeman L, Eichenlaub-Ritter U: MCAK is present at centromeres, midspindle and chiasmata and involved in silencing of the spindle assembly checkpoint in mammalian oocytes. Molecular human reproduction 2010, 16: 665–684. 10.1093/molehr/gaq025
Revenkova E, Jessberger R: Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma 2006, 115: 235–240. 10.1007/s00412-006-0060-x
Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, Hoog C: Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. Journal of cell science 2008, 121: 2445–2451. 10.1242/jcs.033233
Yuncken C: Meiosis in the human female. Cytogenetics 1968, 7: 234–238. 10.1159/000129986
Hultén M, Lawrie NM, Laurie DA: Chiasma-based genetic maps of chromosome 21. American journal of medical genetics 1990, 7: 148–154.
Hultén M, Lindsten J, Ming PM, Fraccaro M: The XY bivalent in human male meiosis. Annals of human genetics 1966, 30: 119–123. 10.1111/j.1469-1809.1966.tb00012.x
Acknowledgements
I am grateful to many colleagues, who have shared my enthusiasm for the cytogenetic analysis of meiotic chromosomes, including in particular Sue Armstrong, Andrew Barlow, Jackie Batanian, Alastair Goldman, Geraldine Hartshorne, Gareth Jones, David Laurie, Jan Lindsten, Martin Lawrie, Roger Palmer, Nina Saadallah, Charles Tease, Maira Tankimanova and Bridgit Wallace.
I am also indebted to William Clocksin and Nigel Dyer for helpful discussions on the COM model.
Author information
Authors and Affiliations
Corresponding author
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hultén, M.A. On the origin of crossover interference: A chromosome oscillatory movement (COM) model. Mol Cytogenet 4, 10 (2011). https://doi.org/10.1186/1755-8166-4-10
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8166-4-10