Abstract
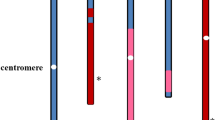
Two partially reconstructed karyotypes (RK1 and RK2) of Arabidopsis thaliana have been established from a transformant, in which four structurally changed chromosomes (α, β, γ, and δ) were involved. Both karyotypes are composed of 12 chromosomes, \( {2}n = {1}\prime \prime + {3}\prime \prime + {4}\prime \prime + {5}\prime \prime + \alpha \prime \prime + \gamma \prime \prime = {12} \) for RK1 and \( {2}n = {3}\prime \prime + {4}\prime \prime + {5}\prime \prime + \alpha \prime \prime + \beta \prime \prime + \gamma \prime \prime = {12} \) for RK2, and these chromosome constitutions were relatively stable at least for three generations. Pairing at meiosis was limited to the homologues (1, 3, 4, 5, α, β, or γ), and no pairing occurred among non-homologous chromosomes in both karyotypes. For minichromosome α (mini α), precocious separation at metaphase I was frequently observed in RK2, as found for other minichromosomes, but was rare in RK1. This stable paring of mini α was possibly caused by duplication of the terminal tip of chromosome 1 that is characteristic of RK1.





Similar content being viewed by others
References
Anastassova-Kristeva M, Nicoloff H, Künzel G, Rieger R (1977) Nucleolus formation in structurally reconstructed barley karyotypes with six satellite chromosomes. Chromosoma 62:111–117
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Hahnenberger KM, Carbon J, Clarke L (1991) Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol 11:2206–2215
Henry IM, Dilkes BP, Young K, Watson B, Wu H, Comai L (2005) Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170:1979–1988
Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H (2002) Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res 9:117–121
Koornneef M (1994) Arabidopsis genetics. In: Meyerowitz EM, Somervilee CR (eds) Arabidopsis. Cold Spring Harbor Laboratory Press, New York, pp 89–120
Koorneef M, Dresselhuys HC, Stree-Ramulu K (1982) The genetic identification of translocations in Arabidopsis. Arabidopsis Inf Serv 19:93–99
Koornneef M, Van der Veen JH (1983) The trisomics of Arabidopsis thaliana (L.) Heynh. and the location of linkage groups. Genetica 61:46–46
Kotani H (2002) The size and genome organization of centromere of Arabidopsis thaliana. In: Murata M, Sakamoto W (eds) Structure and function of plant centromeres: a challenge to the orthodoxy. Kurashiki, Okayama, pp 35–39
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2000) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res 7:315–321
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res 8:285–290
Laufs P, Autran D, Traas JA (1999) Chromosomal paracentric inversion associated with T-DNA integration in Arabidopsis. Plant J 18:131–139
Lysak MA, Fransz PF, Ali HB, Schubert I (2001) Chromosome painting in Arabidopsis thaliana. Plant J 28:689–697
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103:5224–5229
Michaelis A, Rieger R (1971) New karyotype of Vicia faba L. Chromosoma 35:1–8
Murata M, Motoyoshi F (1995) Floral chromosomes of Arabidopsis thaliana for detecting low-copy DNA sequences by fluorescence in situ hybridization. Chromosoma 104:39–43
Murata M, Tsuchiya T, Roos EE (1984) Chromosome damage induced by artificial seed aging in barley. 3. Behavior of chromosomal aberrations during plant growth. Theor Appl Genet 67:161–170
Murata M, Shibata F, Yokota E (2006) The origin, meiotic behavior, and transmission of a novel minichromosome in Arabidopsis thaliana. Chromosoma 115:311–319
Murata M, Yokota E, Shibata F, Kashihara K (2008) Functional analysis of the Arabidopsis centromere by T-DNA-insertion-induced centromere breakage. Proc Natl Acad Sci USA 105:7511–7516
Niwa O, Matsumoto T, Chikashige Y, Yanagida M (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. Embo J 8:3045–3052
Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4:507–516
Sears LMS, Lee-Chen S (1970) Cytogenetic studies in Arabidopsis thaliana. Can J Genet Cytol 12:217–223
Tax FE, Vernon DM (2001) T-DNA-Associated duplication/translocations in Arabidopsis. Implications for mutant analysis and functional genomics. Plant Physiol 126:1527–1538
Warwick SI, Al-Shehbaz IA (2006) Brassicaceae: chromosome number index and database on CD-Rom. Plant Syst Evol 259:237–248
Acknowledgements
The authors are grateful to K. Kashihara for technical assistance. This work was supported in part by CREST/JST, the Ohara Foundation for Agricultural Sciences, and Promotion of Basic Research Activities for Innovative Biosciences, BRAIN, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Shaw
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table 1
List of BAC clones used for chromosome painting (DOC 50 kb)
Fig. 1
Schematic representation of the presumed mechanism producing chromosomes α, β, γ, and δ. RT reciprocal translocation, Dp duplication, CO crossing over, Tl 1/2 presumed translocation between chromosomes 1 and 2, TEL telomere. F3C11, T12C24, F13K23, F14L17: BAC clones. Modified from Murata et al. (2008) (DOC 44 kb)
Rights and permissions
About this article
Cite this article
Yokota, E., Nagaki, K. & Murata, M. Minichromosome stability induced by partial genome duplication in Arabidopsis thaliana . Chromosoma 119, 361–369 (2010). https://doi.org/10.1007/s00412-010-0259-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-010-0259-8