Abstract
Whether chromosome sets inherited from father and mother occupy separate spaces in the cell nucleus is a question first asked over 110 years ago. Recently, the nuclear organization of the genome has come increasingly into focus as an important level of epigenetic regulation. In this context, it is indispensable to know whether or not parental genomes are spatially separated. Genome separation had been demonstrated for plant hybrids and for the early mammalian embryo. Conclusive studies for somatic mammalian cell nuclei are lacking because homologous chromosomes from the two parents cannot be distinguished within a species. We circumvented this problem by investigating the three-dimensional distribution of chromosomes in mule lymphocytes and fibroblasts. Genomic DNA of horse and donkey was used as probes in fluorescence in situ hybridization under conditions where only tandem repetitive sequences were detected. We thus could determine the distribution of maternal and paternal chromosome sets in structurally preserved interphase nuclei for the first time. In addition, we investigated the distribution of several pairs of chromosomes in human bilobed granulocytes. Qualitative and quantitative image evaluation did not reveal any evidence for the separation of parental genomes. On the contrary, we observed mixing of maternal and paternal chromosome sets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spatial organization of the genome in the nucleus has come into focus as an important level of epigenetic regulation (for reviews, see Kosak and Groudine 2004; Lanctôt et al. 2007; Misteli 2007). However, basic essential questions are still unanswered, such as whether chromosomes inherited from mother and father are separated in the nucleus. This question is nearly as old as the discovery of chromosomes itself. Häcker (1895) suggested the autonomy of maternal and paternal genomes after observing that nuclei in the crustacean Cyclops were organized in two lobes (Fig. 1a). Similar conclusions were proposed by Rückert (1895) and Conklin (1901) (Fig. 1b; all cited after Hertwig 1906). Spatial separation of parental genomes does occur in a number of plant hybrids (Leitch et al. 1991; Bennett and Bennett 1992; Lima-Brito et al. 1996; Kikuchi et al. 2007). Fluorescence in situ hybridization (FISH) with whole genomic DNA of the parent species revealed separate areas for maternal and paternal chromosomes in mitotic figures as well as in interphase nuclei (Leitch et al. 1991). In contrast, in Drosophila melanogaster, homologous chromosomes are closely paired (Fung et al. 1998), excluding a separation of parental genomes.
a and b Bilobed nuclei found in some species first suggested the separation of parental genomes. a 16-cell stage of the copepod Cyclops brevicornis (Häcker 1895). b Two-cell stage of the shell Crepidula (Conklin 1901) (both reproduced from Hertwig 1906). c Schemes of potential ways to separate parental genomes in spherical or oval nuclei. Left both genomes are separable by a line or, in 3D, by a plane. Center one parental genome is at the center, the other is at the periphery of the nucleus. Right the spaces occupied by the two genomes are randomly formed
In vertebrates, a radial organization of chromosome territories was identified in human (Croft et al. 1999), primate (Tanabe et al. 2002), mouse (Mayer et al. 2005), and chicken cell nuclei (Habermann et al. 2001). Gene-rich as well as small chromosomes are preferentially at internal positions and gene-poor or large chromosomes at peripheral positions (for reviews, see Foster and Bridger 2005; Cremer et al. 2006; Lanctôt et al. 2007). Homologous association as observed in Drosophila can be excluded as a general theme in mammals, since numerous FISH studies found two distinct chromosome territories in interphase nuclei.
Strong evidence was provided for parental genome separation in the very early vertebrate embryo (<16 cell stage). Moenkhaus (1904) studied hybrids of two fish species, one with large and one with small chromosomes, and found spatial separation during the first and second mitosis but intermingling at later stages. When sperm was radioactively labeled and two and four cell embryos were investigated, the radioactive genome was shown to occupy only a part of the nucleus (Odartchenko and Keneklis 1973). This was later confirmed with higher optical resolution using bromodeoxyuridine-labeled sperm (Mayer et al. 2000b). Separation of parental genomes was also described in human × hamster hybrids before ceasing of development at the two-cell stage (Brandriff et al. 1991).
The investigation of parental genomes in vertebrates is hampered by the difficulty to distinguish maternal and paternal chromosomes. FISH probes for chromosomes or gene loci produce signals on both homologs, even in related species (Wienberg 2005). Repetitive sequences evolve faster and can, therefore, show significant sequence differences between closely related species. In hybrids of Mus musculus and Mus spretus, centromeres were sufficiently diverse to allow their identification by FISH (Mayer et al. 2000a, b). Again, parental genome separation was found up to the 16-cell stage, a separation that was discussed to have functional importance for the distinct methylation properties of maternal and paternal genomes (Haaf 2001). With each cell duplication, the separation progressively diminished (Mayer et al. 2000a, b). However, the authors described “a striking separation” of centromeres of different parental origins in 5–10% of adult fibroblasts (Mayer et al. 2000b). The fibroblasts in that study were trypsinized, hypotonically treated, fixed in suspension with methanol–acetic acid, dropped on slides, and air-dried. We recently showed that this procedure can disturb the structure of centromere clusters in mouse nuclei (Hepperger et al. 2007). It thus appears possible that this procedure leads to an artificial increase or decrease of nonrandom patterns. In a follow-up study, Mayer et al. (2000a) demonstrated that, in late spermatid nuclei, maternal centromeres cluster separately from paternal centromeres.
While these data demonstrate separate genomes in the early mammalian embryo, they also indicate that this separation is gradually reduced during subsequent cell cycles. However, the data on mouse hybrid spermatids and fibroblasts argue that separation can be either retained in a minority of cells or regained at later stages. An ability of adult cells to isolate haploid genomes or even sort out maternal and paternal chromosomes was also deduced from work on rat liver. Diploid metaphase spreads were described to be separable by a single line in two haploid sets of chromosomes while other spreads had only a haploid set of chromosomes (“Genomsonderung”; Gläss 1956, 1957). The latter was also reported for fibroblasts of the mouse Microtus agrestis (Pera and Rainer 1973). A haploid karyotype was found in a human osteosarcoma (Bridge et al. 1997). Assuming that these reports are not based on artifacts, generating haploid cells from a diploid tissue would require a process for haploid genome recognition, an ability which is also a requirement for active parental genome separation. Parental genome recognition may seem highly speculative at first; however, it is well-established for some amphibian and fish interspecies hybrids. During gametogenesis, they eliminate the complete haploid genome of one parent and thus produce only germ cells containing the chromosome set from the other parent, a process called hybridogenesis. Examples are the water frog Rana esculenta (e.g., Spolsky and Uzzell 1984; Guerrini et al. 1997) and the fish Poeciliopsis monacha x lucida (Quattro et al. 1991). In a triploid population of the toad Bufo viridis, in males, one haploid genome is eliminated before the onset of meiosis (Stöck et al. 2002).
Parental genome separation in humans was deduced from FISH experiments on blood and bone marrow cells (Chaudhuri and Walther 2003; Chaudhuri et al. 2005). With a frequency of 0.1% in specimens from healthy donors and 1.5% in specimens from patients with various leukemias or lymphomas, nuclei with a “biphasic” pattern were described, i.e., two regions were stained with very different intensities by the DNA counterstain 4′,6-diamidino-2-phenylindole (DAPI). The authors report for FISH-detected homologous genomic sites that one was within the strongly stained area and the other outside. They concluded that two haploid genomes were condensed to a different degree. Although haploid genomes are not necessarily the maternal or paternal ones, this appeared to the authors as the most likely possibility. A precise antiparallel positioning of chromosome sets was claimed for the mitotic rosette and interphase nuclei, thus advocating a separation of haploid genomes (Nagele et al. 1995, 1998, 1999). While an antiparallel order could not be confirmed by other groups (Allison and Nestor 1999; Cornforth et al. 2002; Bolzer et al. 2005; Mayer et al. 2005), whether or not parental genomes are separated in another pattern remained unclear.
In summary, parental genome separation exists in plant hybrids and in very early embryos and spermatids of mouse hybrids. Available data do not provide a definite view for somatic mammalian cells. Several studies argue for such a separation but data may have been biased by a subconscious selection of microscopic examples.
To test for parental genome distribution in mammals, we resorted to mules, hybrids of a horse mother, and a donkey father. The parent species had a last common ancestor about 2 million years ago (Oakenfull et al. 2000). While chromosomal libraries made from horse chromosomes also paint donkey chromosomes (Raudsepp and Chowdhary 1999; Yang et al. 2004), we developed a method to discriminate donkey and horse centromeres by two-color FISH with whole genomic DNA from the parent species as probes. When performing FISH on primary lymphocytes and fibroblasts, structural preservation of nuclei was essential to allow conclusions on the in vivo distribution, in particular, to exclude loss of genome separation during preparative steps. We used confocal microscopy, qualitative and quantitative image analysis to test for several models of parental genome separation (Fig. 1c). Early assumptions of genome separation were triggered by bilobed nuclei (Häcker 1895; Rückert 1895; Conklin 1901). A tendency for homologous chromosome territories being in separate lobes of bilobed human granulocyte nuclei was described (Bártová et al. 2001). Such a tendency might be caused by separation of parental genomes. Therefore, we additionally examined the distribution of pairs of homologous chromosomes in this human cell type.
Materials and methods
FISH with equine probes
Equine genomic DNA was isolated from liver using standard procedures. Donkey liver was kindly provided by Barbara Bank, Frankfurt am Main. Horse liver was bought at a local horse butchery (Pferdemetzgerei Kaspar Wörle, Munich, Germany). Isolated DNA was digested with RNase A (60 μg/mL) for 2 h at 37°C followed by phenol–chloroform precipitation, according to a protocol by Shirley Reynolds (University of California at San Diego; http://hedricklab.ucsd.edu/Protocol/PhenChlor.html). Purified DNA was amplified with a Genomi-Phi-V2 DNA-Amplification-Kit (Amersham Biosciences, now part of GE Healthcare, Munich, Germany; see Lasken and Egholm 2003). DNA labeling was performed with biotin- or digoxigenin-coupled dUTPs by nick translation. DNA concentration in hybridization mixes was 40 ng/μL for each species. All hybridization mixes were tested on metaphase spreads before hybridization on structurally preserved cell nuclei. Hybridization was as described elsewhere (Hepperger et al. 2007), except that whole genomic DNA probes were hybridized overnight or shorter to minimize single-copy hybridization. Biotin-labeled probes were always detected with Avidin-Alexa488 (Molecular Probes, now part of Invitrogen, Karlsruhe, Germany) and digoxigenin-labeled probes with mouse-α-dig-cy3 (Jackson ImmunoResearch Europe, Suffolk, UK). To exclude that measured nuclear distributions of donkey and horse hybridization signals were biased by labeling and detection schemes, experiments were repeated with inversed schemes, leading to the same results (data not shown).
Equine metaphase spreads and cell nuclei
Mule fibroblasts from a 45-day-old mule fetus were obtained as described elsewhere (Woods et al. 2003). Equine blood samples were obtained during routine diagnostic measures and were used for the generation of metaphase spreads as well as the isolation of unstimulated lymphocytes. Mule blood was kindly provided by Dr. von Rennenkampff, Mountain Pack Animal Centre of the German Army, Bad Reichenhall. Donkey blood samples were kindly provided by Dr. Maltzan, Tierpark Hellabrunn, Munich (Martina-Franca donkey) and by Dr. Cronau (domestic donkey). Horse blood samples of an Appaloosa and an Arabian mare were kindly provided by Dr. Cronau and Dr. Pachten, horse clinic of the Ludwig-Maximilians-Universität München.
For the isolation of lymphocytes, 15 mL whole blood was mixed with 500 μL heparin (Braun, Melsungen) and diluted with 10 mL Hanks buffered salt solution (Sigma-Aldrich, Deisenhofen, H2387). The mixture was transferred to Leucosep tubes (Greiner, bio-one) prepared with 15.5 mL Ficoll (Ficoll-Paque PLUS, Amersham Biosciences). After centrifugation, the buffy coat with mononuclear cells was transferred to a 50-mL tube. Cells were washed twice with Hanks solution and then resuspended in 5 mL RPMI 1640 medium with 15% FCS (S 0115) (both Biochrom, Berlin). Lymphocyte solution (2.5 mL) was stimulated with 100 μL pokeweed mitogen (stock, 4.4 μg/5 mL; Biochrom) and 100 μL phytohemagglutinin (2.4 μg/5 mL; Biochrom). Cells were incubated 3–4 days at 37°C and 5% CO2. After hypotonic treatment with 0.56% KCl, fixation, and washings with methanol–acetic acid (3:1, −20°C) (Hepperger et al. 2007), cells were dropped on glass slides (Deng et al. 2003).
To obtain structurally preserved cell nuclei, fibroblasts or unstimulated lymphocytes were seeded on coverslips, fixed for 10 min in 4% freshly prepared buffered formaldehyde, and permeabilized as described (Hepperger et al. 2007). For unstimulated lymphocytes, coverslips were pretreated with poly-l-lysine to promote adherence (30–60 min incubation). Flattened nuclei for fast visual screening of large amounts of cells were obtained by fixing cells on coverslips in methanol–acetic acid (3:1, −20°C, 30 min) and subsequent air-drying but without hypotonic treatment and dropping (Hepperger et al. 2007).
Human granulocytes
Human granulocytes were isolated from blood samples of female healthy donors by density gradient centrifugation with Histopaque 1077 and Histopaque 1191 (both Sigma-Aldrich). The ring of cells between the two Histopaques contains granulocytes and smaller amounts of monocytes and lymphocytes. Cells were fixed as described above for “flattened” lymphocytes and subjected to FISH with whole chromosome paint probes. Of those cells that could be unequivocally identified as granulocytes by nuclear morphology, about one third had two lobes and were thus suitable for the analysis presented in this study. The remaining had three or more lobes.
Microscopy
All specimens were mounted in VectaShield (Vector Laboratories, Burlingame, CA, USA) and covered with 170-μm thick coverslips (Hecht-Assistent, Sondheim, Germany). Confocal image stacks of structurally preserved preparations and some metaphase spreads were generated on a Leica TCS SP2 microscope with voxel sizes of 80 nm in x, y and 250 nm in z or less with a 63× NA 1.4 oil objective at room temperature. Nuclei were selected randomly. Image stacks of flattened nuclei were generated on a VisiScope Cell Explorer (Visitron Systems, Puchheim, Germany) based on a Zeiss Axiovert 200 MOT microscope and a Spot TR-SE6 CCD Camera with Sony ICX285 chip, controlled by Metamorph Software.
Image processing and analysis
The software ImageJ (W.S. Rasband, U. S. National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2008) was used on confocal image stacks for shift correction of chromatic aberration and image processing where necessary (e.g., erasing neighboring nuclei) as well as determination of signal thresholds for subsequent computerized image analysis. Signal clusters of maternal and/or paternal DNA probes in 3D interphase nuclei were counted manually by opening stacks in ImageJ. Signals that touched each other were considered as belonging to the same cluster. Weak signal of the other color in the same 3D-volume was considered cross-hybridization and in itself did not lead to categorization of the cluster as being of mixed parental origin.
Absolute 3D distance measurements from signal voxels to the nearest surface of the nucleus was performed with the software eADS, written by Tobias Thormeyer, Munich (Albiez et al. 2006), after an algorithm by Johann von Hase, Heidelberg. Briefly, at the determined threshold, a surface around the reference object (nucleus) is computed. Thereafter, for each voxel of a segmented signal, the shortest 3D distance to the surface is calculated. Intensity-weighted frequencies were plotted in classes with 250 nm width as a percentage of the total given signal in a nucleus and the averages over the population of nuclei were calculated. Signals inside the reference object obtain negative distance values, those outside obtain positive values. To statistically compare signals in mule cell nuclei generated by donkey and horse DNA, we used the two-sided Kolmogorov–Smirnov (KS) test. For each class of distances, the average value over the population of investigated nuclei was used to generate the cumulative distribution required for the KS test. p values were calculated according to Young (1977).
For a random distribution of chromosomes in granulocytes with two lobes of equal size, the probability of the first chromosome of a given homologous pair to end up in one of the two lobes is one (100%). The probability for the second chromosome to end up in the other lobe is 0.5 (disregarding the small volume restriction due to the presence of the first chromosome). Thus, the probability for any given pair of homologs to end up in the two different lobes is also 0.5. The probability that three pairs of homologs all end up with one chromosome each in the two lobes is thus 0.5 × 0.5 × 0.5 = 0.125 or 12.5%. For the same reason, 12.5% is also the probability that none of the three pairs of homologs is separated to different lobes. The remaining two options (exactly one pair is separated or exactly two pairs are separated) can be reached each in three ways. Thus, their probability is 37.5% each.
Statistical comparison of the experimentally determined distribution (combined data from both granulocyte FISH experiments) with the random distribution was done using the percentage of the occurrence of zero, one, two, and three separated pairs of homologs in both cases. The chi-square test was applied (Statistica for Windows Ver. 5.1, 1997, StatSoft, Tulsa, OK, USA). This test requires a certain minimum of expected cases in each category. Thus, it could not be applied to the expected distribution for genome separation.
Results
Hybridization with genomic DNA allows distinction of horse and donkey chromosomes
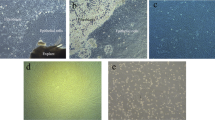
To test whether tandem repetitive sequences from horse (Equus caballus) and donkey (Equus asinus) have accumulated sufficient differences for species-specific labeling by FISH with genomic DNA from horse and donkey, we performed experiments on metaphase chromosome spreads from horse, donkey, and mule (Fig. 2a–e). In overlay images of the horse DNA signal (red) and donkey DNA signal (green) of metaphase spreads from horse (Fig. 2a) and donkey (Fig. 2c), most chromosomes showed bright signals of the expected color at the centromeres albeit weaker signals of the other color were also sometimes present (Fig. 2d, e). Several chromosomes in both species did not show any or a very weak hybridization signal, arguing for small centromeric repeat blocks. The exact number depends on hybridization conditions and threshold setting and is in the range of 10–15%. This is in agreement with a report of a horse centromeric repeat hybridizing on 58 of the 64 horse chromosomes (Wijers et al. 1993). One of 32 pairs of chromosomes in the horse and two of 31 pairs of chromosomes in the donkey (2n = 62; Raudsepp and Chowdhary 1999; Yang et al. 2004) each showed a signal of the “wrong” color in addition to the site that was labeled in the expected color (Fig. 2a, b, arrows).
FISH with a genomic DNA mix on metaphase spreads and flattened nuclei. Hybridization signals from horse DNA are false-colored in red, from donkey DNA in green. a Partial horse metaphase spread. The vast majority of horse chromosomes show clearly identifiable red centromere signals. One horse chromosome shows an additional green band (arrow). b Partial donkey metaphase spread. Chromosomes show predominantly green signals. Some centromeres have no or very weak signals. Two pairs of donkey chromosomes show a strong red signal in addition to green centromeres (arrows), only one homolog of both pairs is within the area shown. The larger of the two is probably chromosome 1 which is known to cross-hybridize with paint probes from several horse chromosomes (Raudsepp and Chowdhary 1999). c Complete mule metaphase spread. Most chromosomes can be readily identified as being of horse or donkey origin. The putative donkey chromosome 1 is also recognizable (arrow). d Clipping of another mule metaphase spread showing color overlay and separate channels for horse (center) and donkey DNA signals (right). All four horse chromosomes (red signal) also show a weaker hybridization signal by donkey DNA (right panel) due to cross-hybridization. e The boxed area in c, supplemented with single-color channels. An example for a donkey chromosome (green signal) that shows a FISH signal also by horse DNA (center panel). Note that, in c and d, determination of chromosomal origin is unequivocal, despite additional weak signals from the other DNA probe. Flattened lymphocytes (f) and fibroblasts (g) for fast screening of nonrandom-appearing patterns. Nuclei in these overviews do not show potentially nonrandom patterns, with the possible exception of the lymphocyte nucleus enlarged in h and i. When actively searched for, a few patterns were found in lymphocytes (h and i) and fibroblasts (j and k) which might be interpreted as nonrandom by some observers
In summary, of 63 mule chromosomes, only three contain sites which generate an extra FISH signal that can be misinterpreted in interphase nuclei and about ten generate no detectable signal. We conclude that FISH with genomic horse and donkey DNA on mule interphase nuclei identifies blocks of repetitive sequences on maternal and paternal chromosomes sufficiently well to identify separated parental genomes.
Centromere distribution in flattened mule cell nuclei
Next, we investigated the distribution of parental genomes in mule lymphocyte (Fig. 2f) and fibroblast (Fig. 2g) nuclei fixed with methanol–acetic acid and air-drying. Unlike other FISH studies, we did not include a hypotonic shock or dropping on slides. Because such nuclei are flattened (Hepperger et al. 2007), they allow rapid screening for unusual patterns by eye. Two hundred randomly recorded fibroblast and forty randomly recorded lymphocyte nuclei were screened for any nonrandom-appearing pattern independently by two observers. This specifically included searching for radial separation and a side-by-side arrangement of paternal and maternal signals (Fig. 1c). No pattern suggesting a separation of genomes was found. When several hundred nuclei were visually screened under the microscope, about 1% showed a pattern that might be interpreted as a nonrandom distribution, while other observers might not have this impression (Fig. 2h–k).
Radial nuclear distribution in structurally preserved mule cell nuclei is the same for donkey and horse chromosomes
To test whether one genome would be at the periphery of the nucleus and the other inside (Fig. 1c, center), we quantitatively measured the 3D radial distribution of FISH signals in nuclei from unstimulated lymphocytes and proliferating fibroblast (Fig. 3a–d). In both cell types, donkey and horse probe-derived signals were distributed very similarly (p > 0.1; Fig. 4). The mean distance from both signals to the nuclear surface averaged over all nuclei differed by only 25 nm in lymphocytes and 31 nm in fibroblasts, well below the resolution of confocal light microscopy.
FISH on structurally preserved mule cells. a Lymphocyte nucleus hybridized with genomic DNA from horse (red and center panel) and donkey (green and bottom panel). DNA counterstain in blue. b Color overlays of three lymphocyte nuclei. c and d Fibroblast nuclei, displayed as in a and b. Small letters in the top row indicate examples of large, medium, and small chromocenters as evaluated for Table 1. Some mixed chromocenters are highlighted by green and red arrows. e Mitotic fibroblast cells in prophase (P) or prometaphase (PM). The third panel includes an interphase nucleus for comparison. Projections of confocal stacks are shown. All scale bars = 5 μm
Radial distribution of donkey and horse chromosomes in lymphocyte (a) and fibroblast (b) nuclei. Values on the x-axis show the absolute distance to the nuclear surface, negative values represent signals inside the nucleus. Segmented nuclei were divided in shells of 0.25 μm thickness. For each nucleus, the percentage of given FISH signals or DNA counterstain in each shell was calculated. Curves show the average over all nuclei. The standard error of the mean is indicated by error bars
The above experiments on metaphase chromosomes showed that FISH-labeled centromeres from both species also contained smaller signal contributions from the other species. Thus, the donkey signal also contains signal from donkey DNA cross-hybridized to horse chromosomes and vice versa. To remove this contamination, we took the 3D image of the donkey probe signal and subtracted the image of the horse probe signal, thus eliminating donkey probe signal at horse centromeres. The reciprocal process resulted in pure horse signals in the other channel. Computation of radial distributions again showed no significant difference (data not shown).
Centromeres from both parents cluster in joint chromocenters
Centromeres in interphase nuclei of many species do not form separate entities, but they cluster to so-called chromocenters (Baccarini 1908), the degree of clustering depending on cell type and nuclear shape (e.g., Hsu et al. 1971; Mayer et al. 2005). In this study, we show that centromere clustering also occurs in equine cell nuclei.
In mule lymphocyte nuclei (Fig. 3a, b), we found, on average, 11 labeled chromocenters. With 22 chromocenters, on average, the clustering was less pronounced in flat fibroblast nuclei (Fig. 3c, d), as it was observed in mouse (Mayer et al. 2005). If maternal and paternal genomes were spatially separated, no common centromere clusters would be present, with the possible exception of some clusters at the interface between the two compartments. However, when we inspected 3D images of lymphocyte nuclei, clusters with both donkey and horse FISH signals were distributed equally through the nuclear volume without any indication of separation. Quantitative evaluation revealed that all large chromocenters and most intermediate-sized and small chromocenters contained centromeres from both species (Table 1), effectively excluding the possibility of genome separation.
In mule fibroblasts, the vast majority of large- and intermediate-sized chromocenters also contained centromeres from both parent species. Only for small chromocenters, a majority of about three quarters contained FISH signals of only one species (Table 1). Given the total numbers of chromocenters and centromeres detected by FISH and the small size of some fibroblast centromere signals, it is likely that many of these small ones contain the centromere signal of only one chromosome. Visual inspection revealed that “large” chromocenters were usually in central areas of fibroblast nuclei, often around nucleoli which were identified as large holes in the DAPI counterstain (e.g., Fig. 3d, top image). We also noted smaller holes in the DAPI staining which corresponded to chromocenters themselves (Fig. 5). DAPI preferentially labels AT-rich sequences (Tanious et al. 1992); thus, this finding suggests a low AT content at the centromeres. This is in agreement with the description of GC-rich satellite DNA in horse (Ryder and Hansen 1979; Wijers et al. 1993) and donkey (Gadi and Ryder 1983). A discrimination of holes caused by nucleoli from holes caused by chromocenters was not always possible. The central location of large chromocenters in fibroblast nuclei is probably due to spatial restrictions that do not allow centromere clustering involving many chromosomes close to the equatorial periphery of this flat cell type, similar to the absence of nucleoli from the periphery. When nucleolar and potentially nucleolar chromocenters were disregarded, the remaining chromocenters also did not show any recognizable deviation from a random distribution. In spherical lymphocyte nuclei, no preferential positioning of large chromocenters to nucleoli was observed.
Nucleoli and chromocenters in mule fibroblasts. Confocal sections of two fibroblast nuclei (a and b) stained with the AT-specific DNA counterstain DAPI (white or blue). In the blow-ups, horse FISH signals are colored in red, while donkey signals are colored in green. Sections and blow-ups shown at the top include large nucleoli. Sections at the bottom include typical holes in the DAPI staining which are filled by the FISH signals, i.e., repetitive DNA, indicating that equine centromeres are AT-poor and thus GC-rich
Parental chromosome sets mix in mitotic fibroblasts
Proliferating fibroblast cells attach relatively well to the substrate. Thirty cells in mitotic stages were still present after FISH and could be recorded. In these, donkey and horse chromosomes did not show any recognizable pattern of separate distribution. Instead, the two parental chromosome sets mixed, as in interphase nuclei (Fig. 3e). The lymphocytes from peripheral blood investigated by 3D FISH were unstimulated, to exclude any possibility of chromatin rearrangements during culture. Thus, no mitotic cells were observed.
Distribution of homologous chromosomes in bilobed human granulocyte nuclei
We next investigated the possibility of genome separation in an example of human cells, granulocytes isolated from peripheral blood. These nuclei can have a variety of shapes, such as oval, or with several lobes of various sizes. We restricted our investigations to nuclei with two similar-sized lobes and investigated whether one such lobe would contain one parental set of chromosomes, as suggested in the nineteenth century for bilobed invertebrate nuclei. We labeled three pairs of homologous chromosomes, 18–19–X or 7–12–X, in female cells (Fig. 6). In the case of parental genome separation, we would expect in both lobes one chromosome of each type in close to 100% of the cells. In the case of random chromosomal distribution, however, the probability for a single pair of homologous chromosomes to end up in different lobes is 50%, the probability that all three pairs are separated is 12.5% (see the “Materials and methods” section).
For single homologous chromosome pairs, we found a separation to different lobes in 35–56% of the cases (Table 2), compatible with fluctuations in a random distribution but not with genome separation. When three chromosome pairs were evaluated together, we found that only 14% of the bilobed nuclei had distributed the homologs of all three pairs to different lobes (Table 3). This result is incompatible with separation of parental genomes to the two lobes of these nuclei. Instead, our experimentally determined distribution does not show a significant difference to the random distribution (p = 0.18).
Discussion
By performing FISH on cell nuclei of mule cells, we provide the first report on the distribution of parental genomes in structurally preserved mammalian cell nuclei. Selection of appropriate hybridization conditions with total genomic DNA from the parent species as probes allowed us to exclusively label blocks of tandem repetitive sequences. Repetitive blocks from horse and donkey were sufficiently different in sequence so that the vast majority of mule chromosomes could be identified by centromere-near FISH signals as horse or donkey chromosomes.
Data from mule lymphocyte and fibroblast nuclei did not reveal any indication of a separation of maternal and paternal centromeres. On the contrary, we observed spatial intermingling in both mule cell types. In particular, we can exclude (1) a radial separation with chromosomes from one parent in the middle and of the other parent at the periphery and (2) a side-by-side arrangement of the two parental chromosomal sets, separated by a line or plane (Fig. 1c, left and center). A third option, a separation to randomly shaped but separate nuclear areas (Fig. 1c, right), is more difficult to eliminate. The regular presence of centromeres from both parents at shared chromocenters is hardly compatible with such an arrangement, however.
It is not possible to label chromosomes of different parental origin discriminatively with whole chromosome paint probes. Our approach allows to circumvent this problem and to assign the vast majority of centromeres to their respective parental origin and to determine their three-dimensional position. Unlike in metaphase spreads, in interphase nuclei, it is not possible to deduce the position of complete chromosomes from centromere labeling. Formally, that leaves the possibility that separation of genomes is limited to nontandem-repetitive regions of the chromosome territories. We were, however, not able to imagine a parental separation of chromosome arms without also necessarily separating the neighboring centromeres.
Lymphocytes and fibroblasts have very different cellular shapes. For humans and primates, it was shown that they have different predominant patterns of chromosome positioning (see the “Introduction”). We, therefore, considered the possibility that these cell types also showed differences in the spatial positioning of the parental genomes. In flat fibroblast nuclei, a pattern that could be interpreted as separation to a noteworthy degree should be immediately obvious to the observer. However, even when actively searched for, we found patterns that might appear as nonrandom in only about 1% of artificially flattened nuclei. This argues for a chance occurrence of such patterns caused by chromosomes distributed through the nuclear volume without regard to their parental origin.
Fibroblasts were investigated earlier with regard to genome separation. That earlier study on fibroblasts from a mouse hybrid described a separation of centromeres from the parent species in “5–10%” of cells (Mayer et al. 2000b). The six nuclei displayed in the article do not show a common pattern of separation. In some, centromeres from the two parent species are separable by a curved line and others show centromeres from one parent in the center and from the other parent at the periphery. The authors do not elaborate on how likely it is to find such a pattern in a large number of nuclei by random chance. They used a preparation method for the nuclei that involves hypotonic treatment, methanol–acetic acid, dropping of cell on a slide, and air-drying, a procedure for which we and others meanwhile showed that it severely impairs nuclear morphology (Hepperger et al. 2007 and references therein). In particular, we demonstrated that this treatment can lead to dispersion of centromere clusters in mouse cells (Hepperger et al. 2007). It thus cannot be excluded that originally clustered centromeres from the two parent species were artificially separated. It is difficult to imagine, however, how such impairment could artificially create a separation of parental centromeres in a subset of cells.
Also, in bilobed human granulocytes, we found a distribution that was compatible with a random segmentation of maternal and paternal chromosomes to the two lobes but no evidence for parental genome separation. Our results differ from those obtained by Bártová et al. (2001). These authors investigated the distribution of 19 chromosome pairs in bilobed human granulocytes and observed one chromosome per lobe in the majority of nuclei (59–75%, depending on the chromosome, average 66%), including chromosomes 7 (70%) and 18 (61%) which were also investigated in the present study. A tendency for one homologous locus in each lobe, albeit less pronounced (59%), was also found for several gene loci (Bártová et al. 2001). When the Abl and Bcr genes were detected together with the centromeres of chromosomes 6 and 17 in the same set of nuclei, the occurrence of all four loci separated to different lobes was indicated as “three times more frequent than predicted …” (no absolute numbers were given) but also a large number of other configurations was found, arguing against a separation of complete parental genomes to lobes (Bártová et al. 2001). Accordingly, these authors do not consider parental genome separation as a cause for their findings but discuss the importance of chromosomal positioning at the onset of the segmentation of the granulocyte nucleus.
In summary, our data are incompatible with a genome separation of the types described previously for somatic mammalian cell nuclei (Nagele et al. 1995; Nagele et al. 1998; Nagele et al. 1999; Mayer et al. 2000b; Chaudhuri and Walther 2003; Chaudhuri et al. 2005). Two options seem possible to explain this discrepancy. Either, there are species- and/or cell type-specific differences concerning parental genome separation. Although there is no respective evidence, the possibility cannot be formally excluded. Or, previous reports were biased by unconscious selection of nuclei with peculiar patterns or hampered by an underestimation of the random occurrence of nonrandom-looking patterns and/or an unclear definition of nonrandom-looking nuclei. In either case, we can exclude parental genome separation as a general motif of nuclear organization of the mammalian genome in somatic cells. The question remains how and why maternal and paternal centromeres are separated in hybrid male haploid mouse germ cells (Mayer et al. 2000a).
References
Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Küpper K, Joffe B, Thormeyer T, von Hase J, Yang S, Rohr K, Leonhardt H, Solovei I, Cremer C, Fakan S, Cremer T (2006) Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 14:707–733
Allison DC, Nestor AL (1999) Evidence for a relatively random array of human chromosomes on the mitotic ring. J Cell Biol 145:1–14
Baccarini P (1908) Sulle cinesi vegetative del “Cynomorium coccineum L”. Nuovo Giornale Botanico Italiano Nuova Serie 15:189–203
Bártová E, Kozubek S, Jirsová P, Kozubek M, Lukášová E, Skalníková M, Cafourková A, Koutná I, Paseková R (2001) Higher-order chromatin structure of human granulocytes. Chromosoma 110:360–370
Bennett ST, Bennett MD (1992) Spatial separation of ancestral genomes in the wild grass Milium montianum Parl. Ann Bot 70:111–118
Bolzer A, Kreth G, Solovei I, Köhler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, Cremer T (2005) Three-dimensional maps of all chromosome positions demonstrate a probabilistic order in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3:e157
Brandriff BF, Gordon LA, Segraves R, Pinkel D (1991) The male-derived genome after sperm–egg fusion: spatial distribution of chromosomal DNA and paternal–maternal genomic association. Chromosoma 100:262–266
Bridge JA, Nelson M, McComb E, McGuire MH, Rosenthal H, Vergara G, Maale GE, Spanier S, Neff JR (1997) Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet 95:74–87
Chaudhuri JP, Walther JU (2003) Separation of parental genomes in human blood and bone marrow cells and its implications. Int J Oncol 23:1257–1262
Chaudhuri JP, Kasprzycki E, Battaglia M, McGill JR, Brogger A, Walther JU, Reith A (2005) Biphasic chromatin structure and FISH signals in reflect intranuclear order. Cell Oncol 27:327–334
Conklin EG (1901) The individuality of the germ nuclei during the cleavage of the egg of Crepidula. Biol Bull 2:257–265
Cornforth MN, Greulich-Bode KM, Loucas BD, Arsuaga J, Vázquez M, Sachs RK, Brückner M, Molls M, Hahnfeldt P, Hlatky L, Brenner DJ (2002) Chromosomes are predominantly located randomly with respect to each other in interphase human cells. J Cell Biol 159:237–244
Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S (2006) Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol 18:307–316
Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
Deng W, Tsao SW, Lucas JN, Leung CS, Cheung AL (2003) A new method for improving metaphase chromosome spreading. Cytometry A 51:46–51
Foster HA, Bridger JM (2005) The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma 114:212–229
Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW (1998) Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol 141:5–20
Gadi IK, Ryder OA (1983) Molecular cytogenetics of the equidae. II. Purification and cytological localization of a (G + C)-rich satellite DNA from Equus hemionus onager and cross-species hybridization to E. asinus chromosomes. Cytogenet Cell Genet 35:124–130
Gläss E (1956) Die Identifizierung der Chromosomen im Karyotyp der Rattenleber. Chromosoma 7:655–699
Gläss E (1957) Das Problem der Genomsonderung in den Mitosen unbehandelter Rattenlebern. Chromosoma 8:468–492
Guerrini F, Bucci S, Ragghianti M, Mancino G, Hotz H, Uzzell T, Berger L (1997) Genomes of two water frog species resist germ line exclusion in interspecies hybrids. J Exp Zool 279:163–176
Haaf T (2001) The battle of the sexes after fertilization: behaviour of paternal and maternal chromosomes in the early mammalian embryo. Chromosome Res 9:263–271
Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584
Häcker V (1895) Über die Selbständigkeit der väterlichen und mütterlichen Kernbestandteile. Archiv f mikrosk Anat XLVI
Hepperger C, Otten S, von Hase J, Dietzel S (2007) Preservation of large-scale chromatin structure in FISH experiments. Chromosoma 116:117–133
Hertwig O (1906) Allgemeine Biologie. Verlag von Gustav Fisher, Jena
Hsu TC, Cooper JE, Mace ML Jr., Brinkley BR (1971) Arrangement of centromeres in mouse cells. Chromosoma 34:73–87
Kikuchi S, Tanaka H, Wako T, Tsujimoto H (2007) Centromere separation and association in the nuclei of an interspecific hybrid between Torenia fournieri and T. baillonii (Scrophulariaceae) during mitosis and meiosis. Genes Genet Syst 82:369–375
Kosak ST, Groudine M (2004) Gene order and dynamic domains. Science 306:644–647
Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8:104–115
Lasken RS, Egholm M (2003) Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol 21:531–535
Leitch AR, Schwarzacher T, Mosgoller W, Bennett MD, Heslop Harrison JS (1991) Parental genomes are separated throughout the cell cycle in a plant hybrid. Chromosoma 101:206–213
Lima-Brito J, Guedes-Pinto H, Harrison GE, Heslop-Harrison JS (1996) Chromosome identification and nuclear architecture in triticale x tritordeum F-1 hybrids. J Exp Bot 47:583–588
Mayer W, Fundele R, Haaf T (2000a) Spatial separation of parental genomes during mouse interspecific (Mus musculus x M. spretus) spermiogenesis. Chromosome Res 8:555–558
Mayer W, Smith A, Fundele R, Haaf T (2000b) Spatial separation of parental genomes in preimplantation mouse embryos. J Cell Biol 148:629–634
Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S (2005) Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol 6:44
Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128:787–800
Moenkhaus WJ (1904) The development of the hybrids between Fundulus heteroclitus and Mendia notata with especial reference to the behavior of the maternal and paternal chromatin. Am J Anat III 29–65
Nagele R, Freeman T, McMorrow L, Lee HY (1995) Precise spatial positioning of chromosomes during prometaphase: evidence for chromosomal order. Science 270:1831–1835
Nagele RG, Freeman T, Fazekas J, Lee KM, Thomson Z, Lee HY (1998) Chromosome spatial order in human cells: evidence for early origin and faithful propagation. Chromosoma 107:330–338
Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, Lee H (1999) Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. J Cell Sci 112(Pt 4):525–535
Oakenfull EA, Lim HN, Ryder OA (2000) A survey of equid mitochondrial DNA: implications for the evolution, genetic diversity and conservation of Equus. Conservation Genetics 1:341–355
Odartchenko N, Keneklis T (1973) Localization of paternal DNA in interphase nuclei of mouse eggs during early cleavage. Nature 241:528–529
Pera F, Rainer B (1973) Studies of multipolar mitoses in euploid tissue cultures. I. Somatic reduction to exactly haploid and triploid chromosome sets. Chromosoma 42:71–86
Quattro JM, Avise JC, Vrijenhoek RC (1991) Molecular evidence for multiple origins of hybridogenetic fish clones (Poeciliidae:Poeciliopsis). Genetics 127:391–398
Raudsepp T, Chowdhary BP (1999) Construction of chromosome-specific paints for meta- and submetacentric autosomes and the sex chromosomes in the horse and their use to detect homologous chromosomal segments in the donkey. Chromosome Res 7:103–114
Rückert J (1895) Über das Selbständigbleiben väterlicher und mütterlicher Kernsubstanz etc. Archiv f mikrosk Anat XLV
Ryder OA, Hansen SK (1979) Molecular cytogenetics of the Equidae. I. Purification and cytological localization of a (G + C)-rich satellite DNA from Equus przewalskii. Chromosoma 72:115–129
Spolsky C, Uzzell T (1984) Natural interspecies transfer of mitochondrial DNA in amphibians. Proc Natl Acad Sci USA 81:5802–5805
Stöck M, Lamatsch DK, Steinlein C, Epplen JT, Grosse WR, Hock R, Klapperstuck T, Lampert KP, Scheer U, Schmid M, Schartl M (2002) A bisexually reproducing all-triploid vertebrate. Nat Genet 30:325–328
Tanabe H, Müller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T (2002) Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci USA 99:4424–4429
Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD (1992) DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 31:3103–3112
Wienberg J (2005) Fluorescence in situ hybridization to chromosomes as a tool to understand human and primate genome evolution. Cytogenet Genome Res 108:139–160
Wijers ER, Zijlstra C, Lenstra JA (1993) Rapid evolution of horse satellite DNA. Genomics 18:113–117
Woods GL, White KL, Vanderwall DK, Li GP, Aston KI, Bunch TD, Meerdo LN, Pate BJ (2003) A mule cloned from fetal cells by nuclear transfer. Science 301:1063
Yang F, Fu B, O’Brien PC, Nie W, Ryder OA, Ferguson-Smith MA (2004) Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: insight into the occasional fertility of mules. Chromosome Res 12:65–76
Young IT (1977) Proof without prejudice: use of the Kolmogorov–Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem 25:935–941
Acknowledgement
We are most grateful to those who made this study possible by providing equine blood samples, Dr. von Rennenkampff, Mountain Pack Animal Centre of the German Army, Bad Reichenhall; Dr. Maltzan, Tierpark Hellabrunn; Dr. Cronau and Dr. Pachten, horse clinic of the Ludwig-Maximilians-Universität München; and to Ms. Bank, Frankfurt am Main, for the gift of donkey liver. We thank our colleagues Alexandra Kölbl for the help with human granulocytes, Jürgen Peters for the statistical advice, Felix Habermann for the advice on animal lymphocyte stimulation, and Thomas Cremer for the ongoing support of our work. This work was financially supported by the Deutsche Forschungsgemeinschaft.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Misteli
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hepperger, C., Mayer, A., Merz, J. et al. Parental genomes mix in mule and human cell nuclei. Chromosoma 118, 335–347 (2009). https://doi.org/10.1007/s00412-008-0200-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-008-0200-6