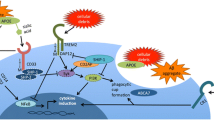
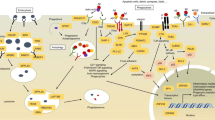
Abstract
In 2011, genome-wide association studies implicated a polymorphism near CD33 as a genetic risk factor for Alzheimer’s disease. This finding sparked interest in this member of the sialic acid-binding immunoglobulin-type lectin family which is linked to innate immunity. Subsequent studies found that CD33 is expressed in microglia in the brain and then investigated the molecular mechanism underlying the CD33 genetic association with Alzheimer’s disease. The allele that protects from Alzheimer’s disease acts predominately to increase a CD33 isoform lacking exon 2 at the expense of the prototypic, full-length CD33 that contains exon 2. Since this exon encodes the sialic acid ligand-binding domain, the finding that the loss of exon 2 was associated with decreased Alzheimer’s disease risk was interpreted as meaning that a decrease in functional CD33 and its associated immune suppression was protective from Alzheimer’s disease. However, this interpretation may need to be reconsidered given current findings that a genetic deletion which abrogates CD33 is not associated with Alzheimer’s disease risk. Therefore, integrating currently available findings leads us to propose a model wherein the CD33 isoform lacking the ligand-binding domain represents a gain of function variant that reduces Alzheimer’s disease risk.

These data (mean ± SD, n = 3) are derived from http://web.stanford.edu/group/barres_lab/brainseq2/brainseq2.html [77]. Other microglial RNAseq studies report similar SIGLEC expression profiles [78]







Similar content being viewed by others
References
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43(5):429–435
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J et al (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5):436–441
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12):1452–1458
Efthymiou AG, Goate AM (2017) Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol Neurodegener 12(1):43
Malik M, Parikh I, Vasquez JB, Smith C, Tai L, Bu G et al (2015) Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol Neurodegener 10:52
Hansen DV, Hanson JE, Sheng M (2018) Microglia in Alzheimer’s disease. J Cell Biol 217(2):459–472
Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF et al (2008) Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet 83(5):623–632
Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC et al (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 51(3):414–430
Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S et al (2019) Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51(3):404–413
Schwarz F, Fong JJ, Varki A (2015) Human-specific evolutionary changes in the biology of siglecs. Adv Exp Med Biol 842:1–16
Salminen A, Kaarniranta K (2009) Siglec receptors and hiding plaques in Alzheimer’s disease. J Mol Med (Berl) 87(7):697–701
Taylor VC, Buckley CD, Douglas M, Cody AJ, Simmons DL, Freeman SD (1999) The myeloid-specific sialic acid-binding receptor, CD33, associates with the protein-tyrosine phosphatases, SHP-1 and SHP-2. J Biol Chem 274(17):11505–11512
Crocker PR, Varki A (2001) Siglecs in the immune system. Immunology 103(2):137–145
Padler-Karavani V, Hurtado-Ziola N, Chang YC, Sonnenburg JL, Ronaghy A, Yu H et al (2014) Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J 28(3):1280–1293
Ishii T, Angata T, Wan ES, Cho MH, Motegi T, Gao C et al (2017) Influence of SIGLEC9 polymorphisms on COPD phenotypes including exacerbation frequency. Respirology 22(4):684–690
Graustein AD, Horne DJ, Fong JJ, Schwarz F, Mefford HC, Peterson GJ et al (2017) The SIGLEC14 null allele is associated with Mycobacterium tuberculosis- and BCG-induced clinical and immunologic outcomes. Tuberculosis (Edinb) 104:38–45
Yamanaka M, Kato Y, Angata T, Narimatsu H (2009) Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19(8):841–846
Varki A (2011) Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21(9):1121–1124
Angata T (2018) Possible influences of endogenous and exogenous ligands on the evolution of human siglecs. Front Immunol 9:2885
Lubbers J, Rodriguez E, van Kooyk Y (2018) Modulation of immune tolerance via siglec–sialic acid interactions. Front Immunol 9:2807
Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A (2009) Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113(14):3333–3336
Angata T, Margulies EH, Green ED, Varki A (2004) Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA 101(36):13251–13256
Cao H, Crocker PR (2011) Evolution of CD33-related siglecs: regulating host immune functions and escaping pathogen exploitation? Immunology 132(1):18–26
Bornhofft KF, Goldammer T, Rebl A, Galuska SP (2018) Siglecs: a journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol 86:219–231
Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M (2006) Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J 20(12):1964–1973
Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD (2008) SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol 38(8):2303–2315
Macauley MS, Crocker PR, Paulson JC (2014) Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol 14(10):653–666
Crocker PR, Paulson JC, Varki A (2007) Siglecs and their roles in the immune system. Nat Rev Immunol 7(4):255–266
Ravetch JV, Lanier LL (2000) Immune inhibitory receptors. Science 290(5489):84–89
Paul SP, Taylor LS, Stansbury EK, McVicar DW (2000) Myeloid specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood 96(2):483–490
Walter RB, Hausermann P, Raden BW, Teckchandani AM, Kamikura DM, Bernstein ID et al (2008) Phosphorylated ITIMs enable ubiquitylation of an inhibitory cell surface receptor. Traffic 9(2):267–279
Perez-Oliva AB, Martinez-Esparza M, Vicente-Fernandez JJ, Corral-San Miguel R, Garcia-Penarrubia P, Hernandez-Caselles T (2011) Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33 M and CD33 m) on lymphoid and myeloid human cells. Glycobiology 21(6):757–770
Ulyanova T, Blasioli J, Woodford-Thomas TA, Thomas ML (1999) The sialoadhesin CD33 is a myeloid-specific inhibitory receptor. Eur J Immunol 29(11):3440–3449
Mizuno K, Tagawa Y, Mitomo K, Arimura Y, Hatano N, Katagiri T et al (2000) Src homology region 2 (SH2) domain-containing phosphatase-1 dephosphorylates B cell linker protein/SH2 domain leukocyte protein of 65 kDa and selectively regulates c-Jun NH2-terminal kinase activation in B cells. J Immunol 165(3):1344–1351
Mizuno K, Tagawa Y, Mitomo K, Watanabe N, Katagiri T, Ogimoto M et al (2002) Src homology region 2 domain-containing phosphatase 1 positively regulates B cell receptor-induced apoptosis by modulating association of B cell linker protein with Nck and activation of c-Jun NH2-terminal kinase. J Immunol 169(2):778–786
Orr SJ, Morgan NM, Elliott J, Burrows JF, Scott CJ, McVicar DW et al (2007) CD33 responses are blocked by SOCS3 through accelerated proteasomal-mediated turnover. Blood 109(3):1061–1068
Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM et al (2006) A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol 79(1):46–58
Sutherland MK, Yu C, Lewis TS, Miyamoto JB, Morris-Tilden CA, Jonas M et al (2009) Anti-leukemic activity of lintuzumab (SGN-33) in preclinical models of acute myeloid leukemia. MAbs 1(5):481–490
Zhao L (2018) CD33 in Alzheimer’s disease—biology, pathogenesis, and therapeutics: a mini-review. Gerontology. https://doi.org/10.1159/000492596
Duan S, Koziol-White CJ, Jester WF Jr, Nycholat CM, Macauley MS, Panettieri RA Jr et al (2019) CD33 recruitment inhibits IgE-mediated anaphylaxis and desensitizes mast cells to allergen. J Clin Investig 129(3):1387–1401
Linnartz B, Neumann H (2013) Microglial activatory (immunoreceptor tyrosine-based activation motif)- and inhibitory (immunoreceptor tyrosine-based inhibition motif)-signaling receptors for recognition of the neuronal glycocalyx. Glia 61(1):37–46
Gratuze M, Leyns CEG, Holtzman DM (2018) New insights into the role of TREM2 in Alzheimer’s disease. Mol Neurodegener 13(1):66
Shi Y, Holtzman DM (2018) Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 18(12):759–772
Jay TR, von Saucken VE, Landreth GE (2017) TREM2 in neurodegenerative diseases. Mol Neurodegener 12(1):56
Sada K, Takano T, Yanagi S, Yamamura H (2001) Structure and function of Syk protein-tyrosine kinase. J Biochem 130(2):177–186
Underhill DM, Goodridge HS (2007) The many faces of ITAMs. Trends Immunol 28(2):66–73
Strzelecka A, Kwiatkowska K, Sobota A (1997) Tyrosine phosphorylation and Fcgamma receptor-mediated phagocytosis. FEBS Lett 400(1):11–14
Hamerman JA, Ni M, Killebrew JR, Chu CL, Lowell CA (2009) The expanding roles of ITAM adapters FcRgamma and DAP12 in myeloid cells. Immunol Rev 232(1):42–58
Barrow AD, Trowsdale J (2006) You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol 36(7):1646–1653
Pasquier B, Launay P, Kanamaru Y, Moura IC, Pfirsch S, Ruffie C et al (2005) Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM. Immunity 22(1):31–42
Ganesan LP, Fang H, Marsh CB, Tridandapani S (2003) The protein-tyrosine phosphatase SHP-1 associates with the phosphorylated immunoreceptor tyrosine-based activation motif of Fc gamma RIIa to modulate signaling events in myeloid cells. J Biol Chem 278(37):35710–35717
Peng Q, Malhotra S, Torchia JA, Kerr WG, Coggeshall KM, Humphrey MB (2010) TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci Signal 3(122):ra38
Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL (2006) Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol 177(4):2051–2055
Billadeau DD, Leibson PJ (2002) ITAMs versus ITIMs: striking a balance during cell regulation. J Clin Investig 109(2):161–168
Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K et al (2013) Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78(4):631–643
Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT et al (2013) CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci 33(33):13320–13325
Walker DG, Whetzel AM, Serrano G, Sue LI, Beach TG, Lue LF (2015) Association of CD33 polymorphism rs3865444 with Alzheimer’s disease pathology and CD33 expression in human cerebral cortex. Neurobiol Aging 36(2):571–582
Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A et al (2014) CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet 23(10):2729–2736
Malik M, Chiles J 3rd, Xi HS, Medway C, Simpson J, Potluri S et al (2015) Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Hum Mol Genet 24:3557–3570
Mortland L, Alonzo TA, Walter RB, Gerbing RB, Mitra AK, Pollard JA et al (2013) Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin Cancer Res 19(6):1620–1627
Lamba JK, Chauhan L, Shin M, Loken MR, Pollard JA, Wang YC et al (2017) CD33 splicing polymorphism determines gemtuzumab ozogamicin response in de novo acute myeloid leukemia: report from randomized phase III children’s oncology group trial AAML0531. J Clin Oncol 35(23):2674–2682
Laszlo GS, Beddoe ME, Godwin CD, Bates OM, Gudgeon CJ, Harrington KH et al (2018) Relationship between CD33 expression, splicing polymorphism, and in vitro cytotoxicity of gemtuzumab ozogamicin and the CD33/CD3 BiTErAMG 330. Haematologica 104:e59–e62
Gbadamosi M, Meshinchi S, Lamba JK (2018) Gemtuzumab ozogamicin for treatment of newly diagnosed CD33-positive acute myeloid leukemia. Future Oncol 14:3199–3213
Laszlo GS, Harrington KH, Gudgeon CJ, Beddoe ME, Fitzgibbon MP, Ries RE et al (2016) Expression and functional characterization of CD33 transcript variants in human acute myeloid leukemia. Oncotarget 7(28):43281–43294
Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A et al (2013) CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci 16(7):848–850
Schwarz F, Springer SA, Altheide TK, Varki NM, Gagneux P, Varki A (2016) Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc Natl Acad Sci USA 113(1):74–79
Siddiqui S, Schwarz F, Springer S, Khedri Z, Yu H, Deng L et al (2017) Studies on the detection, expression, glycosylation, dimerization, and ligand binding properties of mouse siglec-E. J Biol Chem 292(3):1029–1037
Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK et al (2017) A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169(7):1276–1290
Tajuddin SM, Schick UM, Eicher JD, Chami N, Giri A, Brody JA et al (2016) Large-scale exome-wide association analysis identifies loci for white blood cell traits and pleiotropy with immune-mediated diseases. Am J Hum Genet 99(1):22–39
Stein PS, Desrosiers M, Donegan SJ, Yepes JF, Kryscio RJ (2007) Tooth loss, dementia and neuropathology in the Nun study. J Am Dent Assoc 138(10):1314–1322
Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A et al (2019) Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv 5(1):eaau3333
Papageorgiou I, Loken MR, Brodersen LE, Gbadamosi M, Uy GL, Meshinchi S et al (2019) CCGG deletion (rs201074739) in CD33 results in premature termination codon and complete loss of CD33 expression: another key variant with potential impact on response to CD33-directed agents. Leuk Lymphoma. https://doi.org/10.1080/10428194.2019.1569232
Hoenig JM, Heisey DM (2001) The abuse of power: the pervasive fallacy of power calculations for data analysis. Am Stat 55(1):19–24
Wang J, Wu Y, Hu H, Wang W, Lu Y, Mao H et al (2011) Syk protein tyrosine kinase involves PECAM-1 signaling through tandem immunotyrosine inhibitory motifs in human THP-1 macrophages. Cell Immunol 272(1):39–44
Balaian L, Zhong RK, Ball ED (2003) The inhibitory effect of anti-CD33 monoclonal antibodies on AML cell growth correlates with Syk and/or ZAP-70 expression. Exp Hematol 31(5):363–371
Konishi H, Kiyama H (2018) Microglial TREM2/DAP12 signaling: a double-edged sword in neural diseases. Front Cell Neurosci 12:206
Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD et al (2016) Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89(1):37–53
Olah M, Patrick E, Villani AC, Xu J, White CC, Ryan KJ et al (2018) A transcriptomic atlas of aged human microglia. Nat Commun 9(1):539
Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlic A, Rose PW (2018) NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 34(21):3755–3758
Crocker PR, Varki A (2001) Siglecs, sialic acids and innate immunity. Trends Immunol 22(6):337–342
Marczynke M, Groger K, Seitz O (2017) Selective binders of the tandem src homology 2 domains in Syk and Zap70 protein kinases by DNA-programmed spatial screening. Bioconjug Chem 28(9):2384–2392
Acknowledgements
This work was supported by the National Institute on Aging (RF1AG059717 and P30AG028383).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Estus, S., Shaw, B.C., Devanney, N. et al. Evaluation of CD33 as a genetic risk factor for Alzheimer’s disease. Acta Neuropathol 138, 187–199 (2019). https://doi.org/10.1007/s00401-019-02000-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-02000-4