Abstract
Cryptochromes are photolyase-related blue-light receptors acting as core components of the mammalian circadian clock in the cell nuclei. One or more members of the cryptochrome protein family are also assumed to play a role in avian magnetoreception, but the primary sensory molecule in the retina of migratory birds that mediates light-dependent magnetic compass orientation has still not been identified. The mRNA of cryptochrome 2 (Cry2) has been reported to be located in the cell nuclei of the retina, but Cry2 localisation has not yet been demonstrated at the protein level. Here, we provide evidence that Cry2 protein is located in the photoreceptor inner segments, the outer nuclear layer, the inner nuclear layer and the ganglion cell layer in the retina of night-migratory European robins, homing pigeons and domestic chickens. At the subcellular level, we find Cry2 both in the cytoplasm and the nucleus of cells residing in these layers. This broad nucleic expression rather points to a role for avian Cry2 in the circadian clock and is consistent with a function as a transcription factor, analogous to mammalian Cry2, and speaks against an involvement in magnetoreception.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptochromes are blue-light receptors originally discovered in Arabidopsis thaliana (Ahmad and Cashmore 1993) but found throughout the biological kingdoms, from bacteria to insects and mammals (Sancar 2003; Lin and Todo 2005; Chaves et al. 2011). They are evolutionarily related to photolyases, which catalyse the blue-light-dependent repair of UV-damaged DNA (Sancar 1994, 2003). Like their photolyase ancestors, cryptochromes are flavoproteins: They often contain flavin adenine dinucleotide (FAD) as a covalently bound cofactor, and some cryptochromes also have a second light-harvesting chromophore (Cashmore et al. 1999; Sancar 2003). Cryptochromes, though, possess no DNA repair activity; instead, they are involved in the entrainment of circadian rhythms in vertebrates (Miyamoto and Sancar 1998; Cashmore et al. 1999; Sancar 2000, 2004). Circadian rhythms are established and maintained through transcription-translation feedback loops (TTFL), which consist of positive and negative regulatory elements. In mammals, positive elements of the TTFL are Clock, Npas2, Bmal1 and Cycle, whereas Period (Per) and Cryptochrome (Cry) proteins serve as negative elements (Takahashi 2017). Heterodimers of Clock and Bmal1 act as transcription factors to drive the rhythmic expression of Per and Cry genes, which in turn dimerise, translocate back into the nucleus and repress their own Clock/Bmal1-dependent transcription in the main feedback loop (Griffin et al. 1999; Kume et al. 1999; van der Horst et al. 1999). Apart from their core function in the circadian clock, vertebrate cryptochromes have also been proposed to play a role in avian magnetoreception (Ritz et al. 2000; Hore and Mouritsen 2016; Mouritsen 2018), which is thought to be based on a light-dependent (Wiltschko and Wiltschko 1993; Zapka et al. 2009; Wiltschko et al. 2010), quantum–mechanical mechanism involving radical pairs (Schulten et al. 1978; Ritz et al. 2000; Hore and Mouritsen 2016). It takes place in both eyes of birds (Hein et al. 2010, 2011; Engels et al. 2012), and magnetic signals are further processed in the bird’s thalamofugal visual pathway (Heyers et al. 2007; Zapka et al. 2009, 2010; Mouritsen et al. 2016). Since cryptochromes are the only proteins known in the vertebrate retina to be able to form radical pairs upon photoexcitation (Ritz et al. 2000; Maeda et al. 2008, 2012; Biskup et al. 2009; Hiscock et al. 2016), they are suggested to be the primary magnetoreceptors in the magnetic compass of migratory birds (Ritz et al. 2000; Hore and Mouritsen 2016). In mammals, two cryptochrome genes (Cry1 and Cry2) exist and the resulting proteins, Cry1 and Cry2, are both involved in the circadian clock (Miyamoto and Sancar 1998). Birds, in contrast, possess three Cry genes, Cry1, Cry2 and Cry4, coding for at least six different Cry proteins: Cry1a, Cry1b, Cry2a (formerly called Cry2; Yamamoto et al. 2001), Cry2b (Hochstoeger et al. 2020), Cry4a (formerly called Cry4) (Liedvogel and Mouritsen 2010) and Cry4b (Einwich et al. 2020). Both Cry2b and Cry4b have just recently been reported: Cry2b has been identified in the homing pigeon (Hochstoeger et al. 2020) and Cry4b has been found in the night-migratory songbird European robin (Einwich et al. 2020). Deducing from the amino acid identity, avian Cry1a and Cry2a/b are orthologues to mammalian Cry1 and Cry2, respectively. Cry1b, Cry2b and Cry4b seem to be bird-specific (Möller et al. 2004; Bolte et al. 2016; Nießner et al. 2016; Einwich et al. 2020; Hochstoeger et al. 2020), whereas Cry4(a) has been found in fish (Kobayashi et al. 2000), birds (Kubo et al. 2006, Liedvogel and Mouritsen 2010), amphibians (Takeuchi et al. 2014) and reptiles (Liu et al. 2015). During the past few years, the retinal location of the Cry proteins has been reported in several avian species. Cry1a was specifically found in the UV/V cone outer segments (Nießner et al. 2011; Bolte et al. 2021; Pinzon-Rodriguez and Muheim 2021), Cry1b is expressed in the photoreceptor inner segments, ganglion cells, and displaced ganglion cells (Bolte et al. 2016; Nießner et al 2016), and antibodies recognising both Cry4a and Cry4b proteins detected the Cry4 isoforms in the photoreceptor double cones and long-wavelength single cones in the European robin and the chicken (Günther et al. 2018). However, Hochstoeger et al. (2020) localised Cry4a in horizontal cells of the pigeon sretina. Currently, Cry4a is considered as the most probable candidate for being the light-dependent magnetoreceptor in the avian retina, both due to its location in the double cone outer segments (Hore and Mouritsen 2016; Worster et al. 2017; Günther et al. 2018, 2021), its ability to bind FAD (Öztürk et al. 2009; Wang et al. 2018; Zoltowski et al. 2019; Hochstoeger et al. 2020; Xu et al., 2021) and its magnetic-sensitive photochemistry demonstrated in vitro (Xu et al. 2021). Avian Cry2 proteins have been reported to possess a nuclear localisation signal (NLS), which presumably associates them to the nucleus (Möller et al. 2004; Mouritsen et al. 2004), but their location within the avian retina has not been demonstrated yet. On mRNA level, avian Cry2a has been identified in several tissues of both the chicken and the quail, including retina and pineal gland (Bailey et al. 2002; Fu et al. 2002). Specifically, Cry2a mRNA was found in the photoreceptor inner segments, the inner nuclear layer and the ganglion cell layer of the chicken retina (Bailey et al. 2002). A distribution of Cry2 proteins within or associated with the cell nuclei would rather speak against Cry2 as a magnetoreceptor, since a proper alignment that is essential for a directional magnetic field effect can only be achieved by tethering to the cytoskeleton or the membranes (Liedvogel and Mouritsen 2010; Hore and Mouritsen 2016), as outlined in Worster et al. (2017). Instead, both a nuclear location and a widely-distributed expression of the avian retinal Cry2 proteins would point to a role as a transcription factor of the circadian clock, analogous to mammalian Cry2 (Vitaterna et al. 1994; Thresher et al. 1998; Okamura et al. 1999; Griffin et al. 1999; Kume et al. 1999; van der Horst et al. 1999; Selby et al. 2000) since nuclear localisation appears to be a prerequisite for the transcriptional repressor activity of cryptochromes (Hirayama et al. 2003). Therefore, we analysed the subcellular location of the Cry2 proteins in the retina of migratory European robins, homing pigeons and domestic chickens using two different antibodies recognising both Cry2 isoforms. The aim of the present study is to investigate whether the Cry2 proteins, due to their location within the retina, are more likely to be involved in the circadian clock or if they might also play a different role, e.g. in magnetoreception.
Material and methods
Birds
Nine European robins (Erithacus rubecula) were wild-caught in the vicinity of the campus of the Carl von Ossietzky University Oldenburg using mist nets. One homing pigeon (Columba livia domestica) and three domestic chickens (Gallus gallus domesticus) were raised in the animal care facility of the University of Oldenburg. The birds were kept indoors under the natural light–dark cycle. Birds were sacrificed by decapitation to avoid the effects of injected substances on tissues of interest, or by an intramuscular injection of an overdose of Ketamin/Domitor (1 + 1 mix, 50–100 mg/kg bodyweight each). Eyes were prepared by cornea dissection followed by removal of the lens and vitreous bodies.
Cloning
Retinae from freshly prepared eyecups were vortexed for 2 min in TRIzol Reagent (Life Technologies, Carlsbad, CA, USA), placed into liquid nitrogen and stored at − 80 °C. RNA samples were extracted according to the RNA preparation protocol for the TRIzol Reagent (Life Technologies), contaminating genomic DNA was digested with DNase I Amplification Grade (Invitrogen, Carlsbad, CA, USA). The cDNA synthesis was performed with the SmarterTM RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Kalifornien, USA) according to the manufacturer’s protocol. An erCry2 specific primer (5′-AGCCCTGCCCCAAAGTGGAG-3′) was designed based on an already published erCry2 fragment (GenBank accession number AY7772689.1) to amplify the coding region of European robin Cry2a (erCry2a) by 5’RACE PCR. PCR products of the expected length were sub-cloned into the pGEM T easy vector (Promega, Madison, WI, USA) and subjected to sequencing (LGC, Berlin, Germany). By that, almost the entire erCry2a coding region was obtained, with only the first 39 nucleotides lacking (as assumed by alignment with the Cry2 mRNA from other bird species available in BLAST databases). The sequence is available under the GenBank accession number MN656980.2. Cloning primers (sense: 5'-ATTCTCGAGATGTTTTGCCGCTCCGTGCA-3', with an artificial start codon included, and antisense: 5'-TCTAAGCTTGCACAAACTCTTCCCAGGGA-3') including restriction sites for XhoI and HindIII were used to amplify the coding regions with the RT-PCR kit GoTaq Long PCR Master Mix (Promega, Madison, WI, USA). The PCR products were cloned into the expression vector pTurboGFP-N (Evrogen, Moscow, Russia) according to the protocol for the Rapid DNA Dephos & Ligation Kit (Roche Diagnostics, Mannheim, Germany). The information about the missing N-terminal 39 nucleotides of the erCry2a ORF was taken from the published genome assembly of the European robin genome (GenBank accession number LR812108.1) (Feng et al. 2020). The full erCry2a sequence was then amplified with erCry2a-GFP in pTurbo as a template and a sense primer containing the missing 39 nucleotides (underlined) (5′ATATGCTAGCGGCCATTACGGCCATGGCGGCGGCGGCCGCTCTGGGCCCGGCGCCGGCCCTGTGCCGCTCCGTGCAC 3′), and the antisense primer (5′ GATATCTGCAGAATTCTCACAAACTCTTCCCAGGGATCTC 3′) using the CloneAmp HiFi PCR Premix (Takara Bio Inc., Shiga, Japan). The purified PCR product was then cloned into the SfiI and EcoRI linearized mammalian expression vector pcDNA3.1(+), modified to contain a Kozak sequence, an N-terminal deca-histidine tag and an SfiI restricition site (gift from Prof. Karl-Wilhelm Koch, Carl von Ossietzky University Oldenburg), using the In-Fusion Snap Assembly Master Mix (Takara Bio Inc.).
Antibodies
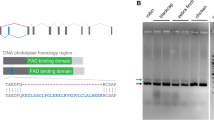
To localise the Cry2 proteins in the avian retina, two different anti-Cry2 antibodies were used to stain the retinae of six European robins, one homing pigeon and three domestic chickens. We produced a mouse monoclonal antibody, erCry2-26E11 (IgG2b/k), by immunisation of a 14 amino acid long peptide near the C-terminal end of erCry2 using hybridoma technology as described in Günther et al. (2018). This peptide sequence (12 out of 14 amino acids in total) is located within the NLS predicted by the NLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) (Fig. 1). A goat-derived polyclonal antibody, mmCry2-A20 from Santa Cruz Biotechnology, Inc. (Dallas, Texas, USA), had been raised against a 51 amino acid long peptide mapping near the C-terminus of the house mouse (Mus musculus) Cry2 (mmCry2, NCBI Reference Sequence: AF156987.1). The C terminal end is much less conserved between mouse and the birds than the core of the protein, but around 50% of the mmCry2 peptide sequence the mmCry2-A20 antibody was raised against is also conserved in the erCry2a protein (Fig. 1). The amino acid sequences of pigeon Cry2a (clCry2a, GenBank accession number: KX168609.1), clCry2b (GenBank accession number: KX168610.1) and chicken Cry2a (ggCry2a, NCBI Reference Sequence: NM_204244.1) are included in Fig. 1 to highlight mismatches to the two antibodies used in this study.
Alignment of the amino acid sequences of erCry2a, clCry2a, clCry2b, ggCry2a and mmCry2. The 14 amino acid long peptide from the European robin that the antibody erCry2-26E11 was raised against is shown in green (amino acid position 544 to 557 in erCry2), the 51 amino acid long peptide for mmCry2-A20 is shown in yellow (amino acid position 520 to 570 in mmCry2). All five Cry2 proteins possess a nuclear localisation signal (blue, bold). The amino acid sequences of clCry2a, clCry2b and ggCry2a have two mismatches to the peptide that the antibody erCry2-26E11 was raised against (marked in red). The multiple sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/)
Tissue preparation for immunohistochemistry
After preparation, eyecups were post-fixed in 4% PFA/PBS for 20–30 min and washed in PBS (pH 7.4). Tissue adaptation to cryoprotectant solution was performed overnight at 4 °C in 30% saccharose solution in 0.1 M PB (pH 7.4) and embedded in a cryoblock at − 20 °C. Vertical retinal sections of 20 µm were cut on a cryostat (CM 1950, Leica, Wetzlar, Germany), collected on gelatinized superfrost coverslips (Carl Roth, Karlsruhe, Germany), or Menzel SuperFrost Plus slides (Thermo Fisher Scientific, Waltham, MA, USA), and heat-fixed at 37 °C for one to three hours.
Immunohistochemistry
After washing in 0.1 M phosphate buffer (PB), cryosections were blocked with 10% normal donkey serum (Sigma-Aldrich) in blocker mix (0.1 M PB containing 0.3% Triton X-100 and 2% normal donkey serum) for 30 to 60 min at room temperature. Incubation with primary antibodies diluted in blocker mix was performed at 4 °C overnight. The affinity-purified polyclonal antibody mmCry2-A20 was used in a dilution of 1:500. Monoclonal antibody erCry2-26E11 was used as unpurified hybridoma supernatant diluted 1:2 for the European robin and the homing pigeon and 1:3 for the domestic chicken. Due to the use of monoclonal hybridoma supernatant instead of affinity purified antibody, a low dilution was applied. For the pre-adsorption control, the antibody erCry2-26E11 was incubated for four hours at room temperature with 500 µg/ml Cry2 or Cry4 peptide (as a control) and then applied to the slices. After washing with 0.1 M PB, the slices were incubated for one hour with the secondary antibodies (Invitrogen, Carlsbad, CA, USA; for the European robin and the homing pigeon: Alexa 488 donkey anti-mouse and Alexa 568 donkey anti-goat, for the domestic chicken: Alexa 568 donkey anti-mouse and Alexa 647 donkey anti-goat), diluted 1:500 in blocker mix. After washing, slices were covered with Aqua-Poly/Mount (Polysciences, Inc., Warrington, PA, USA) and a cover slip. No measures to reduce potential autofluorescence were necessary as it was found to be weak, apart from the well-known fluorescence of oil droplets (e.g., Fig. 4l).
Cell culture and protein expression
N2A and HEK293 cells were maintained in DMEM with 10% fetal calf serum and 1% antibiotics/antimycotics (Sigma-Aldrich). For immunocytochemistry, cells were plated in 24-well plates on glass coverslips that had been treated with poly L-Lysine for 30 min. For the immunoblot, HEK293 cells were seeded in 6-well plates (Roth). ErCry1a (Bolte et al. 2016), erCry2a, lacking the first 39 nucleotides and erCry4a (Günther et al. 2018) were expressed as fusion proteins with GFP from the respective generated pTurbo-GFP-N vectors using 2 µl Lipofectamine 2000 reagent (Invitrogen) per µg DNA in OptiMEM medium (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol.
Immunocytochemistry of transfected cells
Forty hours after transfection, the cells were washed in phosphate buffered-saline (PBS) followed by a fixation with ice-cold methanol (7 min). After washing, the primary antibodies were incubated at 4 °C overnight (erCry2-26E11 diluted 1:3, mmCry2-A20 diluted 1:500 in PBS). After washing, the cells were incubated for two hours with appropriate secondary antibodies (Alexa 555 goat anti-mouse and Alexa 568 donkey anti-goat, diluted 1:500 in PBS) and mounted with Vectashield Antifade Mounting Medium with DAPI (Vector Laboratories Inc.).
Immunoblot
Forty-eight hours after the transfection, the HEK293 cells were harvested from the wells and mechanically homogenised in buffer (50 mM Tris/HCl, pH 7.4, 2 mM EGTA, pH 7.4, 2 mM EDTA, pH 7.4, 0.1 mM sodium orthovanadate, 1 mM DTT, 2 μg/ml leupeptin, 5 μg/ml aprotinin, 2 mM PMSF in DMSO, Roche cOmplete™ Protease Inhibitor Cocktail). Some of it was stored at − 20 °C until usage whereas the rest was centrifuged (15 min, 8,000 rpm, 10 °C) until it had separated into a pellet and a supernatant. Total protein concentration of the total homogenate and the supernatant was measured by the Neuhoff assay (Neuhoff et al. 1979). The pellet was suspended in 8 M urea until the solution had become viscous. Both the supernatant and the pellet solution were used as samples.
For the immunoblot, the samples were mixed with SDS buffer (10% glycerol, 2% SDS, 1% beta-mercaptoethanol, 50 mM Tris–HCl pH 6.8; 12.5 mM EDTA pH 8.0, 0.02% bromophenol blue) and heated for four minutes at 95 °C. Ten to 15 µg of the samples were loaded on a standard SDS-PAGE (7.5% acrylamide, with a 4% acrylamide stacking gel). Gel electrophoresis was performed in an Eco Mini Buffer Tank (Biorad) using a 150 to 160 V separation current. Protein transfer from gel to membrane was performed in a wet blot system (350 mA for 60 min). After blocking of the nitrocellulose membrane (Whatman, Sigma-Aldrich) for one hour in 5% milk powder (Roth) in TBS-Tween, the membranes were incubated with the erCry2-26E11 primary antibody hybridoma supernatant (1:5 dilution) or a polyclonal GFP antibody (rabbit; Evrogen, Cat No AB513, Lot No 51301121268, 1:1000 dilution) diluted in 5% milk powder (Roth) in TBS-Tween overnight at 4 °C. After incubation with HRP-conjugated secondary antibodies (HRP anti-mouse or HRP anti-rabbit, dilutions 1:10,000), a chemiluminescence procedure was performed with Super Signal West Pico chemiluminescent substrates (Thermo Fisher Scientific) and the Image Lab Software version 5.2.1 (BioRad).
Confocal microscopy and image processing
Confocal micrographs of fluorescent retinae and transfected cells were analysed with a confocal laser scanning microscope (TCS SP8, Leica) using the 488, 555 and 647 nm laser lines. Scanning was performed with the oil immersion 40 × HPX PL APO (NA 1.25 or 1.3) objective. Pixel size was adjusted for each experiment. All images of the same experiment were taken with the same microscope settings. Confocal stacks were subtracted in background, median-filtered (stainings with DAPI), and normalised in contrast in Fiji (https:\\fiji.sc) (Schindelin et al. 2012). Unless stated otherwise, all figures show maximum projections of confocal stacks, adjusted for contrast and brightness for presentation purposes.
Results
Due to the recent report of a second isoform of avian Cry2 by Hochstoeger et al. (2020), we use the following terminology throughout this paper: ‘Cry2’ is used when the two isoforms cannot be distinguished, as it is the case for the immunohistological stainings. For the control experiments, however, we only worked with the erCry2a isoform, which we indicate as such.
In the retina of European robin, the two anti-Cry2 antibodies applied in this study, our custom-made mouse monoclonal antibody erCry2-26E11 and the commercially available polyclonal goat antibody mmCry2-A20 (Fig. 1), stained the photoreceptor inner segments as well as the outer nuclear layer, inner nuclear layer and ganglion cell layer (Fig. 2). Interestingly, the two Cry2 antibodies labelled the same cell types, but different regions of them: erCry2-26E11 seemed to stain predominantly the cytoplasm of the outer nuclear layer, inner nuclear layer and ganglion cell layer, whereas mmCry2-A20 staining was restricted to the nuclear part of those cells. The antibodies also showed a divergent signal in the photoreceptor inner segments: erCry2-26E11 signal was more prominent at the apex of the inner segment (the ellipsoid), which is rich in mitochondria in all vertebrate photoreceptors (Cohen 1972; Rodieck 1973; Stone et al. 2008), whereas mmCry2-A20 stained the lower region, called the myoid, where the paraboloid, endoplasmic reticulum, Golgi complex and ribosomes are located (Cohen 1972). A similarly differentiated staining of the Cry2 antibodies could also be observed in the pigeon retina (Fig. 3a–d). Here, erCry2-26E11 labelled the inner segments and, like in the European robin, the cytoplasm of cells in the inner nuclear layer and ganglion cell layer (Fig. 3b). In contrast, mmCry2-A20 recognised the ellipsoid of the inner segments and additionally the outer limiting membrane. Moreover, mmCry2-A20 seemed to stain both the cytoplasm and nuclei in the inner nuclear layer and ganglion cell layer (Fig. 3c). In the chicken, both antibodies recognised the inner segments and the cytoplasm of cells in the inner nuclear layer and the ganglion cell layer (Fig. 3e–h). The staining in the outer nuclear layer was only very faint with erCry2-26E11, whereas mmCry2-A20 seemed to recognise the outer limiting membrane (Fig. 3g). The staining pattern of the photoreceptor inner segments was again slightly diverging in chicken: erCry2-26E11 labelled the entire inner segments (with a stronger signal in the upper, ellipsoid part; Fig. 3f), whereas mmCry2-A20 exclusively and very strongly marked the ellipsoid part (Fig. 3g).
Cry2 localisation in the retina of the European robin. In the European robin retina (a), Cry2 was detected in the photoreceptor inner segments, outer nuclear layer, inner nuclear layer and ganglion cell layer (b–d). Whereas the erCry2-26E11 antibody labelled the ellipsoid of the inner segments as well as the cytoplasm of cells in the outer, inner and ganglion cell layers (b), mmCry2-A20 recognised more prominently the myoid of the photoreceptor inner segments as well as the nuclei of outer, inner and ganglion cell layers (c). Image a is a transmission image, images b–d are maximum projections of confocal stacks (z-size 1.27 µm, 7 sections). Scale bar: 50 µm. OS photoreceptor outer segments, PE retinal pigment epithelium, IS photoreceptor inner segments, ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer, NFL nerve fibre layer
Cry2 location was confirmed in two additional bird species. The antibodies erCry2-26E11 and mmCry2-A20 stained the photoreceptor inner segments as well as the cytoplasm of cells in the inner nuclear layer and ganglion cell layer in the day-flying pigeon (a–d) and the domestic chicken (e–h). mmCry2-A20 was also labelling the outer limiting membrane in the pigeon (c) and the chicken (g). In contrast to the immunosignals observed in the European robin retina, mmCry2-A20 did not label the nuclei but the cytoplasm in pigeon and chicken. Images a and e are bright-field images, images b–d and f–g are maximum projections of confocal stacks (b, c: z-size 4 µm, 20 sections; f–h: z-size 2 µm, 10 sections). Scale bars: d, h 50 µm. PE retinal pigment epithelium, OS photoreceptor outer segments, IS photoreceptor inner segments, OLM outer limiting membrane, ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer, NFL nerve fibre layer
We also included co-stainings of the monoclonal antibody erCry2-26E11 with the nuclear marker DAPI to analyse the subcellular localisation of Cry2 in the retina of the European robin (Fig. S1), the domestic pigeon (Fig. S2) and the domestic chicken (Fig. S3). In all three bird species, labelling of Cry2 and DAPI revealed that Cry2 seems to be more prominently located in the cytoplasm, but was also detected in the nuclei of the ganglion cell layer (Fig. S1-S3e-g), albeit very weakly in the European robin (Fig. S1e-g). The specificity of the Cry2 antibodies was confirmed in several control experiments. After pre-adsorbing the Cry2-26E11 antibody with the respective erCry2 peptide, we obtained no Cry2 signal (Fig. S4c). In contrast, we still obtained a signal when pre-adsorbing the antibody with an erCry4 peptide (Fig. S4d), demonstrating that the Cry2 antibody specifically binds to its target epitope. Immunocytochemical stainings on N2A cells (Fig. 4a-a’’) and HEK293 cells (Fig. 4e-e’’) recombinantly over-expressing either erCry1a-, erCry2a- or erCry4a-GFP fusion proteins (Fig. 4b-b’’, f-f’’) showed that both antibodies, erCry2-26E11 (Fig. 4c-c’’) and mmCry2-A20 (Fig. 4g-g’’), detect the erCry2a-GFP protein (Fig. 4d’, 4 h’), but not the other erCry-GFP proteins (Fig. 4d, d’’, h, h’’). In immunoblot analysis (Fig. 4i, j), erCry2-26E11 detected a single protein in cell lysates of recombinantly expressed erCry2a-GFP but not erCry4a-GFP fusion protein (Fig. 4j). The band was running around 25 kDa higher than the expected size of 934 kDa, which might be due to posttranslational modifications, such as phosphorylation, and/or protein oligomerisation that could alter the molecular weight of the erCry2-GFP protein. By staining with an anti-GFP antibody, we confirmed that the larger than expected band is, in fact, the Cry2a-GFP fusion protein (Fig. S5). We also confirmed that the secondary antibodies did not produce any artefacts in immunohistochemical stainings of the avian retinae (secondary antibody control; here shown for the chicken retina, Fig. 4k, l). With the commercial antibody mmCry2-A20, however, we failed to obtain Cry2 signals on the immunoblot. Unfortunately, this antibody is no longer produced by Santa Cruz Biotechnology, so that we could not determine whether the antibody does not work in immunoblot experiments in general or whether this only affected our antibody lot. We were, therefore, also unable to perform pre-adsorption control experiments for the mmCry2-A20 antibody. Yet, the mmCry2-A20 antibody showed Cry2-specific labelling in immunocytochemical controls (Fig. 4g-g’’) and therefore, the staining in retinal tissue was regarded as specific. It is often the case that antibodies work in immunohistochemistry but not in immunoblot. One potential reason might be that the mmCry2-A20 recognized complex 3D structures that are preserved in fixed tissue but are denatured during sample preparation for immunoblotting.
Controls for the antibodies. Staining of N2A cells (a-a’’) and HEK293 cells (e-e’’) expressing erCry1a-GFP (b, f), erCry2a-GFP (b’, f’) or erCry4a-GFP (b’’, f’’) protein with either our custom-made erCry2-26E11 antibody (c–c’’) or the commercially available mmCry2-A20 (g-g’’), respectively, indicates that both antibodies detected erCry2a-GFP (d’, h’) but not the other erCry-GFP proteins (d, d’’, h, h’’). Since all images are scanned and processed with the same parameters, some of the antibody signals are not visible in the resulting figure image. Immunoblot on recombinantly expressed erCry2a- and erCry4a-GFP fusion proteins (Ponceau staining, i incubated with the erCry2-26E11 antibody showed one single band only in the erCry2a-GFP lane (j). When omitting the primary antibodies in immunohistochemistry, no immunosignal was detected (k, l). Images a-a’’, e-e’’ and k are bright-field images; images b-d’’, f–h’’ and l are single confocal scans. Scale bars: 50 µm. PE retinal pigment epithelium, OS photoreceptor outer segments, OD oil droplets, IS photoreceptor inner segments, ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer. For the immunoblot, the total homogenate of the protein samples was used
Discussion
It is generally accepted that members of the cryptochrome family are involved in the circadian clock (Miyamoto and Sancar 1998; Chaves et al. 2011), but they are also candidates for light-dependent magnetoreception (Ritz et al. 2000; Mouritsen et al. 2004; Hore and Mouritsen 2016; Xu et al. 2021). In this study, we aimed to find out where the Cry2 proteins are expressed in the avian retina since their subcellular location might provide evidence for their probable molecular function. Hence, we included three different bird species with differing magnetosensitive behaviour: night-migratory European robins, day-flying homing pigeons, and non-migratory domestic chickens. Furthermore, as we do not have the possibility to work with knock-out (migratory) birds, it is important to use different antibodies, ideally recognising different peptide epitopes of the same protein to confirm an antibody staining (Uhlen et al. 2016; Günther et al. 2018; Bolte et al. 2021). Here, we applied two different antibodies that were both raised against peptides near the C-terminal end of the European robin or mouse Cry2 proteins, respectively. Both antibodies should recognise both Cry2 protein isoforms, Cry2a and the recently reported bird-specific Cry2b protein (Hochstoeger et al. 2020). The mouse-specific polyclonal mmCry2-A20 antibody was raised against a considerably longer peptide (51 amino acids) than the European robin-specific antibody erCry2-26E11 (14 amino acids), but the sequences partially overlap. With both antibodies, we found Cry2 proteins ubiquitously expressed in the photoreceptor inner segments as well as in most, if not all cells of the nucleic cell layers (outer nuclear, inner nuclear and ganglion cell layers) in the retinae of all three bird species. This pattern is very similar to the pattern seen in mouse, where the expression of several clock proteins (Clock, Bmal1, Npas1, Per1, Per2, and Cry2) was observed prominently in cells of the inner nuclear layer and the ganglion cell layer, but also in a few cells in the outer nuclear layer (Liu et al. 2012). However, how Cry2 protein expression compares to the expression of other clock proteins in the avian retina, we cannot tell because to our knowledge, the expression of the clock genes has only been studied on the mRNA level, but not on the protein level so far (Yoshimura et al. 2000; Bian et al. 2019; Renthlei and Trivedi 2019). In our stainings, though, we detected slight differences between both antibodies and bird species: erCry2-26E11 showed stronger Cry2 labelling in the cytoplasm than in the cell nuclei in all three species (S1-Fig. S3e-g), whereas mmCry2-A20 recognised Cry2 in the cytoplasm of the pigeon and the chicken retina but in the cell nuclei of the European robin retina. A distribution of Cry2 in both cellular compartments is not per se surprising as protein synthesis takes place in the cytoplasm. In the circadian clock of mammals, freshly synthesised Cry2 (as well as Cry1) proteins heterodimerise with Per proteins in the cytoplasm and translocate to the nucleus, where they repress their own transcription (Kume et al. 1999). In accordance with that, human Cry2 protein has been found in both the cytoplasm and the nuclei of the retinal ganglion cell layer (Thompson et al. 2003). The exclusively cytoplasmic location we observed with the mmCry2 antibody in the pigeon and the chicken retina might be explained by masking effects. For instance, the antigenic site of the Cry2 protein may be masked by Per when located in the nucleus as a complex. Mammalian Cry1 and Cry2, however, have been shown to interact with Per1 and Per2 proteins at their photolyase homology region (Miyazaki et al. 2001; Ozber et al. 2010; Tomita et al. 2010), so that an interaction with Per proteins should probably not hide the antigenic site of avian Cry2 either. The two Cry2 antibodies used in this study also labelled the photoreceptor inner segments in all three birds species examined. Since the inner segment serves as the major housekeeping compartment of the photoreceptor cell (Pearring et al. 2013), where protein biosynthesis takes place, the Cry2 antibodies might recognise the freshly synthesised Cry2 proteins there. Since our antibodies are directed against both isoforms of avian Cry2, however, we cannot make any statement about a possible diverging localisation of Cry2a and Cry2b proteins in the bird retina. We observed species- and antibody-specific differences, though: In the European robin and the chicken, erCry2-26E11 showed a stronger signal in the ellipsoid of the inner segments, which is rich in mitochondria in all vertebrate photoreceptors (Cohen 1972; Rodieck 1973; Stone et al. 2008). In contrast, more or less the entire inner segment was stained by erCry2-26E11 in the pigeon retina. The sequence that this erCry2-specific antibody was raised against is well conserved in pigeon and chicken Cry2 (two amino acid mismatches out of 14; see Fig. 1). We also confirmed the specificity of both antibodies in several control experiments, which makes us confident that these stainings are reliable. The antibody mmCry2-A20 also shows species-specific differences in the photoreceptor inner segment stainings, but these differed from the pattern obtained with the erCry2-26E11 antibody: In European robins, mmCry2-A20 labelled the myoid, where paraboloid, endoplasmic reticulum, Golgi complex and ribosomes are located (Cohen 1972), whereas in the pigeon and the chickens, the signal was restricted to the ellipsoid region. The species-specific difference of Cry2 location in European robin versus pigeon and chicken retina could mean that the Cry2 gene functions have diverged slightly between the (night-migratory) European robin and the (non-migratory) pigeon and chicken.
Their distribution within the cell nuclei of nearly every cell type of the retina could be a hint that the Cry2 proteins are involved in the circadian clock and would also speak against a role of Cry2 in avian magnetic compass orientation. Both proper fixation and alignment of the primary sensory protein(s) (which is hardly possible in the cell nuclei) are pre-requisites for light-dependent magnetoreception since this is needed for accurately detecting directional information (Soloy’yov et al. 2010; Hore and Mouritsen 2016; Hiscock et al. 2016; Worster et al. 2017). Furthermore, we would not expect a magnetoreceptive protein to be located within every retinal cell type. Sensory proteins are usually located in specialised sensory cells connected to the brain via a specific neuronal sensory pathway. However, since most proteins possess multiple functions (Jeffery 2020), we cannot entirely exclude that Cry2 acts as a magnetoreceptor in one cell type while it plays another role in other cell types. Moreover, since we could not distinguish between the two Cry2 isoforms with our antibodies, we cannot exclude that one of the isoforms is expressed exclusively in one specific cell type. Yet, currently available findings suggest that Cry2 is unable to bind FAD (Öztürk et al. 2009; Kutta et al. 2017), but FAD is essential for generating radical pairs and for any possible magnetic sensitivity of a Cry protein (Henbest et al. 2008; Maeda et al. 2012; Evans et al. 2013; Zollitsch et al. 2018; Xu et al. 2021). Together with recent independent reports that avian Cry2a/b mRNA displays a rhythmic circadian expression pattern characteristic of a circadian clock protein (Günther et al. 2018; Pinzon-Rodriguez et al. 2018; Hochstoeger et al. 2020), we conclude that the Cry2 proteins most likely act as core components of the circadian clock, not only in mammals but also in birds.
Availability of data and material
The erCry2a mRNA, complete cds, is available under the GenBank accession number MN656980.2.
Abbreviations
- cl:
-
Columba livia domestica (Homing pigeon)
- Cry:
-
Cryptochrome
- er:
-
Erithacus rubecula (European robin)
- FAD:
-
Flavin adenine dinucleotide
- gg:
-
Gallus gallus domesticus (Domestic chicken)
- mm:
-
Mus musculus (House mouse)
- NLS:
-
Nuclear localisation signal
- PB:
-
Phosphate buffer
- PBS:
-
Phosphate-buffered saline
- Per:
-
Period
- TTFL:
-
Transcription–translation feedback loop
References
Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. https://doi.org/10.1038/366162a0
Bailey MJ, Chong NW, Xiong J, Cassone VM (2002) Chickens’ Cry2: molecular analysis of an avian cryptochrome in retinal and pineal photoreceptors. FEBS Lett 513:169–174. https://doi.org/10.1016/s0014-5793(02)02276-7
Bian J, Wang Z, Dong Y, Cao J, Chen Y (2019) Effect of pinealectomy on the circadian clock of the chick retina under different monochromatic lights. Chronobiol Int 36:548–563. https://doi.org/10.1080/07420528.2019.1566740
Biskup T, Schleicher E, Okafuji A, Link G, Hitomi K, Getzoff ED, Weber S (2009) Direct observation of a photoinduced radical pair in a cryptochrome blue-light photoreceptor. Angew Chem Int Ed Engl 48:404–407. https://doi.org/10.1002/anie.200803102
Bolte P, Bleibaum F, Einwich A, Günther A, Liedvogel M, Heyers D, Depping A, Wöhlbrand L, Rabus R, Janssen-Bienhold U, Mouritsen H (2016) Localisation of the putative magnetoreceptive protein cryptochrome 1b in the retinae of migratory birds and homing pigeons. PLoS ONE 11:e0147819. https://doi.org/10.1371/journal.pone.0147819
Bolte P, Einwich A, Seth PK, Chetverikova R, Heyers D, Wojahn I, Janssen-Bienhold U, Feederle R, Hore PJ, Dedek K, Mouritsen H (2021) Cryptochrome 1a localisation in light- and dark-adapted retinae of several migratory and non-migratory bird species: no signs of light-dependent activation. Ethol Ecol Evol 33:248–272. https://doi.org/10.1080/03949370.2020.1870571
Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284:760–765. https://doi.org/10.1126/science.284.5415.760
Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364. https://doi.org/10.1146/annurev-arplant-042110-103759
Cohen AI (1972) Rods and cones. In: Fuortes MGF et al (eds) Physiology of photoreceptor organs. Handbook of Sensory Physiology, vol. 7/2. Springer, Berlin/Heidelberg, pp 63–110
Einwich A, Dedek K, Seth PK, Laubinger S, Mouritsen H (2020) A novel isoform of cryptochrome 4 (Cry4b) is expressed in the retina of a night-migratory songbird. Sci Rep 10:15794. https://doi.org/10.1038/s41598-020-72579-2
Engels S, Hein CM, Lefeldt N, Prior H, Mouritsen H (2012) Night-migratory songbirds possess a magnetic compass in both eyes. PLoS ONE 7:e43271. https://doi.org/10.1371/journal.pone.0043271
Evans EW, Dodson CA, Maeda K, Biskup T, Wedge CJ, Timmel CR (2013) Magnetic field effects in flavoproteins and related systems. Interface Focus 3:20130037. https://doi.org/10.1098/rsfs.2013.0037
Feng S, Stiller J, Deng Y, Armstrong J, Fang Q, Reeve AH et al (2020) Dense sampling of bird diversity increases power of comparative genomics. Nature 587:252–257. https://doi.org/10.1038/s41586-020-2873-9
Fu Z, Inaba M, Noguchi T, Kato H (2002) Molecular cloning and circadian regulation of cryptochrome genes in Japanese quail (Coturnix coturnix japonica). J Biol Rhythms 17:14–27. https://doi.org/10.1177/074873002129002302
Griffin EA Jr, Staknis D, Weitz CJ (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768–771. https://doi.org/10.1126/science.286.5440.768
Günther A, Einwich A, Sjulstok E, Feederle R, Bolte P, Koch KW, Solov’yov IA, Mouritsen H (2018) Double-cone localization and seasonal expression pattern suggest a role in magnetoreception for european robin cryptochrome 4. Curr Biol 28:211-223.e4. https://doi.org/10.1016/j.cub.2017.12.003
Günther A, Dedek K, Haverkamp S, Irsen S, Briggman KL, Mouritsen H (2021) Double cones and the diverse connectivity of photoreceptors and bipolar cells in an avian retina. J Neurosci 41:5015–5028. https://doi.org/10.1523/JNEUROSCI.2495-20.2021
Hein CM, Zapka M, Heyers D, Kutzschbauch S, Schneider NL, Mouritsen H (2010) Night-migratory garden warblers can orient with their magnetic compass using the left, the right or both eyes. J R Soc Interface 7(Suppl 2):S227-233. https://doi.org/10.1098/rsif.2009.0376.focus
Hein CM, Engels S, Kishkinev D, Mouritsen H (2011) Robins have a magnetic compass in both eyes. Nature 471:E11–12; discussion E12–13. https://doi.org/10.1038/nature09875
Henbest KB, Maeda K, Hore PJ, Joshi M, Bacher A, Bittl R, Weber S, Timmel CR, Schleicher E (2008) Magnetic-field effect on the photoactivation reaction of Escherichia coli DNA photolyase. Proc Natl Acad Sci U S A 105:14395–14399. https://doi.org/10.1073/pnas.0803620105
Heyers D, Manns M, Luksch H, Güntürkün O, Mouritsen H (2007) A visual pathway links brain structures active during magnetic compass orientation in migratory birds. PLoS ONE 2:e937. https://doi.org/10.1371/journal.pone.0000937
Hirayama J, Nakamura H, Ishikawa T, Kobayashi Y, Todo T (2003) Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J Biol Chem 278:35620–35628. https://doi.org/10.1074/jbc.M305028200
Hiscock HG, Worster S, Kattnig DR, Steers C, Jin Y, Manolopoulos DE, Mouritsen H, Hore PJ (2016) The quantum needle of the avian magnetic compass. Proc Natl Acad Sci USA 113:4634–4639. https://doi.org/10.1073/pnas.1600341113
Hochstoeger T, Al Said T, Maestre D, Walter F, Vilceanu A, Pedron M, Cushion TD, Snider W, Nimpf S, Nordmann GC, Landler L, Edelman N, Kruppa L, Dürnberger G, Mechtler K, Schuechner S, Ogris E, Malkemper EP, Weber S, Schleicher E, Keays DA (2020) The biophysical, molecular, and anatomical landscape of pigeon CRY4: a candidate light-based quantal magnetosensor. Sci Adv 6:eabb9110. https://doi.org/10.1126/sciadv.abb9110
Hore PJ, Mouritsen H (2016) The radical-pair mechanism of magnetoreception. Annu Rev Biophys 45:299–344. https://doi.org/10.1146/annurev-biophys-032116-094545
Jeffery CJ (2020) Enzymes, pseudoenzymes, and moonlighting proteins: diversity of function in protein superfamilies. FEBS J 287:4141–4149. https://doi.org/10.1111/febs.15446
Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Toh H, Fukuda I, Tsujimura T, Terada N, Kamei Y, Yuba S, Iwai S, Todo T (2000) Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells 5:725–738. https://doi.org/10.1046/j.1365-2443.2000.00364.x
Kubo Y, Akiyama M, Fukada Y, Okano T (2006) Molecular cloning, mRNA expression, and immunocytochemical localization of a putative blue-light photoreceptor CRY4 in the chicken pineal gland. J Neurochem 97:1155–1165. https://doi.org/10.1111/j.1471-4159.2006.03826.x
Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205. https://doi.org/10.1016/s0092-8674(00)81014-4
Kutta RJ, Archipowa N, Johannissen LO, Jones AR, Scrutton NS (2017) Vertebrate cryptochromes are vestigial flavoproteins. Sci Rep 7:44906. https://doi.org/10.1038/srep44906
Liedvogel M, Mouritsen H (2010) Cryptochromes − a potential magnetoreceptor: what do we know and what do we want to know? J R Soc Interface 7(Suppl 2):S147-162. https://doi.org/10.1098/rsif.2009.0411.focus
Lin C, Todo T (2005) The cryptochromes. Genome Biol 6:220. https://doi.org/10.1186/gb-2005-6-5-220
Liu X, Zhang Z, Ribelayga CP (2012) Heterogeneous expression of the core circadian clock proteins among neuronal cell types in mouse retina. PLoS ONE 7:e50602. https://doi.org/10.1371/journal.pone.0050602
Liu C, Hu J, Qu C, Wang L, Huang G, Niu P, Zhong Z, Hong F, Wang G, Postlethwait JH, Wang H (2015) Molecular evolution and functional divergence of zebrafish (Danio rerio) cryptochrome genes. Sci Rep 5:8113. https://doi.org/10.1038/srep08113
Maeda K, Henbest KB, Cintolesi F, Kuprov I, Rodgers CT, Liddell PA, Gust D, Timmel CR, Hore PJ (2008) Chemical compass model of avian magnetoreception. Nature 453:387–390. https://doi.org/10.1038/nature06834
Maeda K, Robinson AJ, Henbest KB, Hogben HJ, Biskup T, Ahmad M, Schleicher E, Weber S, Timmel CR, Hore PJ (2012) Magnetically sensitive light-induced reactions in cryptochrome are consistent with its proposed role as a magnetoreceptor. Proc Natl Acad Sci USA 109:4774–4779. https://doi.org/10.1073/pnas.1118959109
Miyamoto Y, Sancar A (1998) Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci USA 95:6097–6102. https://doi.org/10.1073/pnas.95.11.6097
Miyazaki K, Mesaki M, Ishida N (2001) Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol Cell Biol 21:6651–6659. https://doi.org/10.1128/mcb.21.19.6651-6659.2001
Möller A, Sagasser S, Wiltschko W, Schierwater B (2004) Retinal cryptochrome in a migratory passerine bird: a possible transducer for the avian magnetic compass. Naturwissenschaften 91:585–588. https://doi.org/10.1007/s00114-004-0578-9
Mouritsen H (2018) Long-distance navigation and magnetoreception in migratory animals. Nature 558:50–59. https://doi.org/10.1038/s41586-018-0176-1
Mouritsen H, Janssen-Bienhold U, Liedvogel M, Feenders G, Stalleicken J, Dirks P, Weiler R (2004) Cryptochromes and neuronal-activity markers colocalize in the retina of migratory birds during magnetic orientation. Proc Natl Acad Sci USA 101:14294–14299. https://doi.org/10.1073/pnas.0405968101
Mouritsen H, Heyers D, Güntürkün O (2016) The neural basis of long-distance navigation in birds. Annu Rev Physiol 78:133–154. https://doi.org/10.1146/annurev-physiol-021115-105054
Neuhoff V, Philipp K, Zimmer HG, Mesecke S (1979) A simple, versatile, sensitive and volume-independent method for quantitative protein determination which is independent of other external influences. Hoppe Seylers Z Physiol Chem 360:1657–1670. https://doi.org/10.1515/bchm2.1979.360.2.1657
Nießner C, Denzau S, Gross JC, Peichl L, Bischof HJ, Fleissner G, Wiltschko W, Wiltschko R (2011) Avian ultraviolet/violet cones identified as probable magnetoreceptors. PLoS ONE 6:e20091. https://doi.org/10.1371/journal.pone.0020091
Nießner C, Gross JC, Denzau S, Peichl L, Fleissner G, Wiltschko W, Wiltschko R (2016) Seasonally changing cryptochrome 1b expression in the retinal ganglion cells of a migrating passerine bird. PLoS ONE 11:e0150377. https://doi.org/10.1371/journal.pone.0150377
Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT (1999) Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286:2531–2534. https://doi.org/10.1126/science.286.5449.2531
Ozber N, Baris I, Tatlici G, Gur I, Kilinc S, Unal EB, Kavakli IH (2010) Identification of two amino acids in the C-terminal domain of mouse CRY2 essential for PER2 interaction. BMC Mol Biol 11:69. https://doi.org/10.1186/1471-2199-11-69
Ozturk N, Selby CP, Song SH, Ye R, Tan C, Kao YT, Zhong D, Sancar A (2009) Comparative photochemistry of animal type 1 and type 4 cryptochromes. Biochemistry 48:8585–8593. https://doi.org/10.1021/bi901043s
Pearring JN, Salinas RY, Baker SA, Arshavsky VY (2013) Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retin Eye Res 36:24–51. https://doi.org/10.1016/j.preteyeres.2013.03.002
Pinzon-Rodriguez A, Muheim R (2021) Cryptochrome expression in avian UV cones: revisiting the role of CRY1 as magnetoreceptor. Sci Rep 11:12683. https://doi.org/10.1038/s41598-021-92056-8
Pinzon-Rodriguez A, Bensch S, Muheim R (2018) Expression patterns of cryptochrome genes in avian retina suggest involvement of Cry4 in light-dependent magnetoreception. J R Soc Interface 15:20180058. https://doi.org/10.1098/rsif.2018.0058
Renthlei Z, Trivedi AK (2019) Effect of urban environment on pineal machinery and clock genes expression of tree sparrow (Passer montanus). Environ Pollut 255:113278. https://doi.org/10.1016/j.envpol.2019.113278
Ritz T, Adem S, Schulten K (2000) A model for photoreceptor-based magnetoreception in birds. Biophys J 78:707–718. https://doi.org/10.1016/S0006-3495(00)76629-X
Rodieck RW (1973) The vertebrate retina: Principles of structure and function. WH Freeman, San Francisco
Sancar A (1994) Structure and function of DNA photolyase. Biochemistry 33:2–9. https://doi.org/10.1021/bi00167a001
Sancar A (2000) Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem 69:31–67. https://doi.org/10.1146/annurev.biochem.69.1.31
Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237. https://doi.org/10.1021/cr0204348
Sancar A (2004) Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem 279:34079–34082. https://doi.org/10.1074/jbc.R400016200
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Schulten K, Swenberg CE, Weller A (1978) A biomagnetic sensory mechanism based on magnetic field modulated coherent electron spin motion. Z Phys Chem 111:1–5
Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A (2000) Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc Natl Acad Sci USA 97:14697–14702. https://doi.org/10.1073/pnas.260498597
Solov’yov IA, Mouritsen H, Schulten K (2010) Acuity of a cryptochrome and vision-based magnetoreception system in birds. Biophys J 99:40–49. https://doi.org/10.1016/j.bpj.2010.03.053
Stone J, van Driel D, Valter K, Rees S, Provis J (2008) The locations of mitochondria in mammalian photoreceptors: relation to retinal vasculature. Brain Res 1189:58–69. https://doi.org/10.1016/j.brainres.2007.10.083
Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18:164–179. https://doi.org/10.1038/nrg.2016.150
Takeuchi T, Kubo Y, Okano K, Okano T (2014) Identification and characterization of cryptochrome4 in the ovary of western clawed frog Xenopus tropicalis. Zoolog Sci 31:152–159. https://doi.org/10.2108/zsj.31.152
Thompson CL, Bowes Rickman C, Shaw SJ, Ebright JN, Kelly U, Sancar A, Rickman DW (2003) Expression of the blue-light receptor cryptochrome in the human retina. Invest Ophthalmol vis Sci 44:4515–4521. https://doi.org/10.1167/iovs.03-0303
Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A (1998) Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282:1490–1494. https://doi.org/10.1126/science.282.5393.1490
Tomita T, Miyazaki K, Onishi Y, Honda S, Ishida N, Oishi K (2010) Conserved amino acid residues in C-terminus of PERIOD 2 are involved in interaction with CRYPTOCHROME 1. Biochim Biophys Acta 1803:492–498. https://doi.org/10.1016/j.bbamcr.2010.01.012
Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, Yamamoto T (2016) A proposal for validation of antibodies. Nat Methods 13:823–827. https://doi.org/10.1038/nmeth.3995
van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627–630. https://doi.org/10.1038/19323
Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719–725. https://doi.org/10.1126/science.8171325
Wang X, Jing C, Selby CP, Chiou YY, Yang Y, Wu W, Sancar A, Wang J (2018) Comparative properties and functions of type 2 and type 4 pigeon cryptochromes. Cell Mol Life Sci 75:4629–4641. https://doi.org/10.1007/s00018-018-2920-y
Wiltschko W, Wiltschko R (1993) Navigation in birds and other animals. J Navigs 46:174–191
Wiltschko R, Stapput K, Thalau P, Wiltschko W (2010) Directional orientation of birds by the magnetic field under different light conditions. J R Soc Interface 7(Suppl 2):S163–S177. https://doi.org/10.1098/rsif.2009.0367.focus
Worster S, Mouritsen H, Hore PJ (2017) A light-dependent magnetoreception mechanism insensitive to light intensity and polarization. J R Soc Interface 14:20170405. https://doi.org/10.1098/rsif.2017.0405
Xu J, Jarocha LE, Zollitsch T, Konowalczyk M, Henbest KB, Richert S, Golesworthy MJ, Schmidt J, Déjean V, Sowood DJC, Bassetto M, Luo J, Walton JR, Fleming J, Wei Y, Pitcher TL, Moise G, Herrmann M, Yin H, Wu H, Bartölke R, Käsehagen SJ, Horst S, Dautaj G, Murton PDF, Gehrckens AS, Chelliah Y, Takahashi JS, Koch KW, Weber S, Solov’yov IA, Xie C, Mackenzie SR, Timmel CR, Mouritsen H, Hore PJ (2021) Magnetic sensitivity of cryptochrome 4 from a migratory songbird. Nature 594:535–540. https://doi.org/10.1038/s41586-021-03618-9
Yamamoto K, Okano T, Fukada Y (2001) Chicken pineal Cry genes: light-dependent up-regulation of cCry1 and cCry2 transcripts. Neurosci Lett 313:13–16. https://doi.org/10.1016/s0304-3940(01)02227-3
Yoshimura T, Suzuki Y, Makino E, Suzuki T, Kuroiwa A, Matsuda Y, Namikawa T, Ebihara S (2000) Molecular analysis of avian circadian clock genes. Brain Res Mol Brain Res 78:207–215. https://doi.org/10.1016/s0169-328x(00)00091-7
Zapka M, Heyers D, Hein CM, Engels S, Schneider NL, Hans J, Weiler S, Dreyer D, Kishkinev D, Wild JM, Mouritsen H (2009) Visual but not trigeminal mediation of magnetic compass information in a migratory bird. Nature 461:1274–1277. https://doi.org/10.1038/nature08528
Zapka M, Heyers D, Liedvogel M, Jarvis ED, Mouritsen H (2010) Night-time neuronal activation of Cluster N in a day- and night-migrating songbird. Eur J Neurosci 32:619–624. https://doi.org/10.1111/j.1460-9568.2010.07311.x
Zollitsch TM, Jarocha LE, Bialas C, Henbest KB, Kodali G, Dutton PL, Moser CC, Timmel CR, Hore PJ, Mackenzie SR (2018) Magnetically sensitive radical photochemistry of non-natural flavoproteins. J Am Chem Soc 140:8705–8713. https://doi.org/10.1021/jacs.8b03104
Zoltowski BD, Chelliah Y, Wickramaratne A, Jarocha L, Karki N, Xu W, Mouritsen H, Hore PJ, Hibbs RE, Green CB, Takahashi JS (2019) Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon. Proc Natl Acad Sci USA 116:19449–19457. https://doi.org/10.1073/pnas.1907875116
Acknowledgements
We thank Irina Fomins, Bettina Kewitz, Jessica Schmidt, Anja Günther, Florian Bleibaum, Sina Engler and Corinna Langebrake for excellent technical support. We acknowledge the Service Unit (Core Facility) Fluorescence Microscopy for the use of the imaging facilities. All animal procedures were performed in accordance with local and national guidelines for the use of animals in research and were approved by the governmental authorities (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit/LAVES, Oldenburg, Germany, Az: 33.9-42502-04-13/1263; 33.9-42502-04-12/0766; 33.9-42502-04-11/0423; 33.12-42502-04-10/0121).
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was funded by a Lichtenberg Professorship from the Volkswagen Stiftung to HM, by the Niedersächsisches Ministerium für Wissenschaft und Kultur to HM, by the Deutsche Forschungsgemeinschaft (RTG 1885/1-2: Molecular Basis of Sensory Biology to AE, PKS, KD and HM; MO 1408/1-2 to HM; SFB 1372: Magnetoreception and Navigation in Vertebrates to KD and HM), by a ERC Synergy Grant 810002 (‘Quantum Birds’) to HM, by DARPA (QuBE N66001-10-1-4061) to HM, and by the Air Force Office of Scientific Research (Air Force Materiel Command USAF award FA9550-14-1-0095 to HM).
Author information
Authors and Affiliations
Contributions
AE, PB, RB, KD and HM designed and supervised the experiments; AE, PKS and RB performed the experiments; AE, KD and HM analysed the data; RF generated monoclonal antibody; AE, PKS and KD prepared figures; AE and HM wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interests.
Code availability
Not applicable.
Ethics approval
All animal procedures were performed in accordance with local and national guidelines for the use of animals in research and were approved by the governmental authorities (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit/LAVES, Oldenburg, Germany, Az: 33.9-42502-04-13/1263; 33.9-42502-04-12/0766; 33.9-42502-04-11/0423; 33.12-42502-04-10/0121).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Einwich, A., Seth, P.K., Bartölke, R. et al. Localisation of cryptochrome 2 in the avian retina. J Comp Physiol A 208, 69–81 (2022). https://doi.org/10.1007/s00359-021-01506-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00359-021-01506-1