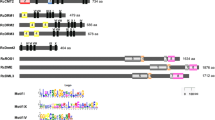
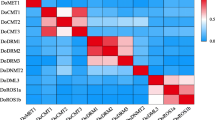
Abstract
DNA methylation is a well-recognized epigenetic modification linked to plant development, and it serves a crucial function in safeguarding the genome, governing gene expression, and enhancing stress resilience in plants. This mechanism is carried out by several conserved cytosine-5 DNA methyltransferases (C5-MTases). In the present study, a total of 10 C5-MTases were identified in rose. Our comprehensive examination, encompassing the study of conserved domains, gene structures, and phylogenetic analysis, led to the categorization of RcC5-MTases into four distinct subgroups: CMT, MET, DRM, and DNMT2. During the present study, numerous cis-elements that play roles in plant growth and development, phytohormone responsiveness, stress adaptation, and light sensitivity were also identified. A significant downregulation in the expression of RcC5-MTase genes (RcCMT1, RcCMT2, RcMET1, RcMET2, RcDRM1, and RcDRM3) were observed under HS, while the heat shock factor RcHsp17.8 was significantly upregulated, contributing to heat tolerance. RcCMT2 expression decreased by 9.8-fold at 6 h, 17.24-fold at 12 h, and 15.88-fold at 24 h under HS. Although RcDRM1 showed non-significant downregulation at 6 and 12 h, a significant decrease of 7.76-fold was observed at 24 h. RcDRM3 consistently showed significant downregulation at all time points, with the highest decrease of 21.79-fold observed at 24 h. Overall, the present investigation provides significant insights into the expression and function of rose C5-MTase-encoding genes. This information will be valuable for future experimental research on the processes of epigenetic control in rose.





Similar content being viewed by others
References
Ahmad F, Huang X, Lan HX, Huma T, Bao YM, Huang J, Zhang HS (2014) Comprehensive gene expression analysis of the DNA (cytosine-5) methyltransferase family in rice (Oryza sativa L.). Genet Mol Res 13:5159–5172
Arora H, Singh RK, Sharma S, Sharma N, Panchal A, Das T, Prasad A, Prasad M (2022) DNA methylation dynamics in response to abiotic and pathogen stress in plants. Plant Cell Rep 41:1931–1944
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:597–603
Ashapkin VV, Kutueva LI, Aleksandrushkina NI, Vanyushin BF (2020) Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int J MolSci 21:7457
Ashapkin VV, Kutueva LI, Vanyushin BF (2016) Plant DNA methyltransferase genes: multiplicity, expression, methylation patterns. Biochem (Mosc) 81:141–151
Babbar R, Tiwari LD, Mishra RC, Shimphrui R, Singh AA, Goyal I, Rana S, Kumar R, Sharma V, Tripathi G, Khungar L (2023) Arabidopsis plants overexpressing additional copies of heat shock protein Hsp101 showed high heat tolerance and endo-gene silencing. Plant Sci 330:111639
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:39–49
Baker NR (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59:89–113
Bartels A, Han Q, Nair P, Stacey L, Gaynier H, Mosley M, Huang QQ, Pearson JK, Hsieh TF, An YQ, Xiao W (2018) Dynamic DNA methylation in plant growth and development. Int J Mol Sci 19:2144
Baxter A, Mittler R, Suzuki N (2014) ROS as important players in plant stress signalling. J Exp Bot 65:1229–1240
Bhattarai K, Poudel MR (2022) DNA methylation and plants response to biotic and abiotic stress. Trends Sci 19:5102
Boyko A, Blevins T, Yao Y, Golubov A, Bilichak A, Ilnytskyy Y, Hollander J, Meins F Jr, Kovalchuk I (2010) Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5:e9514
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Dhar MK, Vishal P, Sharma R, Kaul S (2014) Epigenetic dynamics: role of epimarks and underlying machinery in plants exposed to abiotic stress. Int J Genom 2014:187146
Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, Hetzel J (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151:167–180
Dutta M, Raturi V, Gahlaut V, Kumar A, Sharma P, Verma V, Gupta VK, Sood S, Zinta G (2022) The interplay of DNA methyltransferases and demethylases with tuberization genes in potato (Solanum tuberosum L.) genotypes under high temperature. Front Plant Sci 13:933740
Finnegan EJ, Genger RK, Peacock WJ, Dennis ES (1998) DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol 49:223–247
Fujimoto R, Sasaki T, Nishio T (2006) Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet Syst 81:235–242
Gahlaut V, Samtani H, Khurana P (2020) Genome-wide identification and expression profiling of cytosine-5 DNA methyltransferases during drought and heat stress in wheat (Triticum aestivum). Genomics 112:4796–4807
Garg R, Kumari R, Tiwari S, Goyal S (2014) Genomic survey, gene expression analysis and structural modeling suggest diverse roles of DNA methyltransferases in legumes. PLoS ONE 9:e88947
Ge SX, Jung D, Yao R (2020) ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36:2628–2629
Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23:628–637
Gianoglio S, Moglia A, Acquadro A, Comino C, Portis E (2017) The genome-wide identification and transcriptional levels of DNA methyltransferases and demethylases in globe artichoke. PLoS ONE 12:e0181669
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH (2006) Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395–398
Gu T, Ren S, Wang Y, Han Y, Li Y (2016) Characterization of DNA methyltransferase and demethylase genes in Fragaria vesca. Mol Genet Genom 291:1333–1345
Han Y, Yu J, Zhao T, Cheng T, Wang J, Yang W, Pan H, Zhang Q (2019) Dissecting the genome wide evolution and function of R2R3-MYB transcription factor family in Rosa chinensis. Genes 10:823
Han B, Wu D, Zhang Y, Li DZ, Xu W, Liu A (2022) Epigenetic regulation of seed-specific gene expression by DNA methylation valleys in castor bean. BMC Biol 20(1):57
Henderson IR, Deleris A, Wong W, Zhong X, Chin HG, Horwitz GA, Kelly KA, Pradhan S, Jacobsen SE (2010) The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA–directed DNA methylation in Arabidopsis thaliana. PLoS Genet 6:e1001182
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinform 31:1296–1297
Jurkowski TP, Jeltsch A (2011) On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS ONE 6:e28104
Kanchiswamy CN, Muroi A, Maffei ME, Yoshioka H, Sawasaki T, Arimura GI (2010) Ca2+-dependent protein kinases and their substrate HsfB2a are differently involved in the heat response signaling pathway in Arabidopsis. Plant Biotechnol 27:469–473
Keller TE, Yi SV (2014) DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci USA 111:5932–5937
Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106:10171–10176
Kumar R, Chauhan PK, Khurana A (2016) Identification and expression profiling of DNA methyltransferases during development and stress conditions in solanaceae. Funct Integr Genomics 16:513–528
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547
Kumari P, Gahlaut V, Kaur E, Singh S, Kumar S, Jaiswal V (2023) Genome-wide identification of GRAS transcription factors and their potential roles in growth and development of rose (Rosa chinensis). J Plant Growth Regul 42:1505–1521
Lancíková V, Kačírová J, Hricová A (2023) Identification and gene expression analysis of cytosine-5 DNA methyltransferase and demethylase genes in Amaranthus cruentus L. under heavy metal stress. Front Plant Sci 13:1092067
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:293–296
Li J, Huang Q, Sun M, Zhang T, Li H, Chen B, Xu K, Gao G, Li F, Yan G, Qiao J (2016) Global DNA methylation variations after short-term heat shock treatment in cultured microspores of Brassica napus cv. Topas. Sci Rep 6:38401
Li J, Li C, Lu S (2018) Identification and characterization of the cytosine-5 DNA methyltransferase gene family in Salvia miltiorrhiza. PeerJ 6:e4461
Li ZQ, Xing W, Luo P, Zhang FJ, Jin XL, Zhang MH (2019) Comparative transcriptome analysis of Rosa chinensis ‘Slater’s crimson China’ provides insights into the crucial factors and signaling pathways in heat stress response. Plant Physiol Biochem 142:312–331
Li CH, Fang QX, Zhang WJ, Li YH, Zhang JZ, Chen S, Yin ZG, Li WJ, Liu WD, Yi Z, Mu ZS (2021) Genome-wide identification of the CCCH gene family in rose (Rosa chinensis Jacq.) reveals its potential functions. Biotechnol Biotechnol Equip 35:517–526
Li Z, Liu Q, Zhao K, Cao D, Cao Z, Zhao K, Ma Q, Zhai G, Hu S, Li Z, Wang K (2023) Dynamic DNA methylation modification in peanut seed development. iScience 26:107062
Lin W, Xiao XO, Sun W, Liu S, Wu Q, Yao Y, Zhang H, Zhang X (2022) Genome-wide identification and expression analysis of cytosine DNA methyltransferase genes related to somaclonal variation in pineapple (Ananas comosus L.). Agronomy 12:1039
Liu Z, Friesen T (2012) DAB staining and visualization of hydrogen peroxide in wheat leaves. Bio-Protoc 2:e309
Liu R, Lang Z (2020) The mechanism and function of active DNA demethylation in plants. J Integr Plant Biol 62:148–159
Liu G, Xia Y, Liu T, Dai S, Hou X (2018) The DNA methylome and association of differentially methylated regions with differential gene expression during heat stress in Brassica rapa. Int J Mol Sci 19:1414
Liu Z, Wu X, Liu H, Zhang M, Liao W (2022) DNA methylation in tomato fruit ripening. Physiol Plant 174:e13627
Liu P, Liu R, Xu Y, Zhang C, Niu Q, Lang Z (2023) DNA cytosine methylation dynamics and functional roles in horticultural crops. Hortic Res 10:170
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402-408
López ME, Roquis D, Becker C, Denoyes B, Bucher E (2022) DNA methylation dynamics during stress response in woodland strawberry (Fragaria vesca). Hortic Res 9:174
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:265–268
Marand AP, Eveland AL, Kaufmann K, Springer NM (2023) cis-Regulatory elements in plant development, adaptation, and evolution. Annu Rev Plant Biol 74:111–137
Min L, Li Y, Hu Q, Zhu L, Gao W, Wu Y, Ding Y, Liu S, Yang X, Zhang X (2014) Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol 164:1293–1308
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer EL, Tosatto SC, Paladin L, Raj S, Richardson LJ, Finn RD (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49:412–419
Moglia A, Gianoglio S, Acquadro A, Valentino D, Milani AM, Lanteri S, Comino C (2019) Identification of DNA methyltransferases and demethylases in Solanum melongena L., and their transcription dynamics during fruit development and after salt and drought stresses. PLoS ONE 14:e0223581
Noy-Malka C, Yaari R, Itzhaki R, Mosquna A, Auerbach Gershovitz N, Katz A, Ohad N (2014) A single CMT methyltransferase homolog is involved in CHG DNA methylation and development of Physcomitrella patens. Plant Mol Biol 84:719–735
Pavlopoulou A, Kossida S (2007) Plant cytosine-5 DNA methyltransferases: structure, function, and molecular evolution. Genomics 90:530–541
Pei L, Zhang L, Li J, Shen C, Qiu P, Tu L, Zhang X, Wang M (2019) Tracing the origin and evolution history of methylation-related genes in plants. BMC Plant Biol 19:307
Qian Y, Xi Y, Cheng B, Zhu S (2014) Genome-wide identification and expression profiling of DNA methyltransferase gene family in maize. Plant Cell Rep 33:1661–1672
Raymond O, Gouzy J, Just J, Badouin H, Verdenaud M, Lemainque A, Vergne P, Moja S, Choisne N, Pont C, Carrere S (2018) The Rosa genome provides new insights into the domestication of modern roses. Nat Genet 50:772–777
Saint-Oyant LH, Ruttink T, Hamama L, Kirov I, Lakhwani D, Zhou NN, Bourke PM, Daccord N, Leus L, Schulz D (2018) A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat Plants 4:473–484
Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28:231–234
Sharma R, Mohan Singh RK, Malik G, DeveshwarP TAK, Kapoor S, Kapoor M (2009) Rice cytosine DNA methyltransferases–gene expression profiling during reproductive development and abiotic stress. FEBS J 276:6301–6311
Sharma DK, Andersen SB, Ottosen CO, Rosenqvist E (2015) Wheat cultivars selected for high Fv /Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance, transpiration and dry matter. Physiol Plant 153:284–298
Su Y, Bai X, Yang W, Wang W, Chen Z, Ma J, Ma T (2018) Single-base-resolution methylomes of Populus euphratica reveal the association between DNA methylation and salt stress. Tree Genet Genomes 14:1–11
Sun M, Yang Z, Liu L, Duan L (2022) DNA methylation in plant responses and adaption to abiotic stresses. Int J Mol Sci 23:6910
Thiagarajan D, Dev RR, Khosla S (2011) The DNA methyltransferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics 1:103–113
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F (2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19:900–905
Victoria D, AlikiK VK, Georgios M, Zoe H (2018) Spatial and temporal expression of cytosine-5 DNA methyltransferase and DNA demethylase gene families of the Ricinus communis during seed development and drought stress. Plant Growth Regul 84:81–94
Wadhwa N, Kapoor S, Kapoor M (2024) Arabidopsis T-DNA mutants affected in TRDMT1/DNMT2 show differential protein synthesis and compromised stress tolerance. FEBS J 291:92–113
Wang P, Gao C, Bian X, Zhao S, Zhao C, Xia H, Song H, Hou L, Wan S, Wang X (2016) Genome-wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front Plant Sci 7:7
Wang Q, Qu Y, Yu Y, Mao X, Fu X (2023) Genome-wide identification and comparative analysis of DNA methyltransferase and demethylase gene families in two ploidy Cyclocaryapaliurus and their potential function in heterodichogamy. BMC Genomics 24:287
Weng L, Wang L, Tan M, Wang L, Zhao W, You J, Wang L, Yan X, Wang W (2023) The pattern of alternative splicing and DNA methylation alteration and their interaction in linseed (Linum usitatissimum L.) response to repeated drought stresses. Biol Res 56:12
Wickramasuriya AM, Sanahari WM, Weeraman JW, Karunarathne MA, Hendalage DP, Sandumina KH, Artigala AD (2024) DNA-(cytosine-C5) methyltransferases and demethylases in Theobroma cacao: insights into genomic features, phylogenetic relationships, and protein–protein interactions. Tree Genet Genomes 20:1–22
Wu Q, Huang Q, Guan H, Zhang X, Bao M, Bendahmane M, Fu X (2022) Comprehensive genome-wide analysis of histone acetylation genes in roses and expression analyses in response to heat stress. Genes 13:980
Yadav S, Meena S, Kalwan G, Jain PK (2024) DNA methylation: an emerging paradigm of gene regulation under drought stress in plants. Mol Biol Rep 51:1–6
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins Struct Funct Bioinform 64:643–651
Yu Z, Zhang G, Teixeira da Silva JA, Li M, Zhao C, He C, Si C, Zhang M, Duan J (2021) Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation. BMC Plant Biol 21:1–7
Zhang H, Lang Z, Zhu JK (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19:489–506
Zhang Y, He X, Zhao H, Xu W, Deng H, Wang H, Wang S, Su D, Zheng Z, Yang B, Grierson D (2020) Genome-Wide identification of DNA methylases and demethylases in Kiwifruit (Actinidia chinensis). Front Plant Sci 11:51499
Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y (2013) Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol 31:154–159
Zhu C, Zhang S, Zhou C, Chen L, Fu H, Li X, Lin Y, Lai Z, Guo Y (2020) Genome-wide investigation and transcriptional analysis of cytosine-5 DNA methyltransferase and DNA demethylase gene families in tea plant (Camellia sinensis) under abiotic stress and withering processing. Peer J 8:e8432
Acknowledgements
This study received financial support from the Council of Scientific and Industrial Research (CSIR) under grant MLP-201. V.J. also acknowledges the Science and Engineering Research Board (SERB) for the Early Career Research Award, and VJ and VG express gratitude to the Department of Science and Technology (DST) for the INSPIRE faculty award. HG and PK extend their appreciation to CSIR for the Senior Research Fellowship. This manuscript serves as CSIR-IHBT communication number 5437.
Funding
Funding for this research was provided by the CSIR, India through Grant MLP201, the Department of Science and Technology (DST) through the INSPIRE faculty award, and the Science and Engineering Research Board (SERB) via the Early Career Research Award.
Author information
Authors and Affiliations
Contributions
HG: Formal analysis, Data Curation, Writing-Original draft preparation. PK: Formal analysis, Data Curation, Writing-Original draft preparation. VG: Conceptualization, Methodology, Supervision, Writing-Reviewing and Editing. VJ: Conceptualization, Methodology, Funding acquisition, Writing-Reviewing and Editing, Supervision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Additional information
Handling Editor: Vinay Kumar.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gangwar, H., Kumari, P., Gahlaut, V. et al. Unveiling the Role of Cytosine-5 DNA Methyltransferase Under Heat Stress in Rose (Rosa chinensis). J Plant Growth Regul (2024). https://doi.org/10.1007/s00344-024-11316-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00344-024-11316-9