Abstract
Organic and natural sources of bio-stimulant have a great expectancy to boost green agriculture practices for sustainable, safe, and smart cultivation of crops. In that regard, beneficial endophytic bacteria have great potential. They have unique features in promoting plant growth by colonizing and establishing well in plant roots. In this study, endophytes isolated from the roots of moringa, neem, sesbania, and chilli were screened for crop’s growth-enhancing activities, such as phosphorus (P) solubilization, 1-aminocyclopropane-1-carboxylic-acid deaminase (ACC deaminase) production, and indole-acetic acid (IAA) production. The phosphorus solubilization, indole-acetic acid production, and ACC deaminase production values fall in the range of 55–88 ppm, 20–164 ppm, and 0.317–0.375 mM, respectively. Chilli seeds’ three highest vigor index (VI) values were attained by MR10 (12,457 VI), MR3 (9450 VI), and MR13 (8730 VI). MR13 showed the highest seed germination energy (221%), followed by MR1 (178%) and MR3 (156%). The promising endophytes were tested on chilli seedlings as single and mixed inoculum treatments to study the efficiency of root colonization. Mixed cultures containing CKR8 and MR13 exhibited the highest seedling height (17.0 cm), followed by MR13, MR10, and MR13 (16.8 cm) compared to the control (12.6 cm). A single culture of MR10 (109.0 g and 13.53 cm2) and a mixed culture of MR10 and MR13 (100.0 g and 13.09 cm2) showed the maximum root length and surface area, respectively. The highest relative chlorophyll content was recorded by MR10 and MR13 (40.3 SPAD value), followed by MR13, MR3, and CKR8 (36.8 SPAD value). The non-native endophytic bacteria, MR13, Streptomyces panaciradicis (GenBank accession no. OM001090), and MR3, Bacillus subtilis (GenBank accession no. OM714810), could colonize the roots and improve the growth of chilli at the seedling growth stage.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A plant’s nutrient use efficiency determines its health and quality. Nutrient imbalance in a plant makes it more vulnerable to pathogenic attacks (Marchiol et al. 2020). This may cause a reduction in productivity, low yield and quality, stunted growth, and susceptibility to plant diseases. To boost crop quality, satisfy the global demand for food, and minimize environmental degradation, new strategies have been explored for the sustainable cultivation of crops. One such strategy is the development of bio-stimulant to improve soil fertility and crop productivity (Matthews et al. 2022b). Plant bio-stimulant contains substances and/or microorganisms whose function, when applied to plants or the rhizosphere, is to stimulate natural processes to enhance nutrient uptake, nutrient efficiency, and tolerance to abiotic stresses and crop quality, which indirectly improves the physiological growth, yield, and quality of crops. Apart from that, bio-stimulant consisting of microorganisms could enhance soil fertility by dissolving insoluble nutrients in the soil and mobilizing nutrients to roots for efficient use by plants. It was previously suggested that the new frontier for plant bio-stimulant should include microorganisms and compounds known to develop natural plant–microbe interactions (Woo et al. 2018; Matthews et al. 2022a). It is generally agreed that microorganisms are the future hope to create a sustainable agriculture system. The application of beneficial microorganisms could pose many benefits, including reducing chemical use in arable soils and plants, lessening the cost of agriculture production by decreasing chemical input, increasing soil fertility and recycling of nutrients, and enhancing the food production system. Regulating hormonal and nutritional imbalances of plants, inducing resistance against plant pathogens, and solubilizing nutrients for plant uptake are a few major mechanisms adopted by plant growth-promoting rhizobacteria (PGPR) (Husseiny et al. 2021; Cochard et al. 2022). PGPR has been the general choice of beneficial microorganisms isolated from soil. Among the millions of PGPRs in soil, only a selected group of microorganisms known as endophytes can compete, invade plant roots, and colonize the interior parts of plant tissues successfully without causing any damage to the plants. Endophytes that thrive within the plant play vital roles, such as acquiring nutrients, reducing plant stress, inducing plant resistance, and suppressing plant diseases (Zhang et al. 2021). Endophytic bacteria form mutualistic relationships with host plants in the long term, and they can promote the growth and accumulation of effective components in host plants (Tao et al. 2022). They can promote plant growth and development at different growth stages, such as seed germination, vegetative growth, flowering, and fruiting. Interestingly, the plant immune system does not detect endophytes as a pathogen due to their low cell density compared to pathogenic bacteria (Afzal et al. 2019)
An efficient screening method is essential for isolating PGPR (Shi et al. 2022). Screening rhizobacteria with one or a few PGPR traits is a fundamental criterion for selecting bacteria (Raheem and Ali 2022). In vitro plant growth-promoting activities cannot influence plant growth under controlled conditions (Costa-Gutierrez et al. 2022). Although plant growth-promoting isolates are often selected based on in vitro tests, those do not necessarily correspond to the mechanisms which influence growth promotion in vivo (Bergottini et al. 2015). Mere primary screening of isolates for PGPR traits does not guarantee efficacious plant growth promotion under field conditions, as it is also possible that isolates that exhibit less in vitro growth-promoting activities might possess different plant growth-promotion strategies (Basu et al. 2021). Thus, it is crucial to characterize the plant growth-promoting traits by testing the potential bacteria in the actual plant at various growth stages. Different endophytic isolates with various plant growth-promoting characteristics can integrate and improve plant growth and health by employing diverse mechanisms within different periods of the plant’s life cycle.
Chilli has high economic value and great demand worldwide. Global chilli production is reported to increase by nearly 30% due to its increasing demand and wide utilization in cuisines, pharmaceuticals, cosmetics, and natural coloring agents. Although PGPR has been widely reported to increase crop yield and show enhanced growth on various plants, reports on successful cases of bio-formulation on chilli growth and yield are still limited. A study on the impacts of bio-stimulant during an entire cycle of chilli cultivation showed positive effects on plant growth, fruit quality, and chemical composition at flowering and maturity growth stages (Ertani et al. 2015). Green seaweed extracts have been reported to induce the germination of pepper seeds up to 68% (Chanthini et al. 2019). The inoculation of mineral-solubilizing and nitrogen-fixing bacterial strains resulted in increased growth and physiological parameters in chilli (Devi et al. 2022). In contrast, a report indicated that applying bio-fertilizer in various concentrations did not increase the average total chlorophyll content of curly red chilli plants (Siswanti et al. 2019). Thus, this study assessed the beneficial traits of endophytic bacteria and its indirect contribution to seed germination and seedling growth of chilli.
Plant roots and developmental stages influence the distribution of the rhizosphere microbiome. Thus, plant type is a stronger modulator of microbial richness than soil type (Igiehon and Babalola 2018). Different plant species have various root exudates, which can attract specific types of microbes to the rhizosphere by changing the physical–chemical conditions of the rhizosphere that promote the nutrient acquisition and attract specific beneficial microbial groups (Zhao et al. 2021). This can increase the microbial richness in the soil associated with that particular plant species. Furthermore, some plant species may also release antimicrobial compounds that can inhibit the growth of certain microbes, leading to a reduction in microbial richness. Therefore, the plant type can influence plant leaves, roots, and rhizosphere microbial richness. Consequently, it was expected that the root exudates from medicinal plants could attract novel beneficial bacteria to enter their roots. However, minimal reports have been published on the plant growth-promoting activities of microbial endophytes from medicinal plants (Hassan 2017). Thus, this study aims to isolate plant growth-promoting endophytic bacteria from medicinal plants, particularly their leaves and roots. It also aims to investigate non-native bacteria’s efficiency in chilli growth and development.
Materials and Methods
Plant Sample Collection
Three medicinal plants, Moringa oleifera, Sesbania grandiflora, and Azadirachta indica, and two local varieties of Capsicum annuum, namely Chilli Kulai (MC 11) and L5, were chosen for the sample collection of roots and leaves. The sampling was done from three matured plants/trees of each species/variety with three biological replications of leaves and roots. Leaves from three different branches and roots from three different root branches of a plant were pooled to make it one biological replication. The samples were collected in sterile plastic bags and brought to the lab for immediate analysis or kept at 4 °C overnight before analysis. The details of the sampling are described in Online Resource 1.
Isolation of Endophytic Bacteria from Roots and Leaves
Each plant species/variety consists of three pooled leaf and root samples. One gram of plant sample (leaf/root) was washed in sterile distilled water several times using a shaker until the water became clear. Later, it was washed with 70% ethanol for 5 min, followed by 1% sodium hypochlorite for 2 minutes and finally rinsed with sterile distilled water for 5 min × 4 times. A hundred microliters of the final rinse were inoculated onto triplicate plates of potato dextrose agar (PDA, Oxoid, UK) and tryptic soy agar (TSA, Oxoid, UK) to check the sterilization efficiency. Sterile mortar and pestle were cleaned with 70% ethanol before macerating plant samples with sterile phosphate buffer. Before macerating the plant sample, a small part of the plant tissue was grown on PDA and TSA in triplicates. The macerated plant sample was serially diluted in phosphate buffer until dilution 104. A hundred microliters of each diluent were inoculated onto duplicate plates of PDA and TSA. All the plates were incubated at room temperature for one week. Colonies with different morphologies were selected, purified, and stored at 4 °C. Colonies of identical morphologies from the same plant were discarded to reduce unnecessary repetition of work. Fungal colonies were disregarded in the isolation process.
Qualitative Screening of Plant Growth-promoting Traits in Endophytic Bacteria
Phosphorus solubilization was tested according to the method described by Ringuet et al. (2011) and Gupta and Pandey (2019). Potassium solubilization was done following the method described by Zhang and Kong (2014), and nitrogen fixation was tested according to Baldani et al. (2014). Endophytes were grown overnight in nutrient broth in falcon tubes at 30 °C. Fifty microliters of the culture were inoculated onto Pikovskaya’s agar (PVK) (HiMedia Laboratories, India), Aleksandrov’s agar (HiMedia Laboratories, India), and Asby’s glucose agar (HiMedia Laboratories, India). PVK contains calcium phosphate as the sole insoluble phosphorus source, while Aleksandrov’s agar contains potassium alumino silicate as the sole insoluble potassium source. Asby’s glucose agar does not contain any nitrogen source. The plates were incubated at 28 °C for 1 week. Colonies that produce clear zone were selected as potential endophytes with positive activity. Chitinase production was performed according to the method described by Saima et al. (2013). The hydrogen cyanide test was conducted according to the method (Passari et al. 2016) described. The siderophore production test was done according to the method by Ferreira et al. (2019). Antagonistic activity against Fusarium oxysporum and Colletotrichum sp. was screened using agar plate assays (Yasmin et al. 2020).
Quantitative Screening of Plant Growth-Promoting Traits by Endophytic Bacteria
Quantification analyses of ACC deaminase, indole-acetic acid production, and phosphorus solubilization were conducted for bacterial isolates that showed positive results in the respective qualitative assays.
Phosphorus Solubilization by Endophytic Bacteria via Molybdenum Blue Assay
The microplate colorimetric method was done to quantify orthophosphate or soluble phosphate that was solubilized from insoluble rock phosphates by endophytes. Murphy and Riley adopted this method from an established technique based on the molybdenum blue method (1962). Briefly, the pure culture of potential endophytes was inoculated in 50 mL of Pikovskaya’s broth supplemented with Christmas Island Rock Phosphate (CIRP) as the sole insoluble phosphorus source (Yeast extract, 0.5 g/L; dextrose, 10 g/L; CIRP, 5 g/L; ammonium sulfate, 0.5 g/L; potassium chloride, 0.2 g/L; magnesium sulfate, 0.1 g/L; manganese sulfate, 0.0001 g/L and ferrous sulfate, 0.0001 g/L) in conical flasks. The flasks were incubated in a shaking incubator for one week at room temperature. The samples were analyzed using the Molybdenum Blue Assay after 8 days of incubation. Samples were filtered using 0.22-µm syringe filters to remove particulates before analysis. The absorbance was quantified at 880 nm (UV–VIS Spectrophotometer, Model: Varioskan Flash Multimode Reader, Thermo Fisher Scientific, USA). Standards solutions with potassium phosphate monobasic (KH2PO4) concentrations ranging between 0 and 25 µM were made for the calibration curve.
Indole-Acetic Acid Production by Endophytic Bacteria via Salwoski’s Method
Indole-acetic acid assay was conducted based on the established colorimetric method by Naveed et al. (2013), Passari et al. (2016), and Chandra et al. (2018). Nutrient broth (15 mL) was prepared with 0.1% tryptophan. Potential endophyte isolate was cultured separately into the growth medium and incubated in a shaking incubator at 28 °C, 100 rpm for 48 h. After incubation, indole-acetic acid production was determined by Salwoski’s reagent. A series of standards within the range of 0 ppm to 200 ppm was prepared from a stock solution of 1000 µg/mL of pure indole-acetic acid in acetone in the same culture medium. A hundred microliters of samples and standards were transferred into a 96-well microplate followed by 200 µL of Salwoski reagent. All the microplates were incubated at RT for 25 minutes. The absorbance was read at 530 nm in a UV–Vis Spectrophotometer (Model: Varioskan Flash Multimode Reader, Thermo Fisher Scientific, USA).
ACC Deaminase Production by Endophytic Bacteria via α-Ketobutyrate Production
ACC deaminase production analysis was performed according to the method described by a few authors (Penrose and Glick 2003; Ali et al. 2014). The assay was based on the hydrolysis of ACC which produces α-ketobutyrate. Briefly, bacterial cells were suspended in 5-mL DF salts minimal medium added with 45 µL of 0.5-M ACC solution. After incubation for 24 h at 200 rpm and 30 °C and followed by centrifugation at 4000 rpm for 10 min at 4 °C, the cells were washed with 0.1-M Tris–HCL, pH 7.6. For ACC deaminase assay, bacterial cells were mixed with 1 mL of 0.1-M Tris–HCI, pH 7.6 and centrifuged at 16,000×g for 5 min and re-suspended in 600 µL of 0.1-M Tris–HCI, pH 8.5. Thirty microliters of toluene were added and vortexed, followed by 200 µL of toluenized cells that were mixed with 20 µL of 0.5-M ACC. The mixture was incubated at 30 °C for 15 min and 1 mL of 0.56-M HCI was added, vortexed and centrifuged for 5 min at 16,000×g. One milliliter of supernatant was mixed with 800 µL of 0.56-M HCI and vortexed before adding 300 µL of 2,4-dinitrophenylhydrazine reagent to the glass tube. It was vortexed and incubated at 30 °C for 30 min before mixing with 2 mL of 2-N NaOH. Finally, 300 µL of the sample was used to read absorbance at 540 nm in a 96-well microplate reader. The stock solution of α-ketobutyrate was prepared and diluted with Tris–HCl, 0.1 mol/L to make a 10-mmol/L solution just before use. The concentration of standard α-ketobutyrate ranged between 0.1 and 1 µmol with a total volume of 200 µL of each standard. Three hundred microliters of the 2,4-dinitrophenylhydrazine reagent were added and the contents were vortexed and incubated at 30°C for 30 min. At this time, the α-ketobutyrate is derivatized as phenylhydrazine. The color of phenylhydrazine is developed by the addition of 2 mL of 2-M NaOH. The solution is mixed before measuring the absorbance at 540 nm. A standard curve of absorbance was plotted against standard concentration. The standard curve was used to calculate the concentration of α-ketobutyrate produced by endophytic bacteria.
Preparation of Endophytic Bacterial Culture
Selected pure cultures of bacteria with desired characteristics were inoculated in TSB and incubated for 2 to 5 days at 90 rpm on a shaker incubator at room temperature. The culture was centrifuged at 4000 rpm for 15 minutes and the cells were washed with distilled water. It was then diluted 10 × for seed germination and seedling growth analysis.
Assessment on the Compatibility of the Selected Endophytic Bacteria
Each bacterial endophyte was streaked side by side and crossed with each other on a tryptic soy agar. The bacteria were incubated at room temperature for one week to observe for inhibition of bacterial growth. Incompatible bacteria will hinder the growth of the other bacteria. The compatible bacteria were cultivated in tryptic soy broth for 3 days and re-streaked on tryptic soy agar to check the survival of the bacterial cells in a mixed culture.
Priming of Chilli Seeds with Endophytic Bacteria
Chilli seeds (SAKATA 461) were soaked in 70% ethanol for 1 min, followed by soaking in 1% sodium hypochlorite for 10 min and finally rinsed with sterile distilled water 5 times. After the sterilization, seeds were primed in 10 mL of bacterial cells for 1 h (Roslan et al. 2020) in sealed conical flasks. The seeds were spread out on filter paper to blot out the treatment solution at room temperature for 30 min. Then, the seeds were placed on a petri dish containing sterile Whatman filter paper saturated with 2 mL of sterile distilled water. The Petri dishes were sealed with parafilm and covered with aluminum foil. The plates were kept at 25 ± 2 °C for 1 week. Fifty seeds with 3 replicates were used for each potential endophytic bacteria. Germination was recorded every day and the germination percentage, mean germination time (MGT), seed germination energy, and seed vigor index were estimated based on the formula below.
n = Number of newly germinated seeds at time T (25°C), T = Hours from the beginning of the germination test, ∑n = final germination
Seed germination was analyzed using binomial logistic regression in IBM SPSS Statistics 25. Binomial logistic regression estimates the probability of seed germination. The treatments for seed germination are described in Table 1.
Effect of Selected Endophytic Bacteria on the Vegetative Growth of Chilli Seedling
CKR8, MR3, MR10, and MR13 were selected for the subsequent study based on their potential to enhance seed germination. For the vegetative growth analysis, chilli seeds were germinated into a seedling tray filled up with peat moss in a dark room. It was allowed to grow for about one week or until 2–4 leaves emerged from the seedlings. Then, 2 mL of the treatment (1 × 108 cfu/mL) was applied to the peat moss. Then, the seedlings were transplanted into polybags and kept in the glass house. Each treatment was replicated to five. The treatments are described in Table 2. The growth parameters of the chilli seedlings were measured after 1 month.
Effect of Endophytic Bacteria on the Relative Chlorophyll Content of Chilli Seedlings
Relative chlorophyll content was measured on third, fully expanded leaves from the shoot apex for each treatment. It was measured using the SPAD-502 chlorophyll meter (SPAD 502, Minolta-Camera Co., Osaka Japan). The data points were recorded at five positions along the leaf blade and then the data points were averaged as a single value.
Effect of Endophytic Bacteria on the Length and Surface Area of Chilli Roots
WinRhizo root scanner (EPSON Flatbed Scanner EPSON Expression 11000XL) was used to capture the image of chilli roots. Chilli roots were washed thoroughly in tap water to remove the soil debris and placed in clean water to view under the root imaging system. The instrument automatically calculated the total root length and surface area.
Molecular Identification of the Selected Endophytic Bacteria
Selected endophytic bacteria based on seedling vegetative growth promotion were identified using 16S rRNA gene sequencing. Endophytic bacteria were grown on Luria Bertani agar (MERCK, USA) for a pure colony. One pure colony was picked to inoculate 5 mL of Luria Bertani broth (MERCK, USA) and incubated overnight at 30 °C. Genomic DNA was extracted using Presto Tm Mini gDNA Bacteria Kit (www.geneaid.com). The following primers were used for the amplification of 16 S ribosomal DNA. 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (3′-CTA CGG CTA CCT TGT TAC GA-5′) (Szymańska et al. 2016). Polymerase chain reaction (PCR) was carried out using MyTaqTM Red Mix (Bioline, UK) in 50-µL reaction mixture containing 25 µL of MyTaqTM Red Mix (Bioline, UK), 22 µL of molecular grade water, 1 µL of each primer (10 mM), and 1 µL of genomic DNA. The following thermal conditions were used: 95 °C for 2 min, followed by 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min in 30 cycles, and a final extension step at 72 °C for 5 min. PCR amplification products were purified using NucleoSpin Gel and PCR Cleanup kit (Macherey-Nagel, Germany) according to the manufacturer’s protocol and visualized on a 1% gel red using Syngene (USA). The fragments were outsourced for sequencing service using primers 27F and 1492R (Apical Scientific Sdn. Bhd, Serdang, Malaysia). The acquired sequence was analyzed using reference sequences deposited in the GenBank nucleotide database. The DNA sequences of endophytic bacteria were submitted to GenBank. The 16R rRNA gene sequence was compared with available gene sequences of endophytic bacteria in the National Center for Biotechnology Information (NCBI) GenBank using BLAST Search. A neighbor-joining phylogenetic tree was constructed using MEGA 12.0 with bootstrap analysis based on 1000 replicates.
Root Colonization Analysis by Endophytic Bacteria
Root Colonization by Single and Mixed Cultures of the Selected Endophytic Bacteria by Field Emission Scanning Electron Microscopy Analysis (FESEM)
Root colonization (RC) ability of the selected endophytic bacteria was assessed via an independent experiment. Chilli seeds were germinated and transplanted as described previously. The study was conducted in a glass house for one month after transplanting. Two non-pathogenic endophytic bacteria identified as Bacillus sp. (MR3) and Streptomycetes sp. (MR13) were used for the root colonization study. The bacterial inoculum was prepared as described in the methodology. The treatments were consisting of single and mixed cultures of Bacillus sp. (MR3) and Streptomycetes (MR13). For Streptomycetes, the inoculum was applied during two different growth stages, namely the vegetative stage and the sporulating stages. Streptomycetes undergo a complex developmental process in solid and liquid cultures. Therefore, the growing method and culture medium for Streptomycetes were evaluated in this study. The spores were collected using 0.1% tween 80 from mature colonies of Streptomycetes in solid agar plates, while pellets of hyphae were obtained from the liquid cultures. Fifty microliters of inoculum were applied to the seedlings twice a month after transplanting into polybags. Then, the roots were harvested for analysis. The treatments were as follows:
-
RC (a): Streptomycetes spores
-
RC (b): Streptomycetes mycelium
-
RC (c): Bacillus cells
-
RC (d): Streptomycetes spores and Bacillus cells
-
RC (e): Streptomycetes mycelium and Bacillus cells
-
RC (f): No inoculation (control)
For the observation under a scanning electron microscope, the roots were collected from the soil and washed several times to remove the soil debris. Then, root tips were cut into a bottle filled with sterile distilled water as a final washing step, followed by sonication for 2 min at 8 Hz at room temperature. Then, the roots were dried on tissue paper. Several roots were immersed in 3% glutaraldehyde buffered with 0.1-M phosphate buffer for 24 h. Later, the roots were rinsed 3 times with 0.1 M of phosphate buffer, pH 7.2 for about 10 min per wash. After the fixation process, the root samples were placed in 1% osmium tetroxide in 0.1-M phosphate buffer, pH 7.2 for about 2 h at room temperature in a loose capped bottle. Then, the washing step for 10 min was repeated three times using 0.1-M phosphate buffer, pH 7.2. For the dehydration process, a graded ethanol solution 30%, 50%, 70%, 80%, 90%, and 95% (v/v) were prepared and the roots were placed in each solution for 15 min. Later, the roots were transferred into 1:2 (HMDS:95% ethanol solution) and left for 20 min. This step was followed by immersing the root samples into 2:1 (HMDS:95% ethanol) and 100% HMDS for 20 min each. Finally, the sample was transferred to fresh HMDS and left in a fume hood overnight to evaporate HMDS. The next day, the samples were cut into tiny pieces with a length of 0.3–0.5 cm and placed on double-sided conductive carbon tape. The samples were sputtered with platinum for 30 s before viewing under a Field emission scanning electron microscope (FEI, Quanta 400F/Oxford FESEM).
Total Bacterial Population in Chilli Roots Inoculated with the Selected Endophytic Bacteria
The tap roots were separated from the main root system. Roots were washed several times in sterile water to remove the adhered soil. One gram of tap root was weighed into distilled water and sonicated for 10 min at 19 kHz in an ultrasonicator. The root solution was incubated overnight at 28 °C. The roots were homogenized in a pre-sterilized mortar and pestle before performing serial dilution until 106 and plated on Potato Dextrose Agar.
Statistical Analysis
The data were analyzed using the Turkey HSD test using IBM SPSS Statistics version 25 and 28. The mean values were significant at p˂ 0.05.
Results
Endophytic Bacteria Isolated from the Roots and Leaves of Moringa, Sesbania, Neem, and Chilli
Seventy bacterial endophytes were isolated from the roots and leaves of medicinal crops such as moringa (Moringa oleifera), neem (Azadirachta indica), vegetable hummingbird (Sesbania grandiflora), and healthy chilli plants (Capsicum annuum sp. MC11 and L5 varieties). The endophytes isolated from each part of the plant are detailed in Online Resource 2. The pure colonies were sub-cultured at least five times to ensure survival. The isolates that could not grow after a few sub-culturing were removed from the collection list.
Plant Growth-promoting Traits in Endophytic Bacteria Analyzed via Qualitative Methods
Potential plant growth-promoting endophytes with the ability of phosphorus solubilization, potassium solubilization, nitrogen fixation, siderophore production, ACC-deaminase production, phytohormone production, chitinase production, hydrogen cyanide production, and antagonistic activity against Fusarium oxysporum and Colletotrichum sp. were screened using qualitative agar medium assays. The qualitative assays were conducted to narrow down the number of potential endophytes that will be used to measure specific beneficial activities. In the first round of qualitative assays, endophytes that exhibited clear zones were designated as potential bacteria for a specific activity. The second round of screening was done among the bacteria with positive activities by measuring the diameter of clear zones produced by each endophytic bacteria for the relevant activity. The beneficial activities of all the isolated endophytes were described in Online Resource 3 and the pictures of beneficial traits were depicted in Online Resource 4. Endophytes isolated from sesbania and neem plants could only inhibit the growth of Fusarium sp. and Colletotrichum sp., producing siderophore, chitinase, and ACC deaminase. These beneficial traits were more associated with bio-pesticides and bio-control properties, whereas endophytes from moringa and chilli have both bio-control, bio-fertilizer, and bio-stimulating properties.
In the preliminary assay, Aleksandrov’s medium consisting of potassium aluminium silicate, a complex potassium source was used to select potassium-solubilizing endophytes. Only 19 isolates out of 70 bacteria were able to solubilize insoluble potassium in an agar medium. MR14 (3.3 cm), CKL6 (3.3 cm), CL1 (3.0 cm), and CKR6 (3.0 cm) exhibited higher activity among the potential bacteria for potassium solubilization (Online Resource 3).The clear zone around the colonies indicates potassium solubilization due to organic acid secretion (Online Resource 4b).
In the current study, Asby’s glucose agar medium was used to isolate non-symbiotic nitrogen-fixing endophytes. This medium does not contain a nitrogen source. Endophytes that grow in this medium are expected to fix nitrogen in the air for its growth on the agar plate (Online Resource 4c). About 14 endophytes isolated from chilli and moringa plants were considered potential nitrogen-fixing endophytes. Among these 14 bacteria, MR4, CKL6, and CKR6 showed larger clear zones in an N-free medium, accounting for 3.2 cm, 3.4 cm, and 3.2 cm, respectively (Online Resource 3).
Twenty-nine endophytic bacteria were identified as hydroxamate siderophore producers. Hydroxamate siderophore production is indicated by the formation of orange color colonies on agar blue-dyed CAS medium as a result of iron removal. The diameter of the orange zone produced by the potential endophytic bacteria ranged from 0.7 to 2.0 cm. CKL7, CKR8, MR5 and MR14 were selected for the 96-well microplate assay and it was found that CKL7 produced the highest concentration of siderophore which was reflected by the highest intensity of orange color production (Online Resources 3 and 4d).
Chitinolytic endophytes can be detected by the culture plate’s clearing zone containing chitin. In this study, five isolates from moringa root, three isolates from sesbania (leaf and root), three isolates from chilli (leaf and root), and one isolate from neem root were found to have the ability to produce chitinases as observed by the clearing zone in chitin agar medium (Online Resources 3 and 4e).
In this study, only one bacterial isolate, MR3 was able to change the color of filter paper from yellow to orange which indicates the presence of hydrogen cyanide (Online Resource 3 and 4f).
Bio-control Activity of Endophytic Bacteria Against Colletotrichum sp. and Fusarium oxysporum Using Dual-Culture Assay
All 70 isolates were tested for the antagonistic ability against Colletotrichum sp. and Fusarium oxysporum using a dual-culture agar plate assay. The antagonistic endophytes could produce metabolites that inhibit the growth of fungal pathogens. Control plates grown with the pathogen show full growth in 5–7 days, whereas antagonistic endophytes hinder the growth of fungal mycelium. About 13 endophytes out of 70 isolates have shown inhibition of Colletotrichum sp. in the 45–53% range. Out of 70 endophytes, 25 isolates showed inhibition percentage within the range of 10–63% for Fusarium oxysporum, soil-borne fungi which cause chilli wilt disease (Twenty-nine endophytic bacteria were identified as hydroxamate siderophore producers. Hydroxamate siderophore production is indicated by the formation of orange color colonies on agar blue dyed CAS medium as a result of iron removal. The diameter of the orange zone produced by the potential endophytic bacteria ranged from 0.7 to 2.0 cm. CKL7, CKR8, MR5, and MR14 were selected for the 96-well microplate assay, and it was found that CKL7 produced the highest concentration of siderophore which was reflected by the highest intensity of orange color production (Online Resource 3 and g, h).
Beneficial Traits in Endophytic Bacteria Analyzed via Quantitative Methods
Phosphorus Solubilization by Endophytic Bacteria Using Molybdenum Blue Assay
Potential phosphorus solubilizing endophytes were identified when a clear zone was observed around the colonies in the selective agar plate assay containing insoluble phosphorus (Online Resource 4a). The majority of the bacteria were able to show phosphorus solubilization activity in an agar medium. However, only 14 out of 70 endophytic bacteria were selected for further analysis as the size of the clear zone of all the potential phosphorus-solubilizing bacteria was similar. Among the 14 bacteria, only CKR8 and MR13 showed large clear zones, 2.1 cm, and 1.5 cm, respectively, and the rest exhibited similar sizes of clear zones (Online Resource 3). The standard graph was plotted for orthophosphate to determine the P solubilization activity of endophytic bacterial samples (Online Resource 5). The absorbance reading shows a linear correlation against the concentration of orthophosphate in the solution that was solubilized by endophytes from an insoluble source of phosphate. When tested in molybdenum blue assays, MR13 could produce 4.93 ppm of soluble P followed by 3.74 ppm and 0.19 ppm of soluble P in a control medium without bacteria (Figure 1).
Indole-Acetic Acid Production by Endophytic Bacteria via Salwoski’s Method
The results revealed that ML1and MR10 could produce 164.6 ± 21.3 ppm and 87.6 ± 0.29 ppm of indole-acetic acid, respectively (Fig. 2). Standard graph was plotted for indole-acetic acid to determine the IAA activity of endophytic bacterial samples (Online Resource 6).
ACC Deaminase Production by Endophytic Bacteria via α-Ketobutyrate Production
In this study, all isolates of endophytes were screened for ACC deaminase production using 96-well microplates. The absorbance directly correlates with the concentration of ACC deaminase enzyme. The absorbance directly correlates with the concentration of ACC deaminase enzyme. The color intensity was observed to increase in the microplates as the concentration of standard α-ketobutyrate increased. The standard graph was plotted for α-ketoglutaric (to determine the ACC deaminase activity of endophytic bacterial samples (Online Resource 7) and used to calculate the ACC deaminase concentration produced by the endophytic bacteria. Nineteen isolates produced ACC deaminase; most were isolated from chilli roots, followed by moringa roots. Among them, MR1, SL3, MR4, and MR14 were selected based on yellow color production in the 96-well microplate and absorbance reading. It was found that MR1 produces the highest amount of α-ketobutyrate, 0.375 mM (Fig. 3).
Assessment of the Compatibility Between the Selected Endophytic Bacteria
Among all these 70 endophytic bacteria, 7 bacteria were selected for future experiments. The selection was made following the criteria listed below.
-
(i)
The endophytic bacteria exhibited greater activity based on the diameter of clear zones for qualitative assays.
-
(ii)
Highest solubilization of phosphorus, production of indole acetic-acid and ACC deaminase in the quantitative assays.
The selected 7 endophytic bacteria exhibited compatibility in this experiment. An example of the compatibility test between two bacteria was shown in Fig. 4. No bacteria were found inhibiting the growth of bacteria grown next to it. It was found that all the tested bacteria may co-exist in a mixed culture without harming each other.
Effect of Endophytic Bacterial Treatment on Chilli Seed Germination
The germination percentage was 100% for chilli seeds treated with endophytic bacteria. A binary logistic regression was performed to ascertain the effects of endophytic bacteria, on the germination of chilli seeds. The logistic regression model was not statistically significant, χ2 (1) = 2.809, p = 0.95. The model explained between 0.7% (Cox & Snell R2) to 10% (Nagelkerke R2) of the variance in seed germination and correctly classified 73.4% of cases. Sensitivity was 100%, specificity was 0 h%, positive predictive value was 98.3% and negative predictive value was 0% (Table 3). Mean germination time demonstrates the rate and the time taken for germination while germination energy corresponds to the speed and uniformity of seedling emergence. The lower the mean germination time, the earlier the seeds could germinate. In this study, MR13 and distilled water exhibited the lowest mean germination time (Fig. 5), followed by CKR8, MR3, and MR1; meanwhile, MR13 exhibited the highest germination energy (221%), followed by distilled water (212%), MR1 (178%), MR3 (156%), CKR8 (153%), and MR10 (133%) (Fig. 6). Based on these observations, MR13, MR3, MR10, and CKR8 were selected for the seedling growth analysis. The vigor index (VI) is a measure of indication of the total performance of the seed. Taking into consideration the root and shoot length of the seedlings, MR10 showed the highest vigor index (12 457 VI) followed by MR3 (9450 VI), MR13 (8730 VI), and CKR8 (7120 VI) (Not shown in graph or table). Seed germination analysis also indicates the toxicity effect of endophytes on chilli. It was revealed that the endophytes were not harmful but helped the early germination of seeds and could enhance the overall seed quality.
Graph depicts the germination energy (%) of chilli seeds treated with potential endophytic bacteria. Each value is the mean of four replicates (n = 4), and bars sharing the same letters are not significantly different (p ≤ 0.05) according to Tukey’s HSD. The vertical bar indicates the standard error
Effect of the Selected Endophytic Bacterial Treatment on the Vegetative Growth of Chilli Seedlings
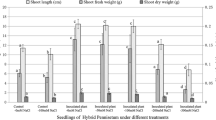
Vegetative growth analysis such as seedlings’ height, root length, root surface area, and relative chlorophyll content was done to select the most efficient bacteria to enhance the vegetative growth of chilli (Table 4). Based on the seed germination analysis, T1(MR13), T2 (CKR8), T3 (MR10), and T4 (MR3) were selected for single inoculant treatment and mixed bacterial culture treatment to determine the efficiency of chilli seedlings’ development. SPAD meter readings revealed that treatment with T11 (MR10 + MR13) resulted in high related chlorophyll readings. The chlorophyll content in a plant determines the photosynthetic capacity, nitrogen level of leaves, and general plant health. Relative chlorophyll content can be measured using a non-destructive method with a SPAD meter. The relative SPAD meter values are proportional to the chlorophyll content of the leaves. All the endophytic bacterial treatments have increased the height of chilli seedlings significantly as compared to the control. The highest height of chilli seedlings was recorded by T8 (CKR8 + MR13) followed by T5 (MR13 + MR10 + MR3) and T6 (MR13 + MR3 + CKR8). Seedling height is a direct parameter that demonstrates the efficiency of bacterial inoculants in promoting plant growth.
The total root length and root surface area of chilli seedlings demonstrated a significant increase in chilli seedlings inoculated with T3 (MR10). The rest of the treatments were not different significantly. T7 (MR13 + MR10 + CKR8) has caused root length and surface area reduction which is lower than the control. The root images depict the effect of the endophytes inoculation on root growth (Fig. 7). It is evident from the root images that inoculation with mixed bacterial cultures shows the development of thick root collar stem and thick roots with high density (Fig. 7g–l) as compared to single bacterial culture and control (Fig. 7a–f).
Molecular Identification of the Selected Endophytic Bacteria and Phylogenetic Analysis
BLAST search revealed the identification of the endophytic bacteria as described in Online Resource 8. Based on 16SrRNA sequence analysis, the endophytic bacterial isolates were classified as the genus Proteus, Enterobacter, Bacillus, and Streptomyces. Due to the ambiguous classification of human pathogenic bacteria and plant growth-promoting bacteria, only MR13 and MR3 could be recommended as potential bio-inoculants for the development of chilli bio-stimulant. The evolutionary history for MR3 and MR13 was determined using the Neighbour-Joining method (Fig. 8). The phylogenetic analysis also confirmed that MR3 closely related to Bacillus sp., while MR 13 is closely related to Streptomyces sp. The DNA sequences were deposited in the NCBI database under the accession numbers OM714810 and OM001090. The published sequences in the National Center for Biotechnology Information (NCBI) can be found at https://www.ncbi.nlm.nih.gov/nuccore/OM714810; https://www.ncbi.nlm.nih.gov/nuccore/OM001090.
Chilli Root Colonization by Single and Mixed Cultures of the Selected Endophytic Bacteria by FESEM
A total plate count was done for all the root samples and the results indicated that the bacterial population was in the range of 108 and 1010 in all the inoculated chilli roots. Non-inoculated chilli roots also exhibited bacterial count at a lower dilution of 106. However, FESEM analysis revealed that non-inoculated roots do not show bacterial colonization in the interior part of the root tips (Fig. 9). Total plate count results indicated that there are some indigenous microorganisms at the exterior part of the root that has dislodged from the root surface during the sonication process. Thus, total plate count analysis alone could not determine bacterial colonization. The slight increase in the inoculated roots is a good indication of a successful colonization process. FESEM images clearly show the colonization of endophytic bacteria in the interior part of the root tips. Since both vegetative and mature growth stages of Streptomyces were used as the inoculum, the mycelium and the spores were spotted in the tap root. FESEM results show that single culture was well established in chilli roots (Fig. 9a–c), but mixed culture colonization was dominated by Streptomyces sp. (Fig. 9d, e).
a Streptomyces spores’ colonization in chilli roots; b Streptomyces mycelium colonization in chilli roots; c Bacillus cells colonization in chilli roots; d mixed culture (Streptomyces spores and Bacillus cells) colonization in chilli roots; e Mixed culture (Streptomyces mycelium and Bacillus cells) colonization in chilli roots; f non-inoculated chilli roots (control)
Discussion
Endophytes are microorganisms that reside within the plant tissues and can benefit their hosts. They could directly or indirectly benefit the plant by increasing the availability of nutrients, suppressing plant pathogens, producing phytohormones, aiding in phytoremediation, and stimulating stress resistance in host plants (Husseiny et al. 2021; Chouhan et al. 2022). Endophytes are generally considered beneficial microorganisms for crops as they thrive within plant cells without causing damage to the plants and establish a community with other beneficial microbes by secreting various bioactive metabolites (Hassan 2017; Chouhan et al. 2022). Although many studies have demonstrated plant growth promotion by endophytes isolated from the same plants, some endophytes were also reported to promote growth in diverse host plants (Afzal et al. 2019). A similar finding was observed in the current study, where non-native bacteria from moringa roots exhibited beneficial effects on the seed germination and vegetative growth of chilli seedlings. It is also reported that plant growth-promoting bacterial endophytes may demonstrate their beneficial activity more efficiently and directly in the host as compared to rhizobacteria, as a lower concentration of metabolites secreted by the bacterial endophytes may exert a greater effect on the plant (Orozco-Mosqueda et al. 2018). Therefore, these bacteria are expected to be more successful in plant growth promotion as they can establish close contact with the host plant (Eid et al. 2021). Only root and leaf endophytes were focused on in this study as the diversity of endophytes in the roots is much higher than in other tissues (Lin et al. 2022). It was reported that leaf surfaces produce exudates that attract microorganisms (Hardoim et al. 2015). This explains the presence of bacterial endophytes in the plant leaves. Additionally, the composition of endophytic populations in leaves and roots can be related to the physical structure, chemical composition, and nutrient conditions of the root and leaves of medicinal and chilli plants. Bacterial endophytes from the rhizosphere colonize root hairs due to the secretion of root exudates and rhizodeposits. This statement could be corroborated by the current study, where different plants harbor different genera of bacteria with distinct abundance. Moringa roots, chilli roots, and chilli leaves showed diverse bacterial isolates with different morphology of colonies. Bacterial isolates from Moringa oleifera have multiple plant growth-promoting characteristics. Moringa is an edible, drought-tolerant wild plant famous for its medicinal and nutritional values. Due to the enormous benefits of the moringa plant, the endophytes that thrive in this plant could exhibit multiple beneficial traits for plants too. The moringa plant’s abundant micronutrients, growth, and health-benefitting compounds have attracted novel and useful endophytes to moringa roots and leaves.
This study noticed bacteria with similar colony morphologies among plant species or parts of the same plant species. This shows that there is a possibility that the same genus of bacteria is distributed in various crops through natural selection. Bacteria with similar morphology in different parts of the same crop species show that endophytes from roots could have traveled to the leaves via a specific colonization route. (Hardoim et al. 2015) stated that the xylem vascular system is the main transport route for the systemic colonization of internal plant compartments. Endophyte colonization systematically travels from roots to shoots to flowers, fruits, and seeds (Chouhan et al. 2022). In contrast, other endophytes colonize the intercellular spaces of plant cells. While this explains the similar microbial genus in different parts of a plant, microbial taxonomic distinction describes the distribution of diverse microbiomes among bacteria associated with a leguminous plant’s stem, leaves, and nodules (Igiehon and Babalola 2018). The nature of the root exudates recruits specific endophytes to live in the host plant. Thus, different plants directly select the type of endophytes associated with them.
In general, bacterial endophytes, MR3 and MR13 that attributed beneficial characteristics could enhance the root colonization and growth of chilli. MR3, Bacillus sp., could produce chitinases, siderophores, ACC deaminase, and hydrogen peroxides, whereas MR13, Streptomycetes sp., could solubilize insoluble phosphorus and potassium; inhibit the growth of Fusarium sp. (pathogen) along with the production of indole acetic acid, siderophores, chitinases, and ACC deaminase. Current findings also confirmed that endophytic bacteria of different host species could benefit the same or different species of test crop. A broad range of hosts for endophytes makes them promising agents as bio-inoculants (Afzal et al. 2019). In the research conducted by Khan et al. (2016), endophytes isolated from Moringa peregrine were found to promote tomato seedlings’ development significantly. Most endophytic bacteria are also rhizospheric bacteria and, thus, could be found in most plant species, especially at the root tips. The endophytic bacteria residing at the interior part of root cells could be unique species, genus, or strain. A wider range of hosts is vital to consider as bio-inoculant for possible agronomic use. This study proves the concept that non-native endophytic bacteria could exert beneficial characteristics of plant growth.
Qualitative analyses exhibited the ability of the endophytic bacteria isolated from medicinal and chilli plants to fix nitrogen, solubilize P and K, secrete phytohormones, siderophores, chitinases, and hydrogen cyanide. Some free-living nitrogen-fixing bacteria can also fix the atmospheric nitrogen into ammonia, making it accessible for plant uptake (Rilling et al. 2018). The nitrogenase enzyme system mediates this conversion. The ability of the nitrogen-fixing bacterial application to stimulate fruit set and fruit development of sweet cherry was proved by a group of researchers (Ahmadi-Rad et al. 2016). Likewise, researchers have isolated endophytes from sugarcane stalks with high nitrogenase activity (Xing et al. 2016). Potassium-solubilizing microorganisms can dissolve K-bearing minerals in soil by secreting organic acids, similar to P-solubilizing microorganisms (Bagyalakshmi et al. 2017). Plants absorb potassium in soluble form, whereas 90–98% of total K in the soil is in unavailable forms, such as feldspar and mica (Meena et al. 2016). Potassium-solubilizing bacteria were used as efficient bio-inoculants for enhancing K solubility in soil (Saha et al. 2016).
Iron is an essential micronutrient for plant development, and it is required by several metalloenzymes involved in photosynthesis and respiration (Ferreira et al. 2019). However, iron has low solubility in its predominant form, Fe3+, in the aerobic environment and forms insoluble hydroxides and oxyhydroxides that plants and microbes do not assimilate. Microbes produce siderophores, low molecular-weight iron chelators that can bind Fe3+ with high affinity and are water soluble (Sinha 2019). When siderophores bind to iron, a complex is formed, which is then absorbed by the microbial cell through specific transport systems. This criterion allows microorganisms to overcome the low solubility of iron in their environment and acquire the necessary iron for their growth and survival (Rana et al. 2020). MR3 and MR13 could successfully colonize chili roots in this study and produce hydroxyamate type of siderophores.
Phosphorus (P) is a major limiting nutrient in arable soils. Many mineral fertilizers applied to soil become unavailable rapidly due to precipitation and fixation with ferric and aluminum ions in acidic soils and calcium ions in alkaline soils. Only 0.1% of the total P in soil is estimated to be available for plant assimilation (Rezakhani et al. 2019). Thus, most of the arable soils are rich with fixed and unavailable P that could be solubilized or released with the help of soil microorganisms. Phosphate-solubilizing endophytic bacteria are p bio-stimulants due to their ability to increase P’s bioavailability, soil fertility, and crop productivity. Microbial phosphate solubilization is an important bio-stimulant trait via various mechanisms such as phytase, phosphatase, organic acids, and siderophore production (De Zutter et al. 2021). Phosphorus-solubilizing bacteria were also suggested as a substitute for mineral phosphate fertilizers to meet the plant’s P demand (Kalayu 2019). In the Molybdenum blue assay, orthophosphate reacts with ammonium molybdate and potassium antimonyl tartrate in an acid medium to form phosphomolybdenum that will be reduced to molybdenum blue by ascorbic acid. The microplate reader-based colorimetric method is reported to be sensitive, simple, and rapid for quantifying orthophosphates (Khati et al. 2018). Previous studies have corroborated the role of organic acids in facilitating phosphorus solubilization (Wei et al. 2018; Mendes et al. 2020). Microorganisms could solubilize unavailable P via a ligand exchange process whereby the hydroxyl and carboxyl groups of organic acids produced by microorganisms chelate the cations bound to P and exchange with P in soil (Tian et al. 2021).
Indole acetic acid, one of the most common phytohormones associated with plant growth, plays an essential role in leaf formation, embryo development, root initiation and development, fruit development, etc. (Chandra et al. 2018). IAA facilitates root elongation and formation of lateral roots, which indirectly enhance the efficiency of water and nutrient uptake by plant root systems (Gupta and Pandey 2019). l-Tryptophan is the direct precursor for IAA synthesis via the indole-3-acetamide pathway. In plants, root exudates secrete l-Tryptophan for rhizospheric microorganisms to enhance IAA biosynthesis in the rhizosphere (Upadhyay et al. 2022). Phytohormone production by endophytes is a typical trait for root-associated endophytes (Hardoim et al. 2015). IAA is the first auxin group of growth stimulants identified to play a central role in plant growth (Pham et al. 2022). Salkowski’s method determined the concentration of IAA produced by endophytic bacteria. In the Salkowski method, the reaction between IAA and sulfuric acid generates a mixture of products that includes indole, which reacts with the ferric ions in the Salicylic acid reagent to produce a pink-to-red color, indicating the presence of IAA. The intensity of the color is directly proportional to the concentration of IAA in the sample.
In addition to these beneficial traits, plant growth-promoting bacteria also employ several mechanisms to alleviate plant stress, with one primary strategy being the reduction of ethylene levels through the hydrolysis of 1-aminocyclopropane-1-carboxylic acid (ACC) by the enzyme ACC deaminase. ACC is a precursor of the plant hormone ethylene. Certain strains of PGPR have been reported to possess ACC deaminase, which can break down ACC into ammonia and α-ketobutyrate, decreasing the level of ethylene within the plant (Chandwani and Amaresan 2022). ACC deaminase production indicates the stress tolerance traits in endophytes that could make it a potential candidate for bio-stimulant.
The bio-control activity of endophytic bacteria could be mediated by various mechanisms, such as the production of hydrogen cyanide, enzymes (chitinases, cellulases, proteases), and antibiotics. Hydrogen cyanide is a secondary metabolite that could inhibit cytochrome C oxidase and other metalloenzymes in microorganisms. Hence, it is considered toxic to most microorganisms. Chitin is a natural polymer of N-acetyl-d-glucosamine monomers linked by β-1-4-covalent bonds. Chitinases found in some microorganisms can decompose chitin, a component of the cell wall of fungi (Starke et al. 2020). This is an essential criterion of a defense method against fungal pathogens. The clearing zone can detect chitinolytic endophytes in a chitin culture plate. A report indicated that endophyte bacteria showed high antifungal activity against F. oxysporum (Wang et al. 2019). Biological control of some soil-borne fungal diseases has been correlated with chitinase production by microorganisms. Endophytic bacteria showed high antifungal activity against F. oxysporum (Wang et al. 2019). Anthracnose caused by Colletotrichum truncatum, formerly known as C. capsici is a major disease of chilli that appears as ripe fruit rot and die-back that causes the infected plants to bear fewer fruits with low quality (Ridzuan et al. 2018). It causes a yield loss of 50% worldwide. It was reported that PGPR-inoculated chilli plants could induce resistance up to 71% against the pathogen Colletotrichum truncatum by utilizing the innate immunity of the chilli plant (Gowtham et al. 2018). Trichoderma has been reported to stimulate induced systemic resistance against the pathogen in chilli (Saxena et al. 2019). This pathogen is distributed worldwide with a great diversity of hosts that causes damage to various crops. Endophytic bacteria were previously reported to show high antifungal activity against F. oxysporum (Wang et al. 2019).
Compatibility is an essential criterion for preparing a mixed culture of microorganisms. Bacteria that showed various beneficial traits might be harmful to other species or a genus of bacteria that co-exist in a solution. The secondary metabolites produced by some bacteria might delay, inhibit, or destroy other bacterial cells. Therefore, the compatibility of selected bacteria was observed in an agar plate assay to ensure that the bacteria do not show antagonistic activity against each other.
In this study, MR13 and distilled water exhibited the lowest mean germination time, followed by CKR8, MR3, and MR1; meanwhile, MR13 demonstrated the highest germination energy, followed by distilled water, MR1, MR3, CKR8, and MR10 (Fig. 5). Seed germination analyses show that MR13 has the potential to trigger the germination of chilli seeds better than other endophytic bacteria. This finding agrees with research that has reported that Streptomycetes (MR13) caused a higher percentage of seed germination due to higher concentrations of IAA production (Vurukonda et al. 2018). The seed vigor test shows the ability of the seeds to produce uniform and rapid seedlings under adverse growth conditions. MR10 showed the highest seed vigor potential but could not be considered a bacterial bio-stimulant candidate based on its molecular identification as Enterobacter cloacae, an opportunistic pathogen.
Root development and the modification of root architecture are the fundamental criteria for beneficial endophytic bacteria where they have a high affinity toward the root exudates and colonize the host plant’s root efficiently. B. subtilis and B. japonicum were reported to improve root growth and positively change soybean’s root architecture to aid the uptake of essential nutrients (Araujo et al. 2021). An extended root surface could increase nutrient acquisition and water uptake by plant roots (Ribeiro et al. 2018). Two-way chemical interactions were observed between endophytes and root hair cells (Chang et al. 2021). Endophytes consume root exudates rich in organic acids, carbohydrates, amino acids, and vitamins, while plants use microorganisms as their source of nutrients. Endophytes also produce ethylene that enhances root hair growth and elongation, which could enhance nutrient acquisition by plant roots. After entering the plant root cells, endophytes become non-walled protoplast due to the degradation of the bacterial cell wall by superoxide produced by plants (White et al. 2018), and the nutrients from the endophytic microorganisms will be extracted by plant cells, while the endophytes proliferate and enhance root elongation and root hair growth inside the root cells. As the nutrients deplete completely, the endophytes will be ejected from the root and scavenge nutrients in the soil. The endophytes can form a new cell wall in the rhizosphere (White et al. 2018). The root images depict the effect of the endophyte inoculation on root growth (Fig. 7). It is evident from the root images that inoculation with mixed bacterial cultures shows the development of thick root collar stem and thick roots with high density (g–l) as compared to single bacterial culture and control (a–f). All bacterial cultures have generally enhanced root development by increasing the root length, hair, density, and thickness compared to non-inoculated seedlings (control). A similar finding was found in the past that co-inoculation of different genera of bacteria promoted the growth of maize and peanut crops (Anzuay et al. 2017). In another study, co-inoculation of B. cepacia and R. palustris was proven to increase the growth peanuts’ growth, quality, and yield.
This study also reveals that the co-culture of microorganisms may have both inhibition and stimulation effects. There is a possibility that the beneficial characteristics genes of interest could be upregulated or downregulated in the mixed culture. T7 (MR13 + MR10 + CKR8) has caused root length and surface area reduction, which is lower than the control (Table 4). This finding shows that there could be antagonistic activity among these three bacteria where the metabolites produced by bacterial culture could have retarded the root growth of the seedlings. Although the compatibility test did not show this antagonism effect in the earlier trial, the active growth stage in the liquid medium could have produced unfavorable metabolites for the growth of the seedlings.
Furthermore, the period from seed germination to seedling growth is deemed a critical growth stage for plants. A small amount of toxicity could be very harmful to the development of the seedlings during this vulnerable growth stage. Previously, it was reported that co-inoculation of Bacillus subtilis and Enterobacter cloacae was not conducive to the growth of B. subtilis. Among all the beneficial traits tested, only siderophore-related genes in Bacillus subtilis were upregulated (Li et al. 2021).
Root colonization is also an essential criterion for the commercialization of bio-inoculants. Streptomycetes sp. (MR13) and Bacillus sp. were selected for co-inoculating chilli seedlings based on their efficiency and bio-safety properties. Bacterial endophytes from the rhizosphere environment colonize root hairs due to the secretion of root exudates and rhizo-deposits. Both endophytic bacteria in this study could colonize the roots when applied as a single culture. In the current study, the establishment of Streptomyces sp. (MR13) supersedes Bacillus sp. (MR3) when the chilli seedlings were treated with a mixed culture. The difference between the colonization competence and the growth performance of crops inoculated with single and mixed cultures indicates that the interaction and mechanism of single and mixed cultures with crops are very complex. Co-inoculation does not guarantee the successful colonization of plant roots, although it could benefit the plant via various mechanisms, such as acidosis and nutrient mobilization (Razzaghi Komaresofla et al. 2019). In contrast to the vegetative growth of the root, FESEM images of root tips revealed that single bacterial culture has better colonization efficiency than mixed culture. Furthermore, the root colonization study shows that mixed cultures of MR3 and MR13 did not show apparent colonization as demonstrated by single cultures. This could be due to the different mechanisms, growth stages, and establishment patterns of bacteria in the root, as they must compete for efficient colonization. The ‛hitchhiking’ model was reported as the possible mechanism of co-inoculation of root-colonizing microbes, the immotile fungus, and motile bacterium (Muok et al. 2021). Streptomyces has a complex lifecycle that involves the formation of aerial hyphae that differentiate into spores, and it was unclear how the spores were transported to plant roots (Muok et al. 2021). It was demonstrated that Streptomyces spores could use the motility machinery of motile microbes by directly attaching to their flagella (Muok et al. 2021). This explains the reason behind the low distribution and establishment of Streptomyces spores inside chilli roots (Fig. 9a, d). Additionally, previous research reported that arbuscular mycorrhizal fungus demonstrated different life cycle stages in different parts of wheat roots from week 2 to week 5 (Fester et al. 1999). In a solid medium, Streptomyces develop into compartmentalized mycelium and produce spores, whereas in liquid cultures, no aerial mycelium or spores appear, but hyphae form pellets and clumps (Manteca and Yagüe 2018). Therefore, the inoculant was prepared in spore and mycelium form to check its colonization capability. It was proven that mycelium grown in liquid culture has better colonization efficiency than spores, as well-established mycelium was observed in chilli root inoculated with single and mixed cultures.
This study has justified that the co-inoculation of MR3 and MR13 mycelium could increase the plant height significantly. Although MR10 (Enterobacter cloacae) exhibited significant results on root length, root surface area, and relative chlorophyll content in the vegetative growth analyses, it could not be regarded as a potential bacterial bio-stimulant due to biosafety concerns. Therefore, co-inoculation of MR3 and MR 13 was suggested based on its ability to promote the relative chlorophyll content and chilli height and its potential to stimulate seed germination. However, it is vital to investigate the co-inoculation’s aptness to enhance the chilli’s yield and quality.
Conclusion
In conclusion, the efficiency of endophytic bacteria was studied, given their potential use for the developing biostimulant. Endophytes with multiple beneficial traits were identified to belong to Streptomycetes, Bacillus, and Enterobacter groups in general. About seven endophytic isolates were proven to enhance seed germination and seedling growth. Mixed culture demonstrated better root development as compared to a single culture. This study has proven that non-native endophytic bacteria from moringa roots could enhance chilli seedlings’ vegetative growth. Although the endophytic bacteria belonging to the Enterobacter group was found to be an efficient bio-stimulating agent for chilli seedlings, they were not suggested to be used as bio-inoculant in field experiments due to biosafety issues. MR3, Bacillus subtilis, and MR13, Streptomycetes panaciradicis were deemed potential endophytes for bio-stimulant formulation exclusively for chilli seedlings
References
Afzal I, Shinwari ZK, Sikandar S, Shahzad S (2019) Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol Res 221:36–49. https://doi.org/10.1016/j.micres.2019.02.001
Ahmadi-Rad S, Gholamhoseini M, Ghalavand A et al (2016) Foliar application of nitrogen fixing bacteria increases growth and yield of canola grown under different nitrogen regimes. Rhizosphere 2:34–37. https://doi.org/10.1016/j.rhisph.2016.08.006
Ali SZ, Sandhya V, Rao LV (2014) Isolation and characterization of drought-tolerant ACC deaminase and exopolysaccharide-producing fluorescent Pseudomonas sp. Ann Microbiol 64:493–502. https://doi.org/10.1007/s13213-013-0680-3
Anzuay MS, Ciancio MGR, Ludueña LM et al (2017) Growth promotion of peanut (Arachis hypogaea L.) and maize (Zea mays L.) plants by single and mixed cultures of efficient phosphate solubilizing bacteria that are tolerant to abiotic stress and pesticides. Microbiol Res 199:98–109. https://doi.org/10.1016/j.micres.2017.03.006
Araujo FF, Bonifacio A, Bavaresco LG et al (2021) Bacillus subtilis changes the root architecture of soybean grown on nutrient-poor substrate. Rhizosphere 18:16–19. https://doi.org/10.1016/j.rhisph.2021.100348
Bagyalakshmi B, Ponmurugan P, Balamurugan A (2017) Potassium solubilization, plant growth promoting substances by potassium solubilizing bacteria (KSB) from southern Indian Tea plantation soil. Biocatal Agric Biotechnol 12:116–124. https://doi.org/10.1016/j.bcab.2017.09.011
Baldani JI, Reis VM, Videira SS et al (2014) The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384:413–431. https://doi.org/10.1007/s11104-014-2186-6
Basu A, Prasad P, Das SN et al (2021) Plant growth promoting rhizobacteria (Pgpr) as green bioinoculants: recent developments, constraints, and prospects. Sustain 13:1–20. https://doi.org/10.3390/su13031140
Bergottini VM, Otegui MB, Sosa DA et al (2015) Bio-inoculation of yerba mate seedlings (Ilex paraguariensis St. Hill.) with native plant growth-promoting rhizobacteria: a sustainable alternative to improve crop yield. Biol Fertil Soils 51:749–755. https://doi.org/10.1007/s00374-015-1012-5
Chandra S, Askari K, Kumari M (2018) Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J Genet Eng Biotechnol 16:581–586. https://doi.org/10.1016/j.jgeb.2018.09.001
Chandwani S, Amaresan N (2022) Role of ACC deaminase producing bacteria for abiotic stress management and sustainable agriculture production. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-022-18745-7
Chang X, Kingsley KL, White JF (2021) Chemical interactions at the interface of plant root hair cells and intracellular bacteria. Microorganisms. https://doi.org/10.3390/microorganisms9051041
Chanthini KM, Senthil-nathan S, Stanley-raja V et al (2019) Biocatalysis and agricultural biotechnology Chaetomorpha antennina (Bory) Kützing derived seaweed liquid fertilizers as prospective bio-stimulant for Lycopersicon esculentum (Mill). Biocatal Agric Biotechnol 20:101190. https://doi.org/10.1016/j.bcab.2019.101190
Chouhan S, Agrawal L, Prakash A (2022) Amelioration in traditional farming system by exploring the different plant growth-promoting attributes of endophytes for sustainable agriculture. Arch Microbiol 204:1–21. https://doi.org/10.1007/s00203-021-02637-4
Cochard B, Giroud B, Crovadore J et al (2022) Endophytic PGPR from tomato roots: isolation, in vitro characterization and in vivo evaluation of treated tomatoes (Solanum lycopersicum L.). Microorganisms. https://doi.org/10.3390/microorganisms10040765
Costa-Gutierrez SB, Adler C, Espinosa-Urgel M, de Cristóbal RE (2022) Pseudomonas putida and its close relatives: mixing and mastering the perfect tune for plants. Appl Microbiol Biotechnol 106:3351–3367. https://doi.org/10.1007/s00253-022-11881-7
De Zutter N, Ameye M, Debode J et al (2021) Shifts in the rhizobiome during consecutive in planta enrichment for phosphate-solubilizing bacteria differentially affect maize P status. Microb Biotechnol 14:1594–1612. https://doi.org/10.1111/1751-7915.13824
Devi R, Kaur T, Kour D et al (2022) Potential applications of mineral solubilizing rhizospheric and nitrogen fixing endophytic bacteria as microbial consortium for the growth promotion of chilli (Capsicum annum L.). Biologia (Bratisl) 77:2933–2943. https://doi.org/10.1007/s11756-022-01127-2
Eid AM, Fouda A, Abdel-rahman MA et al (2021) Harnessing bacterial endophytes for promotion of plant growth and biotechnological applications: an overview. Plants 10:1–33. https://doi.org/10.3390/plants10050935
Ertani A, Sambo P, Nicoletto C et al (2015) The use of organic biostimulants in hot pepper plants to help low input sustainable agriculture. Chem Biol Technol Agric. https://doi.org/10.1186/s40538-015-0039-z
Ferreira CMH, Vilas-Boas Â, Sousa CA et al (2019) Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express. https://doi.org/10.1186/s13568-019-0796-3
Fester T, Maier W, Strack D (1999) Accumulation of secondary compounds in barley and wheat roots in response to inoculation with an arbuscular mycorrhizal fungus and co-inoculation with rhizosphere bacteria. Mycorrhiza 8:241–246. https://doi.org/10.1007/s005720050240
Gowtham HG, Murali M, Singh SB et al (2018) Plant growth promoting rhizobacteria- Bacillus amyloliquefaciens improves plant growth and induces resistance in chilli against anthracnose disease. Biol Control 126:209–217. https://doi.org/10.1016/j.biocontrol.2018.05.022
Gupta S, Pandey S (2019) ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French Bean (Phaseolus vulgaris) plants. Front Microbiol 10:1–17. https://doi.org/10.3389/fmicb.2019.01506
Hardoim PR, van Overbeek LS, Berg G et al (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. https://doi.org/10.1128/mmbr.00050-14
Hassan SED (2017) Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J Adv Res 8:687–695. https://doi.org/10.1016/j.jare.2017.09.001
Husseiny S, Dishisha T, Soliman HA et al (2021) Characterization of growth promoting bacterial endophytes isolated from Artemisia annua L. S Afr J Bot 143:238–247. https://doi.org/10.1016/j.sajb.2021.07.042
Igiehon NO, Babalola OO (2018) Rhizosphere microbiome modulators: contributions of nitrogen fixing bacteria towards sustainable agriculture. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph15040574
Kalayu G (2019) Phosphate solubilizing microorganisms: promising approach as biofertilizers. Int J Agron 2019:1–7. https://doi.org/10.1155/2019/4917256
Khan AL, Halo BA, Elyassi A et al (2016) Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron J Biotechnol 21:58–64. https://doi.org/10.1016/j.ejbt.2016.02.001
Khati P, Pankaj P, Nisha B et al (2018) Effect of nanozeolite and plant growth promoting rhizobacteria on maize. 3 Biotech 8:1–12. https://doi.org/10.1007/s13205-018-1142-1
Li Y, He Y, Wang W et al (2021) Plant-beneficial functions and interactions of Bacillus subtilis SL-44 and Enterobacter cloacae Rs-2 in co-culture by transcriptomics analysis. Environ Sci Pollut Res 28:56333–56344. https://doi.org/10.1007/s11356-021-14578-y
Lin H, Liu C, Peng Z et al (2022) Distribution pattern of endophytic bacteria and fungi in tea plants. Front Microbiol. https://doi.org/10.3389/fmicb.2022.872034
Manteca Á, Yagüe P (2018) Streptomyces differentiation in liquid cultures as a trigger of secondary metabolism. Antibiotics 7:1–13. https://doi.org/10.3390/antibiotics7020041
Marchiol L, Iafisco M, Fellet G, Adamiano A (2020) Nanotechnology support the next agricultural revolution: perspectives to enhancement of nutrient use efficiency, 1st edn. Elsevier, Amsterdam
Matthews S, Ali A, Siddiqui Y (2022) Organic plant bio-stimulant for early. Enhanced Healthy Growth Chilli Seedl Chem Proc 4:1–4. https://doi.org/10.3390/xxxxxwww.mdpi.com/journal/chemproc
Matthews S, Ali A, Siddiqui Y, Supramaniam CV (2022) Plant bio-stimulant: prospective, safe and natural resources. J Soil Sci Plant Nutr. https://doi.org/10.1007/s42729-022-00828-6
Meena VS, Maurya BR, Verma JP, Meena RS (2016) Potassium solubilizing microorganisms for sustainable agriculture. Potassium Solubilizing Microorg Sustain Agric. https://doi.org/10.1007/978-81-322-2776-2
Mendes GdeO, Murta HM, Valadares RV et al (2020) Oxalic acid is more efficient than sulfuric acid for rock phosphate solubilization. Miner Eng 155:106458. https://doi.org/10.1016/j.mineng.2020.106458
Muok AR, Claessen D, Briegel A (2021) Microbial hitchhiking: how Streptomyces spores are transported by motile soil bacteria. ISME J 15:2591–2600. https://doi.org/10.1038/s41396-021-00952-8
Naveed M, Mitter B, Yousaf S et al (2013) The endophyte Enterobacter sp. FD17: a maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol Fertil Soils 50:249–262. https://doi.org/10.1007/s00374-013-0854-y
Orozco-Mosqueda MdelC, Rocha-Granados MdelC, Glick BR, Santoyo G (2018) Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol Res 208:25–31. https://doi.org/10.1016/j.micres.2018.01.005
Passari AK, Chandra P, Zothanpuia, et al (2016) Detection of biosynthetic gene and phytohormone production by endophytic actinobacteria associated with Solanum lycopersicum and their plant-growth-promoting effect. Res Microbiol 167:692–705. https://doi.org/10.1016/j.resmic.2016.07.001
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118:10–15. https://doi.org/10.1034/j.1399-3054.2003.00086.x
Pham T, Dong X, Vu L et al (2022) Isolation of indole-3-acetic acid-producing Azospirillum brasilense from Vietnamese wet rice: co-immobilization of isolate and microalgae as a sustainable biorefinery. J Biotechnol 349:12–20. https://doi.org/10.1016/j.jbiotec.2022.03.007
Raheem A, Ali B (2022) The microphenotron: a novel method for screening plant growth-promoting rhizobacteria. PeerJ 10:1–21. https://doi.org/10.7717/peerj.13438
Rana KL, Kour D, Kaur T et al (2020) Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability. Springer, Cham
Razzaghi Komaresofla B, Alikhani HA, Etesami H, Khoshkholgh-Sima NA (2019) Improved growth and salinity tolerance of the halophyte Salicornia sp. by co–inoculation with endophytic and rhizosphere bacteria. Appl Soil Ecol 138:160–170. https://doi.org/10.1016/j.apsoil.2019.02.022
Rezakhani L, Motesharezadeh B, Tehrani MM et al (2019) Phosphate–solubilizing bacteria and silicon synergistically augment phosphorus (P) uptake by wheat (Triticum aestivum L.) plant fertilized with soluble or insoluble P source. Ecotoxicol Environ Saf 173:504–513. https://doi.org/10.1016/j.ecoenv.2019.02.060
Ribeiro VP, Marriel IE, de Sousa SM et al (2018) Endophytic Bacillus strains enhance pearl millet growth and nutrient uptake under low-P. Braz J Microbiol 49:40–46. https://doi.org/10.1016/j.bjm.2018.06.005
Ridzuan R, Rafii MY, Ismail SI et al (2018) Breeding for anthracnose disease resistance in chili: progress and prospects. Int J Mol Sci 19:1–21. https://doi.org/10.3390/ijms19103122
Rilling JI, Acuña JJ, Sadowsky MJ, Jorquera MA (2018) Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an andisol from southern Chile. Front Microbiol 9:1–13. https://doi.org/10.3389/fmicb.2018.02710
Ringuet S, Sassano L, Johnson ZI (2011) A suite of microplate reader-based colorimetric methods to quantify ammonium, nitrate, orthophosphate and silicate concentrations for aquatic nutrient monitoring. J Environ Monit 13:370–376. https://doi.org/10.1039/c0em00290a
Saha N, Trivedi P, Dutta Gupta S (2016) Surface plasmon resonance (SPR) based optimization of biosynthesis of silver nanoparticles from rhizome extract of Curculigo orchioides Gaertn. and its antioxidant potential. J Clust Sci 27:1893–1912. https://doi.org/10.1007/s10876-016-1050-7
Saima KM, Roohi AIZ (2013) Isolation of novel chitinolytic bacteria and production optimization of extracellular chitinase. J Genet Eng Biotechnol 11:39–46. https://doi.org/10.1016/j.jgeb.2013.03.001
Saxena A, Mishra S, Ray S et al (2019) Differential reprogramming of defense network in Capsicum annum L. plants against colletotrichum truncatum infection by phyllospheric and rhizospheric trichoderma strains. J Plant Growth Regul. https://doi.org/10.1007/s00344-019-10017-y
Shi P, Zhang J, Li X et al (2022) Multiple metabolic phenotypes as screening criteria are correlated with the plant growth-promoting ability of rhizobacterial isolates. Front Microbiol. https://doi.org/10.3389/fmicb.2021.747982
Sinha AK (2019) Effects of growth conditions on siderophore producing bacteria and siderophore production from Indian Ocean sector of Southern Ocean. J Basic Microbiol 59:412–424. https://doi.org/10.1002/jobm.201800537
Siswanti D, Utaminingsih U, Pangestuti N (2019) Capsaicin level and anatomy response of curly red chili (Capsicum annuum L.) to bio fertilizer and sludge biogas application. Eur Union Digit Library 5:371–388. https://doi.org/10.4108/eai.2-5-2019.2284700
Starke R, Morais D, Větrovský T et al (2020) Feeding on fungi: genomic and proteomic analysis of the enzymatic machinery of bacteria decomposing fungal biomass. Environ Microbiol 22:4604–4619. https://doi.org/10.1111/1462-2920.15183
Tao L, Qiuhong L, Fuqiang Y et al (2022) Plant growth-promoting activities of bacterial endophytes isolated from the medicinal plant Pairs polyphylla var. yunnanensis. World J Microbiol Biotechnol 38:1–10. https://doi.org/10.1007/s11274-021-03194-0
Tian J, Ge F, Zhang D et al (2021) Roles of phosphate solubilizing microorganisms from managing soil phosphorus deficiency to mediating biogeochemical p cycle. Biology (Basel) 10:1–19. https://doi.org/10.3390/biology10020158
Upadhyay SK, Srivastava AK, Rajput VD et al (2022) Root exudates: mechanistic insight of plant growth promoting rhizobacteria for sustainable crop production. Front Microbiol. https://doi.org/10.3389/fmicb.2022.916488
Vurukonda SSKP, Giovanardi D, Stefani E (2018) Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int J Mol Sci. https://doi.org/10.3390/ijms19040952
Wang GF, Meng JF, Tian T et al (2019) Endophytic Bacillus velezensis strain B-36 is a potential biocontrol agent against lotus rot caused by Fusarium oxysporum. J Appl Microbiol. https://doi.org/10.1111/jam.14542
Wei Y, Zhao Y, Shi M et al (2018) Effect of organic acids production and bacterial community on the possible mechanism of phosphorus solubilization during composting with enriched phosphate-solubilizing bacteria inoculation. Bioresour Technol 247:190–199. https://doi.org/10.1016/j.biortech.2017.09.092
White JF, Kingsley KL, Verma SK, Kowalski KP (2018) Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms. https://doi.org/10.3390/microorganisms6030095
Woo SL, Pepe O, Fertilizers VSP (2018) Promising probiotics as plant biostimulants for sustainable agriculture. Microb Consortia 9:7–12. https://doi.org/10.3389/fpls.2018.01801
Xing YX, Wei CY, Mo Y et al (2016) Nitrogen-fixing and plant growth-promoting ability of two endophytic bacterial strains isolated from sugarcane stalks. Sugar Tech 18:373–379. https://doi.org/10.1007/s12355-015-0397-7
Yasmin H, Naz R, Nosheen A et al (2020) Identification of new biocontrol agent against charcoal rot disease caused by Macrophomina phaseolina in Soybean (Glycine max L.). Sustain 12:6856. https://doi.org/10.3390/SU12176856
Zhang C, Kong F (2014) Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl Soil Ecol 82:18–25. https://doi.org/10.1016/j.apsoil.2014.05.002
Zhang J, Cook J, Nearing JT et al (2021) Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol Res 245:126690. https://doi.org/10.1016/j.micres.2020.126690
Zhao M, Zhao J, Yuan J et al (2021) Root exudates drive soil–microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ 44:613–628. https://doi.org/10.1111/pce.13928
Accession number (s): The DNA sequences have been deposited in the NCBI database under the accession numbers OM001090 and OM714810.
Author information
Authors and Affiliations
Contributions
SM drafted the experimental design, performed the experiment, analyzed the data, and wrote and revised the manuscript. AA, YS, and CVS reviewed, improvised, and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Handling Editor: Chetan Keswani.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Matthews, S., Siddiqui, Y., Supramaniam, C.V. et al. Boosting Capsicum annuum Growth Through Non-native Endophytic Bacterial Consortium. J Plant Growth Regul 43, 2739–2760 (2024). https://doi.org/10.1007/s00344-024-11302-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-024-11302-1