Abstract
High sensitivity of rice coleoptile elongation length to exogenous gibberellin is a beneficial trait to utilize superior rice cultivars that could not be used originally under water direct-seeded conditions. In the present study, we mined favorable alleles for the trait by combining the phenotypic data of 358 rice accessions with their genotype data of 262 simple sequence repeat (SSR) markers via genome wide association mapping method. Totally, 17 SSR marker loci significantly associated with gibberellin sensitivity index (GSI) of coleoptile elongation length under 10 cm depth of water, were detected by general linear model and mixed linear model across two years, with percent phenotypic variation explained larger than 10%. Twenty nine favorable alleles for GSI on the 17 loci were discovered with phenotypic effect value (PEV) larger than 0.1 cm/cm and RM6869-110 bp showed the largest PEV (0.27 cm/cm). Based on PEV of marker-alleles having positive effects on GSI, seven parental combinations were predicted to improve GSI. In addition, 7 loci for GSI were co-located with loci associated with coleoptile elongation length per se, and one locus (RM1182 on chromosome 5) was co-located with that associated with coleoptile elongation length after gibberellin-soaked seed, under germination condition of 10 cm depth of water. These favorable allele(s) could be used to improve two target traits simultaneously.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is the most important cereal crop in the world. Due to a lack of manpower and higher wages, rice growers turn to the direct seeding method (Angaji et al. 2010). Direct-seeded rice is a common production method in southern Louisiana and areas in Texas and California State, USA (Hardke and Scott 2013). In the same time, rice plants suffer from submergence (flooding) and poor seedling establishment.
Flooding is one of the serious problems which affect rice production in South and Southeast Asia, where the majority of the world’s rice is grown, about 20 million hectares of rice land is prone to flooding. Flooding creates hypoxic or anoxic condition resulting in poor germination and seedling establishment, even in some cases leads to plant death within few days of full submergence (IRRI 2016; Singh et al. 2017). There are different categories of flooding; we are interested in submergence during germination also known as anaerobic germination. On this condition, rapid seedling elongation can provide successful establishment, and escape from submergence stress, hence provides required oxygen for normal growth.
For successful establishment and escape from submergence stress, priming technique is involved to enhance the start of germination processes (Silva and Silva 2016). Doley et al. (2018) studied priming effect on 243 rice genotypes for anaerobic germination under 10 cm of flooding. They found that priming rice seeds for 24 h with different solutions enhanced anaerobic germination under flooding compared to control. In addition, priming three rice cultivars for 48 h was the best seed invigoration treatment under well watered condition (Mulbah and Adjetey 2018). Furthermore, Sarkar 2012 studied two near isogenic lines under flooding and non-flooding conditions. His result revealed that seed priming improved the seedling establishment under anaerobic conditions. Recently, it was observed that rice seed priming followed by sun drying can improve anaerobic germination (Senapati et al., 2019).
Angaji et al. (2010) identified a few tolerant genotypes of over 8000 genotypes screened for the tolerance of flooding during germination. Under submergence, successful rice coleoptile elongation depends on hydrolases induction to mobilize endosperm; α-amylases play a central role in this process. Gibberellic acid (GA3) is an important hormone induces α-amylases expression resulting in germination and seedling growth in rice under anaerobic conditions (Lee et al. 2014). Kaneko et al. (2002) also found that active GA3 is important for α-amylases expression in rice endosperm. Moreover, in barly the expression of the α-amylase gene is up-regulated by exogenous GA3 (Gubler et al. 2002). Rice cultivars have different sensitivity to exogenous gibberellin concentrations via seed treatment, reflecting upon seedling performance (Guadagnin et al. 2017). Likewise, a study on rice showed that the most effective concentration was 2000 ppm GA3, which enhances seedlings length of BW196 (Mutinda et al. 2017).
Mining favorable alleles for coleoptile length (CL), coleoptile length gibberellic acid sensitivity (CLGS) and its gibberellic acid sensitivity index (GSI) for water direct-seeded rice would provide breeders to improve traits. In 2004, Jiang et al. (2004) detected five QTLs for anoxia germinability from 81 RILs with phenotypic variation ranged from 10.5 to 19.6% on chromosomes 1, 2, 5 and 7, respectively. Furthermore, they detected three pairs of epistasis loci located on chromosomes 2, 3, 5 and 11 with significant effects ranging from 16.7 to 48.8%. Five putative QTLs controlling flooding tolerance during germination in rice were detected on chromosomes 1, 3, 7 and 9, explaining 17.9–33.5% of the phenotypic variation (Angaji et al. 2010). Septiningsih et al. (2013) identified six QTLs of mapping 175 F2:3 families genotypes, using 118 SSR markers, on chromosomes 2, 5, 6 and 7 associated with a survival rate of seedling under 10 cm depth of water. Baltzar et al. (2014 detected two major QTLs associated with the survival rate of seedling while analysis 300 lines F2:3 derived from the cross of IR64 and the aus landrace Nanhi. One QTL derived from Nanhi detected on chromosome 7 explained 22.3% phenotypic variance, while the other one was detected on chromosome 2 from IR64 with increased effect. Recently, three QTLs associated with anaerobic germination detected by analysis 285 F2:3 genotypes derived from a cross between Tai Nguyen and Anda using 6 K SNP chip (Kim and Reinke 2018). Two QTLs were detected on chromosome 1 and one QTL on chromosome 8 with variance explained percentage ranged from 5.49 to 14.14%. Taking all together, the QTLs reported up to now for anoxic (flooding) conditions are 4, 4, 2, 3, 1, 6, 1, 1, and 1 on chromosomes 1, 2, 3, 5, 6, 7, 8, 9 and 11 respectively.
In our study, two points are new compared with previous research. One is the QTL detection method (we use GWAS for this trait), the other is gibberellin-treated seeds and germinated under 10 cm depth of water. It is the first report mined favorable alleles of coleoptile elongation and its sensitivity to gibberellic acid for water direct-seeded rice by association mapping using 262 SSR markers from the natural population The aims of the present study were to (1) investigate the phenotypic variation of CL, CLGS and GSI under anoxic condition; (2) identify QTLs and mine the favorable alleles for CL, CLGS and GSI by genome-wide association mapping; (3) predict parental combinations for improve CLGS with high GSI according to superior accessions screened in this study.
Materials and Methods
Plant Materials
The seeds of the 358 rice genotypes were collected, stored, and supplied by State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China (Supplementary Table 1).
Field Planting
All the seeds of the tested materials were sowed in the seedling nursery of paddy field in Jiangpu Experimental Station, Nanjing Agricultural University, in mid-May and transplanted in mid-June in 2017. The experiment was evaluated in a randomized block design with three replications. All the recommended package of practices was followed. In 2018, the dates of sowing and transplanting and field managements were equivalent to 2017. The purpose of field planting was to harvest fresh seeds for germination experiments.
Evaluation of Coleoptile Elongation Length and Its Sensitivity to Gibberellic Acid Under 10 cm Depth of Water Condition
Fifty seeds of each accession were used for each treatment (0 ppm-GA3 and 2000 ppm-GA3). Under the control treatment, the seeds were soaked in distilled water for 24 h; while under GA3 treatment, the seeds were soaked in GA3 solution (2000 ppm) for 24 h. Thirty uniformed soaked seed were visually selected out of the 50 and transferred to a paper towel, lined up on 3 cm from the lower edge, covered with two layers of moist filter paper and rolled the paper up, sailed with a rubber band and placed vertically in plastic box (44 cm × 31 cm × 15 cm) and submerged under 10 cm depth water. The plastic boxes were put under the natural conditions for 13 days to allow the seeds germinate and grow (Supplementary Fig. 1). On the fourteenth day, the coleoptile elongation lengths of 10 seedlings in each replicate of each treatment in each accession were measured with a ruler, and recorded as CL (cm) for the distilled water treatment and CLGS (cm) for the GA3 treatment. Coleoptile elongation length sensitivity of an accession to GA3 was designated as gibberellin sensitivity index (GSI) and was determined using the following formulas:
where CLGS is coleoptile elongation length (cm) under GA3 treatment, and CL is coleoptile elongation length (cm) under distilled water treatment (control).
Phenotypic Data Analysis
The mean, standard deviation, maximum, minimum and coefficient of variation for the CL and CLGS trait were calculated by using XLSTAT: Statistical software for Excel (Version 20.6.5) available from https://www.xlstat.com/en/. Microsoft Excel software 2016 was used to compute the broad-sense heritability using the following formula (Wang et al. 2007):
where \({\delta }_{g}^{2}\) is genetic variance,\({\delta }_{e}^{2}\) is error variance, and n is a number of replicates.
The correlation coefficient was calculated between each of CL, CLGS and GSI by using SPSS statistics 19 (Weaver and Wuensch 2013).
SSR Marker Genotyping
Based on the existing data published on rice molecular mapping, as well as microsatellite data (Temnykh et al. 2000; McCouch et al. 2002; Varshney et al. 2005), 262 pairs of SSR primers distributed on the 12 chromosomes of rice were utilized in genotyping. Leaf blade tissue of a single individual plant in each accession was used to extract genomic DNA using the method described by Dang et al (2019). DNA amplification primers were synthesized by Shanghai Generay Biotech Co. Ltd., Shanghai, China. Every 10 μl PCR mixture contained 1 µl genomic DNA, 0.7 µl of the forward primer and the same amount (0.7 µl) of reverse primer, 10 × Buffer (free MgCl2) 1 µl, dNTPs 0.2 µl, 0.1 µl of Taq polymerase, 0.6 µl MgCl2, and 5.7 µl ddH2O. PCR amplification was performed on a Peltier Thermal Cycler (PTC-100™, MJ Research™ Incorporated, USA) under denaturation of 94℃ for 5 min; 34 cycles of denaturation at 94℃ for 30 s, annealing at 55 ~ 61℃ (depending on the primer used) for 1 min, with extension at 72℃ for 1 min, and, finally, an extension at 72℃ for 10 min. Visualization of the resultant PCR products was done on an 8% polyacrylamide gel run for 1 h at 150 V and observed through silver staining.
Population Genetic Structure Analysis
Using STRUCTURE version 2.2 (Falush et al. 2007) the genetic clusters in the 358 accessions were identified. A mean log-likelihood value over five runs set each K (K from 2 to 10) with random starting points. The length of the burn-in period was set to 50,000 iterations and defined a run of 100,000 Markov Chain Monte Carlo (MCMC) replicates after burn-in was used. If the mean log-likelihood value was positively correlated with the model parameter K; a suitable value for K could not be determined. In this situation, the optimal K value was determined through an ad hoc statistic (∆K) based on the rate of change in [LnP (D)] between successive K values (Evanno et al. 2005). Non- admixed individuals in each genetic group were determined using a Q-matrix assignment greater than 0.9. Power Marker version 3.25 (Liu and Muse 2005) was used to determine the number of alleles per locus, major allele frequency, genetic diversity per locus, and polymorphism information content (PIC) values. The genetic distance was calculated based on 262 molecular markers using Nei’s distance (Nei et al. 1982) and phylogenetic reconstruction was performed using a neighbor-joining method as implemented in Power Marker with the tree viewed using MEGA 4.0 (Tamura et al. 2007). Locus-by-locus analysis of molecular variance (AMOVA) (Weir and Cockerham 1984) based on genetic groups delimited by the Bayesian clustering method in the program Arlequin 3.5 (Excoffier and Lischer 2010) was performed to statistically verify the geographical structure using SSR and standard multi-locus frequency data. The genetic differentiation coefficient or fixation index (Fst) between subpopulations was calculated using the method proposed by Weir and Hill (2002). The calculation process was performed in Arlequin 3.5 software.
Linkage Disequilibrium Analysis
To evaluate the linkage disequilibrium (LD) level, TASSEL 2.1 (Bradbury et al. 2007) software was used in which each pair of SSR loci was evaluated, in all rice accessions and clusters arising from STRUCTURE analysis. The D' value was used to measure the degree of LD between sites (non-alleles). The formula for calculating the D' value is given as (Hedrick 1987):
where u and v represent the number of alleles of the two loci, pi and qj the frequency of the i-th allele at position A and the frequency of the j-th allele at position B, respectively.
\(\left|{D}_{ij}^{^{\prime}}\right|\) is the absolute value of Lewontin’s (1964) normalized measure \({D}_{ij}^{^{\prime}}\)
where \({D}_{ij}^{max}\) is the maximum amount of disequilibrium possible between the i-th allele at locus A and the j-th allele at locus B.
Genome Wide Association Mapping
Genome wide association mapping using General Linear Model (GLM, Q) and Mixed Linear Model (MLM, Q+K) was performed using TASSEL 3.0 to calculate the associations between the target trait and markers (Bradbury et al. 2007). The Q matrix was obtained from the analysis results of Structure 2.2, and genetic relatedness (K) matrix was obtained by the software TASSEL 3.0. A false discovery rate (FDR) of 0.001 was used as a threshold for multiple testing according to the correction method published by Benjamini and Hochberg (1995). In this study, marker loci with phenotypic variation explained (PVE) > 7% were considered for further analysis. The phenotypic effect values of the alleles amplified were calculated based on the null allele (not amplified) method described by Breseghello and Sorrells (2006).
Results
Phenotypic Variations of CL, CLGS and GSI
The phenotypic data of the CL and CLGS followed a normal distribution as showed in Fig. 1, which is also confirmed by Kurtosis and Skewness values for both years (Table 1). The mean value for CL over 358 accessions was 2.59 cm with a range from 0.82 to 3.82 cm in 2017. The coefficient of variance was 20.62% and broad sense heritability was 98.50%. In 2018, the results for CL were similar to those of the previous year (Table 1). On the other hand, the mean value for CLGS was 3.04 cm with a range from 1.25 to 4.76 cm in 2017. The coefficient of variation was 19.61% with H2b of 95.92%. Also, the results for CLGS obtained in 2018 were similar to those of the previous year (Table 1). The broad-sense heritability for CL and CLGS was higher than 90% in both years, indicating that the phenotypic variations of the two traits were mainly controlled by genetic factors.
Gibberellic acid treatment increased the coleoptile elongation length by 0.45 cm/cm and 0.46 cm/cm averaged over 358 accessions in 2017 and 2018, respectively, compared with those of water treatment. The GSI ranged from 0 to 2.12 cm/cm in 2017, while the range in 2018 was from 0 to 2.39 cm/cm (Fig. 2), indicating there exist variations in coleoptile elongation length sensitivity to GA3 among the 358 genotypes used. According to the performances of both CL and GSI grown in the 10 cm depth of water, 6 accessions were considered as superior germplasms for water direct-seeded rice (Table 2). The most sensitive accession to GA3 is Gaoliangqing with GSI of 2.26 cm/cm, followed by Wuxiangjing14 (0.91 cm/cm), Changdaotou (0.87 cm/cm), Hongdao35 (0.79 cm/cm), Zhenghan2 (0.71 cm/cm) and Huajing5 (0.61 cm/cm).
Figure 3 shows the difference in coleoptile elongation length between 0 ppm-GA3 treatment and 2000 ppm- GA3 treatment under 10 cm depth of water in accessions Changdaotou. It can be seen from Fig. 3 that the deference between CL and CLGS are clear.
The correlation coefficients between CL, CLGS and GSI are presented in Table 3. The result revealed positive and highly significant between CLGS and GSI. While the correlation coefficient between CL and GSI was negative and highly significant.
Genetic Diversity of the Entire Population Revealed by SSR Markers
The genetic diversity of the 358 accessions was determined using 262 SSR markers distributed on the 12 chromosomes in rice. Totally 2474 marker alleles were identified with average of 9.443 alleles per locus (ranged from 2 to 25) (Supplementary Table 2). The gene diversity value averaged over 262 loci was 0.731 with a range from 0.100 (RM7163 on chromosome 11) to 0.937 (RM7545 on chromosome10). The polymorphic information content (PIC) value averaged over 262 loci was 0.702 with a range from 0.095 (RM7163 on chromosome 11) to 0.933 (RM7545 on chromosome 10). While 33 markers showed PIC value less than 0.5, the PIC value of 92 markers were more or equal 0.8, and 137 markers were in between 0.5 and 0.8. These results indicate high genetic diversity in the population used.
Population Genetic Structure
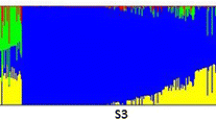
Genetic structure analysis of the entire populations showed an increase in likelihood function LnP (K) value with the increase of subpopulations (Supplementary Fig. 2a). Supplementary Fig. 2b shows that ΔK value reached maximum at K = 6. Therefore, the entire population can be divided into 6 sub-populations. A neighbour-joining tree of the 358 accessions was constructed based on Nei’s genetic distance (Supplementary Fig. 2d), and the results were consistent with the results from the Structure analysis. Using the criterion of Q value > 0.9, each accession was sorted into the corresponding subpopulation. 325 accessions entered into 6 subpopulations (known as SP1, SP2, SP3, SP4, SP5 and SP6) (Supplementary Fig. 2), and the remaining 33 accessions entered into an admixture subpopulation. The numbers of accessions SP1, SP2, SP3, SP4, SP5 and SP6 were 52, 75, 38, 24, 70 and 66, respectively (Supplementary Table 1).
By checking the resources of the 358 accessions, it was found that the 6 subpopulations divided above had different geographic origins or ecotypes. Accessions in SP1 were all from Vietnam (Indica rice). SP2 contains accessions from middle china and a few numbers of northeast accessions (Temperate japonica). Most of the accessions in SP3 are modern cultivars bred in the north-central of Jiangsu province (Temperate japonica). SP4 has accessions from middle-east China (Temperate japonica). SP5 accessions were mainly from south Jiangsu province (Temperate japonica) and SP6 had tall, late-maturing accessions and a small number of northeast accessions in the Taihu Lake Basin (Temperate japonica), as showed in Supplementary Table 1.
The results of the analysis of molecular variance (AMOVA) indicated that 46.2% of the total genetic variation occurred between the subpopulations, whereas 53.8% occurred within the subpopulations (Table 4). These results indicate a high degree of genetic differentiation across the six subpopulations.
Genetic Diversity of the Six Subpopulations
The basic genetic information of each subpopulation is shown in Table 5. SP6 has the highest number of alleles per locus (4.057), the highest genetic diversity (0.524), d while SP3 has the lowest numbers of alleles per locus (2.031), the lowest genetic diversity (0.276), among the 6 subpopulations (Table 5). Compared with the entire population, the genetic parameters of each subpopulation were significantly reduced, indicating that the alleles of partial loci were fixed during the process of differentiation of each subpopulation.
Pairwise Fst Values and Nei’s Genetic Distance Among the Subpopulations
The \({F}_{st}\) values, which reflected the genetic differentiation extent between two subpopulations, for the 15 pairs of subpopulations are shown below the diagonal (Table 6). The \({F}_{st}\) value between SP2 and SP5 was the lowest (0.376), while that between SP3 and SP4 was the highest (0.632). Nei’s genetic distance between SP2 and SP5 was short (0.528), while the distance between SP3 and SP4 was long (0.771) (Table 6). The results in Table 6 indicate that the pairwise \({F}_{st}\) value can reflect the genetic distance between subpopulations.
Ratios of Significant Linkage Disequilibrium Pairwise Loci and Decay Distances in the 6 Subpopulations
The ratio of significant linkage disequilibrium (LD) pairwise loci (P˂0.01) was the lowest (0.17%) in SP4 and was the highest (3.33%) in SP6 (Table 7). The highest mean of D' value was 0.61 (SP4) and the lowest value was 0.57 in both SP5 and SP6, suggesting that the accessions of these subpopulations have been subjected to extreme artificial selection. The decay rate of D' in each subpopulation (Supplementary Fig. 3) follows the logarithmic regression equation \(y=b\mathrm{ln}x+c\). The LD decay distance was 82.21, 98.33, 85.12, 79.46, 93.19 and 92.69 cM for subpopulations SP1, SP2, SP3, SP4, SP5 and SP6, respectively. The shortest distance was 79.46 cM in SP4, whilst SP2 has the longest distance (98.33 cM). These results indicated that the accessions in SP4 have been subjected to more recombination and the accessions in SP2 have been subjected to extreme artificial selection.
SSR Marker Loci Associated with CL, Favorable Alleles and Their Carrier Accessions
Twenty three marker loci were detected using the GLM model and two SSR loci were detected using MLM model in both years with PVE more than 7% (one SSR marker locus common between the two models). All markers were distributed on all chromosomes except chromosome 5 and chromosome 7 (Table 8). The range of PVE was from 7.19% (RM1013 on chromosome 9) to 18.22% (RM6327 on chromosome 11) in 2017 and the results were similar in 2018.
Table 9 shows the top 39 positive favorable alleles of the significant association loci with PEV more than 0.5 cm and their typical carrier materials (which carrying the desired alleles with the highest phenotypic value) for CL in both years. The PEV for those alleles ranged from 0.504 cm of RM3754-80pb (typical carrier accession Songjing12) to 1.594 cm of RM6327-215pb (typical carrier accession Wanqu429).
Six marker loci showed positive average allele effect (AAE +), without negative allele effect (PVE more than 7%); RM6327 was the highest with AAE + equal to1.396 cm, followed by RM106 with AAE + 0.627 cm, RM3513 with AAE + 0.503 cm, RM1358 with AAE + 0.371 cm, RM5356 with AAE + 0.296 cm and RM128 with AAE + 0.274 cm (Table 10).
Based on phenotypic effect value of marker-alleles which have positive effect on CL, the best parental combinations were selected from the top 20 accessions. Seven parental combinations predicted to improve CL; and the predicted phenotypic effect ranged from 0.850 cm to 0.940 cm (Table 11).
SSR Marker Loci Associated with CLGS, Favorable Alleles and Their Carrier Accessions
Twenty-one SSR loci for CLGS were detected using GLM mode and two SSR loci using MLM model in both years with PVE more than 7% (one SSR marker locus common between the two models). Overall, 22 SSR markers were distributed on all chromosomes except chromosome 12 (Table 12). The range of PVE was from 7.06% (RM3688 on chromosome 2) to 17.24% (RM3773 on chromosome 10) in 2017 and the results were similar in 2018.
Table 13 shows the top 56 positive favorable alleles of the significant association loci with the PEV more than 0.5 cm (PVE more than 7%) and their typical carrier accessions for CLGS. The PEV for those alleles ranged from 1.087 cm for RM562-180 (typical carrier accession Xiaoqingmang) to 0.506 cm for RM283-150 (typical carrier accession Zhenghan2).
Three markers showed positive average allele effect (AAE), without negative allele effect (PVE more than 7%); RM562 was the highest one with AAE 0.721 cm, followed by RM434 with AAE 0.538 cm and RM3453 with AAE 0.494 cm (Table 14).
Comparing the association analysis for CL and CLGS, the result showed that RM3754 (chromosome 8) was co-associated with both CL and CLGS analysis with PVE 8.62% and 7.91% for CL and CLGS in 2017, respectively; and the result was similar in 2018. RM3754-80 bp shows phenotypic effect value 0.504 cm for CL and the typical carrier is Songjing12. While RM3754-100 bp shows PEV 0.550 cm for CLGS and the typical carrier is Changdaotou, and RM3754-85 bp shows PEV 0.519 cm and typical carrier Zhenghan2. In 2017, positive AAE value of RM3754 was 0.155 cm and 0.211 cm for CL and CLGS, respectively; and the result was similar in 2018.
Based on PEV of marker-alleles which have positive effects, the best parental combinations were selected from the top 20 accessions for CLGS (Table 15). Seven parental combinations were predicted to improve CLGS ranged from 0.814 to 0.922 cm. Among all, Changdaotou and Hongdao35 were selected before as superior accessions.
SSR Marker Loci Associated with GSI, Favorable Alleles and Their Carrier Accessions
Seventeen SSR loci for GSI were detected using GLM and MLM model in years 2017 and in 2018 with PVE more than 10%. The 17 SSR marker loci were distributed on all chromosomes except chromosome 7 and 8 (Table 16). The range of PVE was from 10.19% (RM112 on chromosome 2) to 36.69% (RM297 on chromosome 1) in 2017 and the results were similar in 2018.
Table 17 shows the top 29 positive favorable alleles of the significant association loci with PEV more than 0.1 cm/cm and their typical carrier accessions for GSI in years 2017 and in 2018. The PEV for those alleles ranged from 0.100 cm/cm for RM6869-125pb (typical carrier accession Gaoliangqing) to 0.270 cm/cm for RM6869-110pb (typical carrier accession Yangdao).
Two markers showed positive average allele effect (AAE), without negative allele effect (PVE more than 10%); RM304 was the highest one with AAE 0.212 cm/cm, followed by RM297 with AAE 0.105 cm/cm. Among 17 markers, the marker RM304 was showing the highest positively AAE (Table 18).
The results showed that 7 markers were associated with both GSI and CL traits. Among all positive favorable alleles, RM3513-80 bp shows phenotypic effect value 0.616 cm for CL and the typical carrier is Haidongqing, and the same marker allele shows phenotypic effect value 0.104 cm/cm for GSI and the typical carrier is Gaoliangqing.
Furthermore, we found that RM1182 was associated with both GSI and CLGS traits. Among all positive alleles, RM1182-145 bp showed phenotypic effect value 0.729 cm for CLGS and the typical carrier was Maijieqing. RM1182-150 bp showed phenotypic effect value 0.578 cm for CLGS and the typical carrier was Shuangchengnuo. While RM1182-165 bp showed phenotypic effect value 0.106 cm/cm for GSI and the typical carrier was Wuxiangjing14.
Based on PEV of marker-alleles which have positive effects on GSI, the best parental combinations were selected from the top 20 accessions for GSI Seven parental combinations were predicted to improve GSI from 0.154 to 0.160 cm/cm (Table 19).
Comparing the parental combination accessions which selected for CL, CLGS and GSI, the accession Hongdao35 had been found to share in both CLGS and GSI parental combinations; also it was one of the superior accessions. In addition to, all parental combination accessions selected were temperate japonica; and these accessions were categorized under three subpopulations Sp2, Sp3 and SP5.
Discussion
Treating the seeds with gibberellic acid can enhance the coleoptile elongation length under submergence condition, which is considered as key of survival under anoxic conditions for water direct-seeded rice (Gubler et al. 2002; Kaneko et al. 2002; Lee et al. 2014; Mutinda et al. 2017).
In this study, there were great variations in the traits under investigation. The mean value for CL ranged from 0.82 to 3.82 cm in 2017, as well as 2018, the same result was obtained by Hsu and Tung (2015). While mean value for CLGS ranged from 1.25 to 4.76 cm 2017 and the results were similar in 2018, which is consistent with the results of others (Guadagnin et al. 2017; Mutinda et al. 2017). Furthermore, there was a positive correlation between CLGS with GSI, moreover, a wide range for GSI indicating the existence of genotype sensitivity. Hence, the broad-sense heritability was higher than 90% in both years for CL and CLGS, which means that the genetic effect is mainly controlling both CL and CLGS comparing to the environmental effect (Visscher et al. 2008).
The six superior accessions were found in this study belonging to temperate japonica. Previous studies reported that coleoptile performance of temperate japonica varieties (as sub-species of japonica varieties) was better than indica varieties under anaerobic conditions (Lasanthi-Kudahettige et al. 2007; Hsu and Tung 2015).
The AMOVA results (46.16% genetic variability among subpopulations and 53.84% within subpopulations) revealed that the rice genotypes under our study highly variable and suitable for conducting association mapping as demonstrated in previous studies (Adeyemo et al. 2005; Agrama and Eizenga 2008; Jaiswal et al. 2012; Bergamaschi and Lama 2015). These accessions probably had a complex breeding history involving intercrossing and introgression between germplasm from diverse backgrounds, overlaid with strong selection pressure for agronomic and quality characteristics (Mather et al. 2004).
In association mapping, the LD used is present in the germplasm set under study. As well, LD might not only be influenced by recombination but also by various other forces (Flint-Garcia et al. 2003). Contrasting to the previous studies, LD was decaying in our study at more than 70 cM, this can be attributed due to outcrossing and recombination events that have been used in breeding programs (Garris et al. 2003; Lu et al. 2005; Olsen et al. 2006; Dang et al. 2014).
Association mapping is a very prevalent method for the explanation of the genetic basis of complex traits in plants. Different statistical approaches had been designed to deal with the superior marker-phenotype association that could be caused by the population structure. GLM depends only on Q matrix generated during the study of population structure while MLM accounts for both population structure and the kinship. Generally GLM will detect a higher number of significant marker-trait associations than in MLM; while MLM is more accurate in claiming associations than GLM (Korte and Farlow 2013). QQ plots (GLM & MLM) were generated to demonstrate that population structure is only controlling the confounding factors that could bias the results (Wei et al 2017) as shown in Fig. 4.
For coleoptile elongation length under control treatment (CL), RM6327 on chromosome11 explained the maximum phenotypic variation, 16 accessions out of 358 (4.47%) showed an excellent alleles RM6327-215 bp, with the largest phenotypic effect values (1.609 cm in 2017 and 1.578 cm in 2018) and the typical carrier accession was Wanqu428.
Exogenous GA3 plays an important role in rice coleoptile elongation under submergence, anoxia or hypoxia (Kota- Noguchi et al. 2008). In this study, for CLGS, RM 562 on chromosome 1 explained the maximum phenotypic value, 14 accessions out of 358 (3.92%) possesses the excellent alleles RM562-180 bp; with the largest phenotypic effect (1.087 cm in both 2017 and 2018) and the typical carrier accession was Xiaoqingmang.
The differences detected between GSI and CLGS verified that they are functioned differently (Zhang et al. 2017). This result indicated that GSI explained the different genetic mechanisms of coleoptile treated with GA3 under anoxic condition. Additionally, RM297 was detected with the highest PVE (36.69% and 34.72% in 2017 and in 2018, respectively), indicated that this was chromosome segment controlling GSI and considered as a promising marker which can increase GSI. Wang et al. (2012) and Zhao et al. (2017) had been detected similar results with PVE value exceeded 20%.
Pleiotropy is the well-established phenomenon of a single gene affecting multiple traits. It has long played a central role in theoretical, experimental, and clinical research in genetics, development, molecular biology, evolution, and medicine (Paaby and Rockman 2012). Seven markers were detected in this study to have a pleiotropic effect for CL and CLGS. A similar result had been found in wheat by Chai et al. (2019); while Ookawa et al. (2010) used the pleiotropy phenomenon for improving rice lodging resistance and yield.
Taking all together, 24, 22 and 17 marker loci were associated significantly with CL, CLGS and GSI. Among all, 34 marker loci were novel in current study; the rest (20) of marker loci were detected in previous studies (Itoh et al. 2001; Kikuchi et al. 2003; Sakai et al. 2003; Toojinda et al. 2003; Huang et al. 2003; Jan et al. 2004; Ling et al. 2004; Angaji 2008; Lo et al. 2008; Narsai et al. 2009; Angaji et al. 2010; Magome et al. 2013; Septiningsih et al. 2013; Hsu and Tung 2015; Tomita and Ishii 2018).
Improving rice coleoptile length under anaerobic condition, all favorable alleles might be pyramided as much as possible into one variety. Crosses between accessions which have favorable alleles (as hybridization parents) should improve target trait. Pyramiding best favorable alleles into new cultivar might need multi round crossing (Cheng et al. 2015). The results of this study provided basic marker information and accession information for breeding cultivars suitable for anaerobic conditions (water direct-seeded rice).
In conclusion, there is a phenotypic variation for coleoptile length under control treatment (CL), coleoptile length under GA3 treatment (CLGS) and molecular marker allele diversity among 358 accessions. Twenty four markers loci significantly associated with CL and 22 markers loci associated with CLGS (PVE > 7%). Thirty nine favorable alleles for CL and 56 favorable alleles for CLGS (PEV > 0.5 cm) were detected across two years by GLM and MLM analysis models. While, 17 markers loci significantly associated with GSI (PVE > 10%), with 29 favorable alleles were detected across two years by GLM and MLM analysis models. Twelve, thirteen and twenty three typical carrier accessions for CL, CLGS and GSI, respectively, possessing the favorable alleles could be used to improve those traits under anoxic condition.
References
Adeyemo AA, Chen G, Chen Y, Rotimi C (2005) Genetic structure in four West African population groups. BMC Genet 6:38. https://doi.org/10.1186/1471-2156-6-38
Agrama HA, Eizenga GC (2008) Molecular diversity and genome-wide linkage disequilibrium patterns in a worldwide collection of Oryza sativa and its wild relatives. Euphytica 160:339–355. https://doi.org/10.1007/s10681-007-9535-y
Angaji S (2008) Mapping QTLs for submergence tolerance during germination in rice. Afr J Biotechnol 15:2551–2558
Angaji SA, Septiningsih EM, Mackill DJ, Ismail AM (2010) QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172:159–168. https://doi.org/10.1007/s10681-009-0014-5
Baltazar MD, Ignacio JCI, Thomson MJ, Ismail AM, Mendioro MS, Septiningsih EM (2014) QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica 197:251–260. https://doi.org/10.1007/s10681-014-1064-x
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300
Bergamaschi ACB, Lama MAD (2015) Gene variation and genetic differentiation among populations of the solitary mud dauber wasp Trypoxylon (Trypargilum) albitarse Fabricius 1804 (Hymenoptera, Crabronidae). Genet Mol Biol 4:519–526. https://doi.org/10.1590/S1415-475738420150097
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Chai L, Chen Z, Bain R et al (2019) Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor Appl Genet. https://doi.org/10.1007/s00122-019-03318-z
Cheng J, Cheng X, Wang L, He Y, An C, Wang Z et al (2015) Physiological characteristics of seed reserve utilization during the early seedling growth in rice. Braz J Bot 38(4):751–759
Dang X, Thi TGT, Dong G, Wang H, Edzesi WM, Hong D (2014) Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 239:1309–1319. https://doi.org/10.1007/s00425-014-2060-z
Dang X, Li Y, Zhang Y et al (2019) Identification of favorable alleles for rice seedling anoxic tolerance using natural and bi-parental populations. Euphytica 215(8):140. https://doi.org/10.1007/s10681-019-2463-9
Doley D, Barua M, Sarma D, Batua PK (2018) Screening and enhancement of anaerobic germination of rice genotypes by pre-sowing seed treatments. Curr Sci 115:1185–1190. https://doi.org/10.18520/cs/v115/i6/1185-1190
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54(1):357–374
Garris A, McCouch SR, Kresovich S (2003) Population structure and its eVect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165:759–769
Guadagnin CMI, Schuch LOB, Venske E, Zimmer PD, Aumonde TZ (2017) Seedling growth of irrigated rice as a function of seed treatment with gibberellic acid. Sci Agrar Parana 16:237–245. https://doi.org/10.18188/1983-1471/sap.v16n1p237-245
Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV (2002) Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol 129:191–200. https://doi.org/10.1104/pp.010918
Hardke J, Scott B (2013) Water-seeded rice. In: Hardke J (ed) Arkansas rice production handbook, 4th edn. University of Arkansas Division of Agriculture, Arkansas, pp 41–44
Hedrick PW (1987) Gametic diequilibrium measures: proceed with caution. Genetics 117:331–341
Huang SB, Greenway H, Colmer TD (2003) Anoxia tolerance in rice seedlings: exogenous glucose improves growth of an anoxia-’intolerant’, but not of a ‘tolerant’ genotype. J Exp Bot 54:2363–2373
Jiang L, Hou M, Wang C, Wan J (2004) Quantitative trait loci and epistatic analysis for seed anoxia germinability in rice (Oryza sativa). Rice Sci 11(5–6):238–244
Hsu SK, Tung CW (2015) Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice. Rice 8:38. https://doi.org/10.1186/s12284-015-0072-3
IRRI (2016) Climate-smart rice. https://books.irri.org/Smart_rice_brochure.pdf. Accessed 8 March 2019
Itoh H, Ueguchi M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M (2001) Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. PNAS 98:8909–8914. https://doi.org/10.1073/pnas.141239398
Jaiswal V, Mir RR, Mohan A, Balyan HS, Gupta PK (2012) Association mapping for pre-harvest sprouting tolerance in common wheat (Triticum aestivum L.). Euphytica 188:89–102. https://doi.org/10.1007/s10681-012-0713-1
Jan A, Yang G, Nakamura H et al (2004) Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol 136:3670–3681. https://doi.org/10.1104/pp.104.052274
Kaneko M, Itoh H, Ueguchi M, Ashikari M, Matsuoka M (2002) The α-Amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol 128:1264–1270. https://doi.org/10.1104/pp.010785
Kikuchi S, Satoh K, Nagata T et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from Japonica Rice. Science 301:376–379. https://doi.org/10.1126/science.1081288
Kim SM, Reinke RF (2018) Identification of QTLs for tolerance to hypoxia during germination in rice. Euphytica 214:160–170. https://doi.org/10.1007/s10681-018-2238-8
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review plant. Methods 9:29. https://doi.org/10.1186/1746-4811-9-29
Kota- Noguchi H, Sasaki R, Yasuda Y (2008) Anoxia tolerance and a-amylase activity in four rice cultivars. Plant Growth Regul 55:35–41. https://doi.org/10.1007/s10725-008-9255-x
Lasanthi-Kudahettige R, Magneschi L, Loreti E et al (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144:218–231. https://doi.org/10.1104/pp.106.093997
Lee KW, Chen PW, Yu SM (2014) Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant, Cell Environ 37:2234–2244. https://doi.org/10.1111/pce.12311
Lewontin RC (1964) The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49:49–67
Ling J, Hou M, Wang C, Wan J (2004) Quantitative trait loci and epistatic analysis for seed anoxia germinability in rice (Oryza sativa). Rice Sci 11(5–6):238–244
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Lo SF, Yang SY, Chen KT et al (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20:2603–2618. https://doi.org/10.1105/tpc.108.060913
Lu H, Redus MA, Coburn JR, Rutger JN, McCouch SR, Tai TH (2005) Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR marker analysis. Crop Sci 45:66–76
Magome H, Nomura T, Hanada A et al (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. PNAS 110:1947–1952. https://doi.org/10.1073/pnas.1215788110
Mather DE, Hyes PM, Chalmers KJ, et al (2004) Use of SSR marker data to study linkage disequilibrium and population structure in Hordeum vulgare: prospects for association mapping in barley. In: International barley genetics symposium, Brno, Czech Republic, 20–26 June 2004. pp. 302–307.
McCouch SR, Teytelman L, Xu Y et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207. https://doi.org/10.1093/dnares/9.6.199
Mulbah Q, Adjetey J (2018) Effect of water seed priming on establishment of direct seeded rice in well watered conditions and aerenchyma formation under varying water regimes. J Agric Sci 40(1):45–54. https://doi.org/10.17503/agrivita.v40i1.1457
Mutinda YA, Muthomi JW, Kimani JM, Cheminigwwa GN, Olubayo FM (2017) Viability and dormancy of rice seeds after storage and pre-treatment with dry heat and chemical agents. J Agric Sci 9:175–185. https://doi.org/10.5539/jas.v9n7p175
Narsai R, Katharine AH, Adam C, Aneta I, Millar HA, James W (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151:306–322. https://doi.org/10.1104/pp.109.142026
Nei M, Tajima F, Tajima F, Tateno Y (1982) Accuracy of estimated phylogenetic trees from molecular data II. J Mol Evol 18(6):387–404
Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD (2006) Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173:975–983. https://doi.org/10.1534/genetics.106.056473
Ookawa T, Hobo T, Yano M et al (2010) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1:132. https://doi.org/10.1038/ncomms1132
Paaby AB, Rockman MV (2012) The many faces of pleiotropy. Trends Genet 29(2):66–73. https://doi.org/10.1016/j.tig.2012.10.010
Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M (2003) Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res 116:161–164. https://doi.org/10.1007/s10265-003-0080-z
Sarkar RK (2012) Seed priming improves agronomic trait performance under flooding and non-flooding conditions in rice with QTL SUB1. Rice Sci 19:286–294. https://doi.org/10.1016/S1672-6308(12)60053-5
Senapati S, Kuanar SR, Sarkar RK (2019) Improvement in anaerobic germination potential and grain yield of rice (Oryza sativa) through seed priming. SAARC J Agric 17:37–48. https://doi.org/10.3329/sja.v17i1.42760
Septiningsih EM, Ignacio JCI, Sendon PMD, Sanchez DL, Ismail AM, Mackill DJ (2013) QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor Appl Genet 126:1357–1366. https://doi.org/10.1007/s00122-013-2057-1
Silva VN, Silva JR (2016) Rice seed priming. Biosci J 32:89–97. https://doi.org/10.14393/BJ-v32n1a2016-29610
Singh A, Septiningsih EM, Balyan HS, Singh NK, Rai V (2017) Genetics, physiological mechanisms and breeding of flood-tolerant rice (Oryza sativa L.). Plant Cell Physiol 58:185–197. https://doi.org/10.1093/pcp/pcw206
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. https://doi.org/10.1093/molbev/msm092
Temnykh S, Park WD, Ayres N et al (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712. https://doi.org/10.1007/s001220051342
Tomita M, Ishii K (2018) Genetic performance of the semidwarfing allele sd1 derived from a Japonica rice cultivar and minimum requirements to detect its single-nucleotide polymorphism by MiSeq whole-genome sequencing. Biomed Res Int 2018:7. https://doi.org/10.1155/2018/4241725
Toojinda T, Siangliw M, Tragoonrung S, Vanavichit A (2003) Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann Bot 91:243–253. https://doi.org/10.1093/aob/mcf072
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55. https://doi.org/10.1016/j.tibtech.2004.11.005
Visscher PM, Hill WG, Wray NR (2008) Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet 9(4):255–266. https://doi.org/10.1038/nrg2322
Wang LQ, Liu WJ, Xu Y et al (2007) Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor Appl Genet 115:463–476. https://doi.org/10.1007/s00122-007-0580-7
Wang Z, Yu C, Liu X et al (2012) Identification of Indica rice chromosome segments for the improvement of Japonica inbreds and hybrids. Theor Appl Genet 124(7):1351–1364. https://doi.org/10.1007/s00122-012-1792-z
Weaver B, Wuensch KL (2013) SPSS and SAS programs for comparing pearson correlations and OLS regression coefficients. Behav Res 45:880–895. https://doi.org/10.3758/s13428-012-0289-7
Wei W, Mesquita ACO, Figueiró AA et al (2017) Genome-wide association mapping of resistance to a Brazilian isolate of Sclerotinia sclerotiorum in soybean genotypes mostly from Brazil. BMC Genomics 18:849–864. https://doi.org/10.1186/s12864-017-4160-1
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Weir BS, Hill WG (2002) Estimating F-statistics. Annu Rev Genet 36:721–750. https://doi.org/10.1146/annurev.genet.36.050802.093940
Zhang M, Lu Q, Wu W et al (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in Indica rice. Front Plant Sci 8:678–678. https://doi.org/10.3389/fpls.2017.00678
Zhao J, Huang L, Ren R et al (2017) Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) using single-locus simple sequence repeat markers. Front Plant Sci 8:2105. https://doi.org/10.3389/fpls.2017.02105
Acknowledgements
We are grateful to thank the National Natural Science Foundation of China for funding support (31671658). Also, we would like to thank Zia Ul-Haq (PhD student), Muhammed Fiaz (PhD student) and undergraduate students for their help in the experimental field.
Author information
Authors and Affiliations
Contributions
DH, DA designed the research; DA, NA and MSE carried out the field experiment; DA, EL and DX carried out the molecular experiment; DA, NA analyzed data; and DA wrote the manuscript; DH revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest among authors and in the research work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
344_2020_10196_MOESM1_ESM.jpg
Supplementary file1 (JPG 4521 kb) Supplementary Fig. 1 Plant growth Part of procedure for germination in submerged conditions; a) dry seeds and label showing variety code and treatment; b) seeds wrapped by tissue paper were put into enamel tray containing water or 2000 ppm GA3 solution for 24h; c) the soaked seeds were lined on a dry filter paper; d) the soaked seeds were rolled by filter papers and put into plastic boxes (44 cm × 31 cm × 15 cm) keeping 10 cm depth of standing tap water for 14 days.
344_2020_10196_MOESM2_ESM.jpg
Supplementary file2 (JPG 1750 kb) Supplementary Fig. 2 Population genetic architecture analysis of the 358 accessions studied; a) changes of log-likelihood value with increase of subpopulation number; b) changes of ∆K values with increase of subpopulation number; c) model based population structure of 358 accessions belonging to six subpopulations; each accession is represented by a vertical bar. The colored subsections within each vertical bar indicate membership coefficient (Q) of the accession to different clusters; d) the neighbor-joining tree for 358 accessions based on Nei’s genetic distance.
344_2020_10196_MOESM3_ESM.jpg
Supplementary file3 (JPG 1836 kb) Supplementary Fig. 3 Relationship between D′ value and genetic distance of syntenic marker pairs in the 6 subpopulations
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdulmajid, D., Ali, N., Eltahawy, M.S. et al. Mining Favorable Alleles for Rice Coleoptile Elongation Length Sensitivity to Exogenous Gibberellin Under Submergence Condition. J Plant Growth Regul 40, 1422–1439 (2021). https://doi.org/10.1007/s00344-020-10196-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10196-z