Abstract
The endemic Antarctic nematode Scottnema lindsayae is described from specimens collected in Taylor Valley, McMurdo Dry Valleys, Victoria Land. The recently collected material is compared with the original description and other subsequent descriptions of the species. A more complete scanning electron microscopy (SEM) study of the species is presented. The phylogenetic position of S. lindsayae is inferred using a secondary structure-based alignment of a partial sequence of nuclear Large Subunit (LSU) ribosomal DNA. Phylogenetic trees were inferred using base-paired substitution models implemented in PHASE 2 software and Bayesian inference, and show S. lindsayae as the sister group to Stegelletina taxa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The endemic nematode Scottnema lindsayae Timm, 1971 is one of only a few soil nematode species found in the extreme environmental conditions of the Antarctic Dry Valleys, South Victoria Land (77–78°S). It has also been reported from the Syowa Station in East Antarctica (~71°S), on the other side of the continent, by Shishida and Ohyama (1986), from the Antarctic Peninsula (~62°S) by Mouratov et al. (2001), and recently as far south as the Transantarctic Mountains (~83°S) by Adams et al. (2007). Its life history, spatial distribution (horizontal and vertical), dispersal and different aspects of its ecology have been investigated in several studies (e.g. Overhoff et al. 1993; Powers et al. 1995; Treonis et al. 1999; Courtright et al. 2001; Porazinska et al. 2002a, b; Moorhead et al. 2002, 2003; Gooseff et al. 2003; Weicht and Moorhead 2004; Nkem et al. 2006a, b; Adams et al. 2007; Ayres et al. 2008, 2010; Barrett et al. 2008; Simmons et al. 2009). The species was originally described by Timm (1971) from a locality near La Croix Glacier, Taylor Valley in Victoria Land. He also reported presence of the species at Ross Island and several other localities in Taylor Valley. Other populations from Ross Island and from many localities in Victoria Land have subsequently been characterized by morphological (Andrássy 1998; Vinciguerra 1994) and molecular methods (Courtright et al. 2000). Courtright et al. (2000) did not observe any fixed differences in nuclear rRNA sequences between populations, but found variation in mitochondrial DNA sequences defining several haplotypes. They concluded that: “The nuclear and mitochondrial variation suggests populations of S. lindsayae represent a single polymorphic species with some restriction of gene flow between geographic populations.”
Scottnema lindsayae is a microbivorous nematode feeding on bacteria, yeast and algal cells. It has been found in several types of habitats: sand or mineral soils with mixed fractions of sand, gravel and stones; moss; green algal mats (e.g. Prasiola); and lake bottom detritus. It has a comparatively long life cycle (218 days at 10°C), indicating that more than one austral summer is required for its completion, and a low fecundity in comparison with many other genera of Cephalobidae (Overhoff et al. 1993). During periods of extreme temperature and/or desiccation, the nematodes will survive by anhydrobiosis entering a cryptobiotic stage (Treonis et al. 2000; Weicht and Moorhead 2004; Treonis and Wall 2005).
In this paper, a population of S. lindsayae from Taylor Valley (77°S) is described by using light (LM) and scanning electron microscopy (SEM) and compared with the original and subsequent descriptions of the species. The phylogenetic position of S. lindsayae is inferred from partial Large Subunit (LSU) nuclear ribosomal DNA sequences.
Methods
Samples were collected by Olle Karlsson on 19 January 2009 in Taylor Valley, Victoria Land, close to Lake Hoare and Lake Chad. The area is characterized by a moraine with sand, gravel and larger blocks of stone, with permafrost beginning at 35–50 cm depth. The samples were collected in the area 77°37.6′S, 162°52.1′E to 77°38.6′S, 162°44.2′E and generally consisted of dry or somewhat moist mixed fractions of sand, coarse sand and gravel.
Specimens were recovered from soil samples using a modified Baermann funnel extraction technique, where the individual samples weighed between 11 and 24 gram dry weight (gdw). The nematodes were relaxed by gentle heat and fixed in cold TAF for morphological studies and in DESS (Yoder et al. 2006) for molecular analysis. For LM, nematodes were transferred to pure glycerine by a slow evaporation method and mounted on permanent slides in glycerine with paraffin wax as support for the coverslip. Measurements are given for individual specimens or a range of specimens, as indicated. For SEM studies, some specimens were postfixed in 1% osmium tetroxide (OsO4) and transferred to pure acetone through an acetone/distilled water series. Specimens were critical point dried in liquid CO2, mounted on stubs, gold-plated under vacuum to a thickness of 200 Å in a sputter coater, and examined in a Hitachi S-4300 SEM at an accelerating voltage of 10 kV. DESS-preserved specimens were used for DNA extraction and PCR as described in Holovachov et al. (2009b) and deposited in GenBank (Accession #HM439773 and HM439774). Eight individual nematodes from each sample were processed in bulk for PCR and sequencing. Sequences generated from specimens from two samples showed no evidence of polymorphism and were identical to each other. Phylogenetic analyses were performed in a framework of selected sequence data (see Table 1) available in public databases for Cephaloboidea and selected representatives from the order Rhabditida sensu De Ley and Blaxter (2004), using a secondary structure-based alignment created as described in Holovachov et al. (2009b) with 4SALE software (Seibel et al. 2006). Furthermore, eight new cephalobid sequences were added to the dataset, from our most recent sampling in Sweden and southern California desert habitats (Table 2). These species were also preserved in DESS and processed in the same way for PCR and sequencing as Scottnema lindsayae. The resulting alignment was analysed with Bayesian phylogenetic inference using the mcmcphase program in the PHASE package (Gowri-Shankar and Jow 2006; Version 2.0). For this analysis, the dataset was partitioned into “stems” (paired sites) and “loops” (non-paired sites) to account for the potential phylogenetic importance of compensatory substitutions. The REV nucleotide substitution model (Tavare 1986) was used for non-paired sites, whereas RNA7A (Higgs 2000), RNA7D (Tillier and Collins 1998) and RNA16A (Gowri-Shankar and Jow 2006) nucleotide substitution models were used for paired sites in three independent analyses. For each model combination, we performed two independent runs; and for each run, chains were allowed to burn in for 500,000 generations, followed by 5 million generations (total 5.5 million generations) during which tree topologies, branch length and model parameters were sampled every 200 generations.
Results
Of the 11 samples collected, two were devoid of metazoans. Nine samples contained S. lindsayae and in two of them single specimens of Eudorylaimus were also found. One sample contained a single tardigrade of the genus Macrobiotus. In the samples where Scottnema was found, the mean number of specimens varied between 0.05 and 10.2 per gram dry weight (gdw) of the extracted sample. Samples with few nematodes contained only females and juveniles, whilst those with higher densities appeared to be active growing populations with females, males and juveniles. The abundance of nematodes was consistent with that recorded by Courtright et al. (2001) from Taylor Valley. They found 2.1–8.6 specimens per gdw when using a sugar centrifugation extraction method with individual samples of 100 g soil.
In many specimens, a string of mineral material that appeared to be stuck together was observed in the intestine, from just behind the cardia almost to rectum. Timm (1971) also recorded that the intestine was filled with fine detritus.
Scottnema lindsayae Timm, 1971 (Figs. 1, 2, 3, 4).
Scottnema lindsayae Timm, 1971. a Male tail, b Female tail, c Anterior end, surface view, d Pharyngeal region, e Female reproductive system, anterior part. Scale bar 20 μm
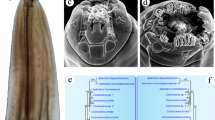
Scottnema lindsayae Timm, 1971. a Anterior end, oblique view (ventral on left side down), b anterior end, lateral view (ventral on right side), c anterior end, lateral view (ventral on left side), d anterior end, lateral view (ventral on right side), e anterior end, apical view (ventral up), f anterior end, oblique view (ventral on right side). Scale bars 10 μm (a, c–f), 5 μm (b)
Scottnema lindsayae Timm, 1971. a–b Vulval region, ventral view, c Excretory pore, d Lateral field and deirid, e Vulval region, lateral view, f Postvulval dorsosublateral papilla (arrowheads in e and f point at papilla). Scale bars 10 μm (a–c), 20 μm (d, e), 5 μm (f)
Scottnema lindsayae Timm, 1971. a Male tail, subventral view, b Female tail, subventral view, c Female tail, ventral view, d Male posterior end, lateral view, e Male tail, lateral view, f Male tail terminus, subdorsal view. Scale bars 20 μm (a–c, e), 50 μm (d), 10 μm (f)
Measurements
See Table 3.
Description
Adult
Body straight to slightly arcuate ventrad in females, ventrally curved posteriorly in males. Cuticle strongly annulated, annuli 1.9–2.8 μm wide at midbody, 3.2–3.8 μm anteriorly. Fine longitudinal striae give the cuticle a tiled appearance. Lateral field consisting of two alae, i.e. three incisures of which the outer two are crenate; occupying about 15–20% of body diameter. Lip region with six labial and four cephalic papillae. Amphid openings rounded, at base of lateral lips. Lip region strongly offset, consisting of six asymmetrical, flattened lips arranged in three pairs: one dorsal and two subventrals. Pairs of lips separated by primary axils with two triangular and elongate-pointed guarding processes originating from the base of the lip region; secondary axils demarcated by incisures separating the two lips of each pair. Cephalic probolae with four tines: one short tine flanking the primary axils; one long slender process, 8–10 μm long from lip surface, directed forward or somewhat inwards; one two-pointed (biacute) tine surrounding the labial papilla; and one triangular-pointed tine in the secondary axils. Three short, broadly triangular and flattened labial probolae, joined basally and separated by U-shaped indentations. Stoma 1.2–1.4 times as long as lip region diameter, divided into cheilostom, gymnostom and stegostom. Cheilostom wide with narrow, curved, grain-like rhabdia; gymnostom short and funnel-shaped with weakly sclerotized rhabdia; stegostom with well-developed rhabdia divided into: prostegostom constituting about half of stegostom length, and meso-, meta- and telostegostom constituting the other half of stegostom length; metastegostom with a dorsal denticle. Pharynx cephaloboid. Pharyngeal procorpus cylindrical; isthmus narrower than metacorpus, often clearly delineated from it by a break in the musculature. Basal pharyngeal bulb oval, with strongly developed valves. Cardia distinct, conoid, enveloped by intestinal tissue. Nerve ring encircling isthmus; excretory pore opens posterior to nerve ring; deirid at level of isthmus-bulb junction or at level with bulb.
Female
Reproductive system monodelphic, prodelphic; ovary branch antidromously reflexed, generally on right-hand side of intestine (dextral). Part of ovary posterior to vulva straight. Spermatheca offset, directed anteriorly between oviduct and uterus, generally filled with rounded spermatozoa. Vulva a transverse slit with slightly protruding lips, vulval opening located in a somewhat left-ventrosublateral position. A pair of dorsosublateral papillae present about 1.5 body diameters posterior to vulva. Postvulval uterine sac moderately developed, generally shorter than one vulval body diameter long. Vagina straight. Rectum 1.1–1.7 times longer than anal body diameter. Phasmids 16–23 μm posterior to anal opening, at 40–49% of tail length. Tail conoid with 19–24 ventral annuli, tapering to a pointed terminus.
Male
Reproductive system monorchic; testis generally on right-hand side of intestine (dextral), reflexed anteriorly, reflexed part 48–77 μm long; a ventral reflex was observed in 60% of the studied specimens, but ventro-lateral, ventro-dorsal (“pretzel”-shaped), dorsal and dorso-lateral reflexes were also observed. Spicules cephaloboid, paired and symmetrical, curved ventrad; with narrow rounded manubrium and conoid shaft. Gubernaculum wedge-shaped. Tail conoid, arcuate ventrad, tapering to a pointed terminus, terminal part constituting a 7- to 9-μm-long “spike”. All three lateral incisures extend posteriorly to lateral papillae at midtail and the two outer incisures extend almost to tail terminus. Phasmid located 21–27 μm posterior to cloacal opening, at 39–56% of tail length. Male genital papillae arranged as follows: three pairs of ventrosublateral precloacal papillae (one pair adcloacal, one pair at about 40–60 μm and one pair at about 80–105 μm anterior to cloaca); a single midventral papilla on anterior cloacal lip, two pairs at midtail (one subventral and one lateral) and three pairs at base of “spike” (one subventral, one lateral and one subdorsal).
Taxonomic remarks
The specimens described agree well in general morphology with the original description of S. lindsayae by Timm (1971) and subsequent descriptions by Vinciguerra (1994) and Andrássy (1998). A comparison of the ranges of some measurements and ratios among the specimens described by these authors and those described here is made in Table 4. The comparison is somewhat restricted because many measurements made in this study were not recorded in all previous descriptions and also the number of specimens measured varies. The specimens described by Timm (1971) and Vinciguerra (1994) are larger than those described by Andrássy (1998) and in this study (L = 0.72–0.86 mm vs. L = 0.57–0.73 mm for females and L = 0.64–0.88 mm vs. 0.54–0.73 mm for males, respectively), although there is some overlap of the ranges. The total length of the specimens will consequently affect some other measurements and ratios. The somewhat right-ventrosublaterally displaced position of vulval opening was not mentioned in any previous description and might not be evident unless specimens are studied by SEM. Timm (1971) recorded a pair of dorsosublateral papillae present somewhat posterior to vulva, but they were not mentioned by Vinciguerra (1994) or Andrássy (1998). Although the papillae are difficult to observe, their presence could be confirmed both by LM and by SEM (Fig. 3e–f) in this study. The number and position of male genital papillae and position of phasmid agree well with the original description (Timm 1971). Andrássy (1998), apart from the adcloacal pair (considered as caudal papillae), recorded two or three pairs of ventrosublateral precloacal papillae (one pair at 41–43 μm, one pair at 75–98 μm and one pair at 140–170 μm anterior to cloaca), of which the most posterior pair is not always present.
Phylogenetic analysis
The phylogenetic relationships of the Scottnema lindsayae isolates were inferred from the secondary structure-based alignment of the 5′ end (D1–D3) region of nuclear large subunit rDNA sequences. Bayesian inference was performed using three different substitution models that treated non-paired (“loops”) and paired (“stems”) sites differently. Two independent mcmc analyses were conducted for each of the three models. Bayesian posterior probabilities and tree topologies were produced by constructing majority-rule consensus trees after discarding trees from the burn in phase. Tree topologies from the two independent “runs” of each were identical, but clade posterior probabilities varied up to 5% between runs. All three models yielded identical results regarding the sister-taxon relationship of S. lindsayae; the Bayesian consensus tree based on the RNA16A model showed slightly greater overall resolution and was therefore selected for presentation (Fig. 5) and discussion. Bayesian consensus tree topologies were identical for the RNA7A and RNA7D models, whereas the topology recovered for the RNA16A model was different with respect to the position of two species. First, with the RNA16A model, Penjatinema novaezeelandiae Holovachov, Boström, Tandingan De Ley, Nadler & De Ley, 2009 was the sister taxon to the clade composed of Cervidellus sp. JB138 and Nothacrobeles borregi Poiras, Baldwin, Mundo-Ocampo & Bumbarger, 2002 (PP 67%). In contrast, when using the RNA7A and RNA7D models, this part of the phylogeny was unresolved. The second difference involved the position of Placodira lobata Thorne, 1937. With the RNA16A model, P. lobata was the sister taxon to a clade composed of Zeldia spannata Waceke, Bumbarger, Mundo-Ocampo, Subbotin & Baldwin, 2005 and Chiloplacus sp. JB81 (PP 50%). In contrast, for the RNA7A and RNA7D models, this part of the phylogeny was unresolved. For all three substitution models, Bayesian analyses recovered S. lindsayae as a monophyletic sister group to the genus Stegelletina Andrássy, 1984 with higher posterior probability; maximum likelihood analysis yielded the same result with high bootstrap support (Fig. 5). This clade of Scottnema plus Stegelletina is nested within the “grade” of taxa with complex labial morphology. A separate maximum likelihood (ML) analysis of the same alignment, but without stem-loop partitioning was implemented using RAxML (Stamatakis et al. 2008). The resulting ML bootstrap tree showed low support for many clades, including several clades that received moderate to high posterior probabilities in the Bayesian analyses. Although there was topological agreement between certain clades in the ML bootstrap and Bayesian analyses (Fig. 5), many other nodes in the Bayesian tree were not resolved in the ML bootstrap (50% majority-rule) consensus tree. By comparison, using a partitioned dataset and base-paired substitution models in a Bayesian context provided greater resolution, although Bayesian posterior probabilities and bootstrap values are not directly comparable (Alfaro et al. 2003). In addition, using a more complex sixteen-state model (RNA16A) produced only a slightly different phylogenetic hypothesis compared to less complex seven-state models (RNA7A and RNA7D). The major difference between seven-state and sixteen-state models is how they “treat” non-complementary pairs (“mismatches”) in stem regions of RNA. We conclude (and visual inspection of the alignment confirms) that the number of “mismatches” in paired sites (“stems”) is too small for these different models to differentially affect the phylogenetic analysis. Previous studies showed that neither using more of the LSU rDNA gene (Nadler et al. 2006; Smythe and Nadler 2007), or using more taxa but shorter LSU rDNA sequences (Holovachov et al. 2010) are sufficient to satisfactorily resolve the phylogenetic relationships within Cephalobidae, and additional phylogenetically informative genes will be required to achieve this goal.
Phylogenetic position of Scottnema lindsayae Timm, 1971 within the family Cephalobidae. Majority-rule consensus tree of the Bayesian phylogenetic analysis (RNA16A model) of Cephalobomorpha LSU rDNA (D1–D3 region). Tree rooted using Geraldius sp. as the outgroup; branch lengths represent the mean posterior estimates of the expected number of substitutions per site. Bayesian posterior probabilities (BI) and bootstrap support (ML) are shown for each branch in the following order: RNA16A/RNA7A/RNA7D/ML bootstrap. Clades that were resolved in ML bootstrap analysis (bootstrap of 50% or higher) are also shown in bold (branches) in the tree
References
Adams BJ, Wall DH, Gozel U, Dillman AR, Chaston JM, Hogg ID (2007) The southernmost worm, Scottnema lindsayae (Nematoda): diversity, dispersal and ecological stability. Polar Biol 30:809–815
Alfaro ME, Zoller S, Lutzoni F (2003) Bayes or Bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol 20:255–266
Andrássy I (1998) Nematodes in the sixth continent. J Nematode Morph Syst 1:107–186
Ayres E, Nkem JN, Wall DH, Adams BJ, Barrett JE, Broos EJ, Parsons AN, Powers LE, Simmons BL, Virginia RA (2008) Effects of human trampling on populations of soil fauna in the McMurdo Dry Valleys, Antarctica. Conserv Biol 22:1544–1551
Ayres E, Nkem JN, Wall DH, Adams BJ, Barrett JE, Simmons BL, Virginia RA, Fountain AG (2010) Experimentally increased snow accumulation alters soil moisture and animal community structure in a polar desert. Polar Biol 33:897–907
Baldwin JG, De Ley IT, Mundo-Ocampo M, De Ley P, Nadler SA, Gebre M (2001) Acromoldavicus mojavicus n. sp. (Nematoda: Cephalobidae) from the Mojave Desert, California. Nematology 3:343–353
Barrett JE, Virginia RA, Wall DH, Adams BJ (2008) Decline in a dominant invertebrate species contributes to altered carbon cycling in a low-diversity soil ecosystem. Glob Change Biol 14:1734–1744
Courtright EM, Wall DH, Virginia RA, Frisse LM, Vida JT, Thomas WK (2000) Nuclear and mitochondrial DNA sequence diversity in the Antarctic nematode Scottnema lindsayae. J Nematol 32:143–153
Courtright EM, Wall DH, Virginia RA (2001) Determining habitat suitability for soil invertebrates in an extreme environment: the McMurdo Dry Valleys, Antarctica. Antarct Sci 13:9–17
De Ley P, Blaxter ML (2004) A new system for Nematoda: combining morphological characters with molecular trees, and translating clades into ranks and taxa. Nematol Monogr Perspect 2:633–653
De Ley P, Felix MA, Frisse LM, Nadler SA, Sternberg PW, Thomas WK (1999) Molecular and morphological characterization of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae). Nematology 1:591–612
Gooseff MN, Barrett JE, Doran PT, Fountain AG, Lyons WB, Parsons AN, Porazinska DL, Virginia RA, Wall DH (2003) Snow-patch influence on soil biogeochemical processes and invertebrate distribution in the McMurdo Dry Valleys, Antarctica. Arct Antarct Alp Res 35:91–99
Gowri-Shankar V, Jow H (2006) PHASE: a software package for phylogenetics and sequence evolution. University of Manchester, Manchester
Higgs PG (2000) RNA secondary structure: physical and computational aspects. Quart Rev Biophys 33:199–253
Holovachov O, Boström S, Nadler SA, De Ley P (2009a) Systematics and phylogenetic position of the genus Tricirronema Siddiqi, 1993 (Cephalobomorpha). J Nematode Morph Syst 12:133–143
Holovachov O, Boström S, Tandingan De Ley I, Nadler SA, De Ley P (2009b) Description of Penjatinema novaezeelandiae sp. n. (Rhabditida: Cephalobidae) from New Zealand—a second species of a rare genus. J Nematode Morph Syst 12:7–18
Holovachov O, Boström S, Robinson C, Tandingan De Ley I, Nadler SA (2010) Redescription of Placodira lobata Thorne, 1937 (Rhabditida: Cephalobidae) with a discussion of the systematic position of the genus. Nematology (in press)
Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, Fitch DH, Felix MA (2007) Trends, stasis, and drift in the evolution of nematode vulva development. Current Biol 17:1925–1937
Moorhead DL, Wall DH, Virginia RA, Parsons AN (2002) Distribution and life-cycle of Scottnema lindsayae (Nematoda) in Antarctic soils: a modeling analysis of temperature responses. Polar Biol 25:118–125
Moorhead DL, Barrett JE, Virginia RA, Wall DH, Porazinska DL (2003) Organic matter and soil biota of upland wetlands in Taylor Valley, Antarctica. Polar Biol 26:567–576
Mouratov S, Lahav I, Barness G, Steinberger Y (2001) Preliminary study of the soil nematode community at Machu Picchu Station, King George Island, Antarctica. Polar Biol 24:545–548
Nadler SA, De Ley P, Mundo-Ocampo M, Smythe AB, Stock SP, Bumbarger D, Adams BJ, Tandingan De Ley I, Holovachov O, Baldwin JG (2006) Phylogeny of Cephalobina (Nematoda): molecular evidence for recurrent evolution of probolae and incongruence with traditional classifications. Mol Phylogen Evol 40:696–711
Nkem JN, Virginia RA, Barrett JE, Wall DH (2006a) Salt tolerance and survival thresholds for two species of Antarctic nematodes. Polar Biol 29:643–651
Nkem JN, Wall DH, Virginia RA, Barrett JE, Broos EJ, Porazinska DL, Adams BJ (2006b) Wind dispersal of soil invertebrates in the McMurdo Dry Valleys, Antarctica. Polar Biol 29:346–352
Overhoff A, Freckman DW, Virginia RA (1993) Life cycle of the microbivorous Antarctic Dry Valley nematode Scottnema lindsayae (Timm, 1971). Polar Biol 13:151–156
Porazinska DL, Wall DH, Virginia RA (2002a) Population age structure of nematodes in the Antarctic Dry Valleys: perspectives on time, space, and habitat suitability. Arct Antarct Alp Res 34:159–168
Porazinska DL, Wall DH, Virginia RA (2002b) Invertebrates in ornithogenic soils on Ross Island, Antarctica. Polar Biol 25:569–574
Powers LE, Freckman DW, Virginia RA (1995) Spatial distribution of nematodes in polar desert soils of Antarctica. Polar Biol 15:325–333
Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M (2006) 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform 7:498
Shishida Y, Ohyama Y (1986) A note on the terrestrial nematodes around Syowa Station, Antarctica (extended abstract). Mem Natl Inst Polar Res Special Issue 44:259–260
Simmons BL, Wall DH, Adams BJ, Ayres E, Barrett JE, Virginia RA (2009) Long-term experimental warming reduces soil nematode populations in the McMurdo Dry Valleys, Antarctica. Soil Biol Biochem 41:2052–2060
Smythe AB, Nadler SA (2007) Molecular phylogeny of Acrobeloides and Cephalobus (Nematoda: Cephalobidae) reveals paraphyletic taxa and recurrent evolution of simple labial morphology. Nematology 8:819–836
Sonnenberg R, Nolte AW, Tautz D (2007) An evaluation of LSU rDNA D1–D2 sequences for their use in species identification. Frontiers Zool 4:6
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web-Servers. Syst Biol 57:758–771
Stock SP, Campbell JF, Nadler SA (2001) Phylogeny of Steinernema Travassos, 1927 (Cephalobina: Steinernematidae) inferred from ribosomal DNA sequences and morphological characters. J Parasitol 87:877–889
Tavare S (1986) Some probabilistic and statistical problems on the analysis of DNA sequences. Lect Math Life Sci 17:262–272
Tillier ERM, Collins RA (1998) High apparent rate of simultaneous compensatory basepair substitutions in ribosomal RNA. Genetics 148:1993–2002
Timm RW (1971) Antarctic soil and freshwater nematodes from the McMurdo sound region. Proc Helminthol Soc Wash 38:42–52
Treonis AM, Wall DH (2005) Soil nematodes and desiccation survival in the extreme arid environment of the Antarctic Dry Valleys. Integr Comp Biol 45:741–750
Treonis AM, Wall DH, Virginia RA (1999) Invertebrate biodiversity in Antarctic Dry Valley soils and sediments. Ecosystems 2:482–492
Treonis AM, Wall DH, Virginia RA (2000) The use of anhydrobiosis by soil nematodes in Antarctic Dry Valleys. Funct Ecol 14:460–467
Vinciguerra MT (1994) Metacrolobus festonatus gen. n. sp. n., Scottnema lindsayae Timm, 1971 (Nemata, Cephalobidae) from Subantarctic and Antarctic regions with proposal of the new subfamily Metacrolobinae. Fundam Appl Nematol 17:175–180
Weicht TR, Moorhead DL (2004) The impact of anhydrobiosis on the persistence of Scottnema lindsayae (Nematoda): a modeling analysis of population stability thresholds. Polar Biol 27:507–512
Yoder M, Tandingan De Ley I, King IW, Mundo-Ocampo M, Mann J, Blaxter M, Poiras L, De Ley P (2006) DESS: a versatile solution for preserving morphology and extractable DNA of nematodes. Nematology 8:367–376
Acknowledgments
We acknowledge the support for fieldwork by the Swedish Polar Research Secretariat and the USA. National Science Foundation (NSF) Office of Polar Programs to Olle Karlsson who conducted the field sampling in Taylor Valley during the expedition SWEDARP 2008/09. This study was supported in part by an award from the NSF Partnerships for Enhancing Expertise in Taxonomy (PEET) program (DEB-0731516).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Boström, S., Holovachov, O. & Nadler, S.A. Description of Scottnema lindsayae Timm, 1971 (Rhabditida: Cephalobidae) from Taylor Valley, Antarctica and its phylogenetic relationship. Polar Biol 34, 1–12 (2011). https://doi.org/10.1007/s00300-010-0850-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-010-0850-8