Abstract
Background
The genus Cissophyllus (Cosmocercoidea: Kathlaniidae) is a rare group of nematodes parasitic in turtles and lizards. To date, only four species have been reported in Asia and North America. However, most of them are inadequately described. The species Cissophyllus leytensis has never been reported since it was originally described by Tubangui and Villaamil in 1933 from the Philippine sailfin lizard Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata). Furthermore, the systematic status of Cissophyllus/Cissophyllinae in the family Kathlaniidae of the superfamily Cosmocercoidea remains under debate.
Methods
The detailed morphology of C. leytensis was studied using light microscopy (LM) and, for the first time, scanning electron microscopy (SEM), based on newly collected specimens from the type host H. pustulatus. Six different genetic markers, including nuclear sequences [small ribosomal subunit (18S), internal transcribed spacer (ITS) and large ribosomal subunit (28S)], plus mitochondrial genes [cytochrome c oxidase subunit 1 (cox1), cytochrome c oxidase subunit 2 (cox2) and 12S small subunit ribosomal RNA gene] of C. leytensis were sequenced. Additionally, in order to test the validity of the subfamily Cissophyllinae and clarify the phylogenetic relationships of Cissophyllus and the other genera in the family Kathlaniidae, phylogenetic analyses based on 18S + 28S and ITS sequence data were performed using maximum likelihood (ML) and Bayesian inference (BI) analyses, respectively.
Results
Our observations using LM and SEM revealed some previously unreported morphological features, necessitating the redescription of this poorly known species. The presence of remarkable morphological variation in the isthmus and the position of excretory pore among different individuals was found. Molecular analysis showed no intraspecific nucleotide divergence detected in the 18S, ITS, 28S, cox2 and 12S regions among different individuals of C. leytensis, but a low level of intraspecific genetic variation was found in the cox1 (0.52%). Our phylogenetic results showed the representatives of the Cosmocercoidea divided into four large clades (Cosmocerca + Aplectana + Cosmocercoides representing the family Cosmocercidae, Cruzia representing the subfamily Cruzinae of Kathlaniidae, Falcaustra + Cissophyllus + Megalobatrachonema representing the subfamily Kathlaniinae of Kathlaniidae, and Orientatractis + Rondonia representing the family Atractidae). The genus Cissophyllus clustered together with the genus Megalobatrachonema in both the ML and BI trees using ITS sequence data, but displayed a sister relationship to the genus Falcaustra in the ML tree and to the genera Falcaustra + Megalobatrachonema in the BI tree using 18S + 28S sequence data.
Conclusions
Molecular phylogenetic results further confirmed that the family Kathlaniidae is not a monophyletic group. The subfamily Cruziinae should be moved from the hitherto-defined family Kathlaniidae and elevated as a separate family Cruziidae. The present phylogenetic results also negated the validity of the subfamily Cissophyllinae and supported the genus Cissophyllus assigned in the subfamily Kathlaniinae. Molecular analysis indicated that the morphological variation in the isthmus and position of excretory pore among different individuals should be considered as intraspecific variation. Moreover, some characters important for the specific diagnosis of C. leytensis are reported for the first time: the number of acuminate denticles (lamellae) on each lip, the chitinized pharynx with three flabellate pharyngeal plates, the presence of single medioventral precloacal papilla and the detailed morphology of caudal papillae. The present study is only the second record of C. leytensis.
Graphical Abstract

Similar content being viewed by others
Background
The genus Cissophyllus (Cosmocercoidea: Kathlaniidae) is a rare group of nematodes parasitic in turtles and lizards [1, 2]. To date, only four species have been reported in Asia and North America, including Cissophyllus laverani Railliet & Henry, 1912 from the Asian forest tortoise Manouria emys (Schlegel & Müller, 1844), the European pond turtle Emys orbicularis (Linnaeus, 1758) and the Malayan flat-shelled turtle Notochelys platynota (Gray, 1834) (Reptilia: Testudines) in India, Malaysia and Indonesia; Cissophyllus roseus (Leidy, 1851) from E. orbicularis (Linnaeus, 1758) (Reptilia: Testudines) in Indonesia; Cissophyllus leytensis Tubangui & Villaamil, 1933 from the Philippine sailfin lizard Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in the Philippines and Cissophyllus penitus (Leydi, 1886) from the red-eared slider turtle Trachemys scripta elegans (Wied-Neuwied, 1792) (Reptilia: Testudines) in the USA [2, 3]. However, most of them have been inadequately described, especially regarding the details of the cephalic structures.
The systematic status of Cissophyllus in the family Kathlaniidae of the superfamily Cosmocercoidea remains under debate. Railliet and Henry [4] established the genus Cissophyllus in 1912. In 1926, Yorke and Maplestone [5] erected the subfamily Cissophyllinae for this genus, due to the unique structure of its lips. The validity of the Cissophyllinae was accepted by Skrjabin et al. in 1964 [6] but rejected in 1978 by Chabaud [7], who placed Cissophyllus in the subfamily Kathlaniinae.
In the present study, the detailed morphology of C. leytensis was studied using light and, for the first time, scanning electron microscopy (SEM), based on newly collected specimens from the Philippine sailfin lizard H. pustulatus. The molecular characterization of nuclear sequences [small ribosomal subunit (18S), internal transcribed spacer (ITS) and large ribosomal subunit (28S)], plus mitochondrial genes [cytochrome c oxidase subunit 1 (cox1), cytochrome c oxidase subunit 2 (cox2) and 12S small subunit ribosomal RNA gene] of C. leytensis are provided for the first time. Additionally, in order to test the validity of the subfamily Cissophyllinae and clarify the phylogenetic relationships of Cissophyllus and the other genera in the family Kathlaniidae, phylogenetic analyses were performed based on 18S + 28S and ITS sequence data using maximum likelihood (ML) and Bayesian inference (BI) analyses, respectively.
Methods
Parasite collection
Nematode parasites were collected from a Philippine sailfin lizard H. pustulatus during a regular anthelmintic treatment by the veterinary surgeon in a zoo in Tangshan, Hebei Province, China. Specimens were washed in physiological saline and then fixed and stored in 75% ethanol, after which they were sent to the corresponding author’s lab for species identification.
Morphological observations
For LM studies, nematodes were placed in temporary mounts and cleared in lactophenol. Photomicrographs were recorded using a Nikon® digital camera coupled to a Nikon® optical microscope (Nikon ECLIPSE Ni-U, Nikon Corporation, Tokyo, Japan). For SEM, the anterior and posterior ends of specimens were re-fixed in a 4% formaldehyde solution, post-fixed in 1% OsO4, dehydrated via an ethanol series and acetone, and then critical-point-dried. Samples were coated with gold and examined using a Hitachi S-4800 scanning electron microscope at an accelerating voltage of 20 kV. Measurements (the range, followed by the mean in parentheses) are given in micrometers (μm) unless otherwise stated. Voucher specimens were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
Molecular procedures
The midbody of one male (isthmus slightly inflated and excretory pore more or less at anterior edge of isthmus) and two females (one individual with isthmus slightly inflated and excretory pore at level of esophageal bulb, one individual with isthmus nearly as wide as corpus and excretory pore at level of esophageal bulb) were chosen for molecular analysis. Genomic DNA was extracted from each sample using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer’s instructions. DNA was eluted in elution buffer and kept at −20 °C until use. For amplifying these target sequences, the following published primers were used: the near-complete 18S ribosomal DNA (rDNA) by the primers 18SF and 18SR [8], the partial ITS region by the primers A and B [9], the partial 28S rDNA by the primers 28SF and 28SR [10], the partial cox1 by the primers CO1F and CO1R [11], the partial cox2 by the primers CO2F and CO2R [12], and the partial 12S by the primers 12SF and 12SR [13]. The cycling conditions were as described previously [14]. Polymerase chain reaction (PCR) products were checked on GoldView-stained 1.5% agarose gels and purified with the Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing of each sample was carried out for both strands. The DNA sequences obtained herein were compared (using the BLASTn algorithm) with those available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Sequences of C. leytensis obtained herein were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov, accession numbers 18S: OM414722, OM414723; 28S: OM414718, OM414719; ITS: OM414724–OM414726; cox1: OM416530, OM416531; cox2: OM436778, OM436779, 12S: OM414720, OM414721).
Phylogenetic analyses
Phylogenetic trees were constructed based on the 18S + 28S and ITS sequence data using ML inference with IQ-TREE and BI with MrBayes 3.2., respectively. Ascaris lumbricoides Linnaeus, 1758 (Ascaridida: Ascaridoidea) was chosen as the out-group. The in-group comprises 22 cosmocercoid species representing all three families in the superfamily Cosmocercoidea according to the current classifications [7, 15], including Cosmocercidae, Atractidae and Kathlaniidae. The detailed information of nematode species included in the phylogenetic analyses is provided in Table 1. Sequences were aligned using ClustalW2. We used a built-in function in IQ-TREE to select a best-fitting substitution model for the sequences according to the Bayesian information criterion [16]. The TIM3e + G4 model and the TVMe + I + G4 model were identified as the optimal nucleotide substitution model for 18S + 28S and ITS sequence data, respectively. Reliability for the ML tree was tested using 1000 bootstrap replications, and the BI tree was tested using 50 million generations. The bootstrap values over 70% are shown in the phylogenetic trees.
Results
Morphology of Cissophyllus leytensis Tubangui & Villaamil, 1933 (Figs. 1, 2, 3, 4, Table 2)
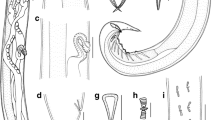
General. Medium-sized, whitish nematodes. Body cylindrical, maximum width at about region of middle body. Cuticle with fine transverse striations. Lateral alae absent. Oral aperture dorsoventrally elongate, surrounded by three small chitinized lips (Figs. 1a, c, 2a). Dorsal lip with one pair of large double papillae, one pair of small triangular cuticular projections (inner ridge armed with 3–5 acuminate denticles (lamellae), single quadrate cuticular plate and large trilobed tooth plate (Figs. 2a, 3b). Subventral lips each with single large double papillae, small papilla and amphid; inner ridge of each subventral lip armed with three clusters of acuminate denticles (lamellae) (smallest cluster with 6–9 denticles, largest cluster with about 80 denticles, medium one with 12–15 denticles) (Figs. 1b, c, 2a). Esophagus divided into short chitinized pharynx with three flabellate pharyngeal plates (Figs. 3a, b, 4a, b), cylindrical corpus, slightly inflated isthmus (Figs. 3a, 4a) (isthmus also nearly as wide as corpus in some specimens) and ovoid posterior bulb with valves (Figs. 3a, 4a). Nerve-ring situated at about 1/3 of total esophageal length. Position of excretory pore varied from anterior edge of isthmus to level of middle of esophageal bulb (Figs. 3a, 4a). Deirids not observed. Tail of both sexes conical, with blunt tip (Figs. 1e, 2b, c, 3e, f, i, j, 4d–f).
Scanning electron micrographs of female Cissophyllus leytensis collected from Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in China: a anterior part of body, ventrolateral view; b magnified image of cephalic end, lateral view; c cephalic end, apical view; d magnified image of vulva; e tail (black arrow showing phasmid), ventral view; f magnified image of phasmid. am amphid, dp large double papillae, lc largest cluster of acuminate denticles (lamellae), mc medium cluster of acuminate denticles (lamellae), sc smallest cluster of acuminate denticles (lamellae), sl subventral lip, sp small papilla
Scanning electron micrographs of male Cissophyllus leytensis collected from Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in China: a cephalic end, apical view; b tail (white arrows showing postcloacal papillae, black arrow showing phasmid), ventral view; c posterior part of body (white arrows showing precloacal papillae), ventral view; d magnified image of precloacal papilla; e magnified image of precloacal medioventral papilla; f magnified image of paracloacal papilla; g magnified image of postcloacal papilla. DL dorsal lip, dp large double papillae, pp paracloacal papilla, pvp precloacal medioventral papilla, qp single quadrate cuticular plate, SL subventral lip, tp small triangular cuticular projection, ttp large trilobed tooth plate
Cissophyllus leytensis collected from Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in China: a anterior part of female, lateral view; b magnified image of cephalic end, dorsal view; c region of vulva, lateral view; d egg; e, f tail of female, lateral view; g spicules, ventral view; h gubernaculum, ventral view; i posterior end of male, ventral view; j posterior end of male, lateral view. ep excretory pore, nr nerve ring, ph phasmid
Photomicrographs of Cissophyllus leytensis collected from Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in China: a anterior part of male, lateral view; b magnified image of cephalic end, lateral view; c region of vulva, lateral view; d posterior end of female, lateral view; e posterior end of male, lateral view; f posterior end of male, ventral view. ep excretory pore, gu gubernaculum, lc largest cluster of acuminate denticles (lamellae), mc medium cluster of acuminate denticles (lamellae), nr nerve ring, php pharyngeal plates, sc smallest cluster of acuminate denticles (lamellae)
Male (based on 10 specimens). Body 14.0–18.0 (16.6) mm long, maximum width 976–1220 (1068). Esophagus 2.00–2.39 (2.13) mm in total length, representing 11.1–17.1 (13.0) % of body length; pharynx + corpus + isthmus 1.70–2.04 (1.83) mm long, size of bulb 250–350 (305) × 260–400 (357). Nerve ring 522–807 (672) and excretory pore 1.82–2.00 (1.94) mm from anterior extremity, respectively. Posterior end of body slightly curved ventrally. Precloacal sucker absent. Spicules alate, equal in length, 600–900 (749) long, distal end sub-pointed, representing 3.89–5.63 (4.55) % of body length (Figs. 3g, j, 4e, f). Gubernaculum present, triangular, 149–248 (206) long (Figs. 3h, 4f). Caudal papillae 10 pairs in total, distributed as: six pairs of precloacal papillae (last three pairs close to each other), one pair of paracloacal papillae and three pairs of postcloacal papillae (one pair ventrolateral, two pairs ventral) (Figs. 2b–g, 3i, j). Single median ventral papilla present (Figs. 2b, c, 3i). Tail 150–260 (228) long, with rounded tip (Figs. 2b, c, 3i, j, 4e, f). Phasmids present, between last two pairs postcloacal papillae (Figs. 2b, 3i, j).
Female (based on five specimens). Body 14.0–18.0 (17.0) mm long, maximum width 976–1293 (1112). Esophagus 2.15–2.49 (2.28) mm in total length, representing 12.1–15.3 (13.5) % of body length; pharynx + corpus + isthmus 1.78–2.07 (1.92) mm long, size of bulb 293–415 (361) × 341–463 (390). Nerve ring 604–894 (753) and excretory pore 1.70–1.96 (1.86) mm from anterior extremity, respectively. Vulva transverse slit, 9.40–12.3 (11.1) mm from anterior extremity, representing 63.0–68.2 (65.3) % of body length (Figs. 1d, 3c, 4c). Vagina muscular (Figs. 3c, 4c); egg oval, with smooth surface, 97–111 (105) × 53–63 (57) (n = 20) (Fig. 3d). Tail 250–350 (296) long, with rounded or more or less finger-like tip (Figs. 1e, 3e, f, 4d). Phasmids present (Figs. 1e, f, 3e, f).
Taxonomic summary
Host and locality
Philippine sailfin lizard H. pustulatus (Eschscholtz, 1829) (Reptilia: Squamata) in a zoo in Tangshan, Hebei Province, China.
Level of infection
Single lizard infected with 15 nematodes.
Voucher specimen deposition
Ten males (HBNU–N-2021R0013L), five females (HBNU–N-2021R0014L), College of Life Sciences, Hebei Normal University, Hebei Province, China.
Genetic characterization
Partial 18S region
Two 18S sequences of C. leytensis obtained herein were both 1749 base pairs (bp) in length, with no nucleotide divergence detected. In the Kathlaniidae, the 18S sequence data are available in GenBank for Megalobatrachonema terdentatum (MG594352–MG594364), Megalobatrachonema wangi (MW325957), Cruzia americana (U94371), Cruzia tentaculata (MN873564–MN873566, MN873570), Cruzia sp. (MT809125–MT809126), Falcaustra ararath (MT160412), Falcaustra araxiana (KM200715), Falcaustra catesbeianae (AB818380), Falcaustra sp. (MN727387, MN727389, MN727390) and Spectatus spectatus (KR139827). Pairwise comparison of 18S sequences of C. leytensis and these 10 kathlaniid species displayed 4.77% (F. araxiana) to 9.61% (C. tentaculata) nucleotide divergence.
Partial ITS (ITS1–5.8S-ITS2) region
Three ITS sequences of C. leytensis obtained herein were all 837 bp in length, with no nucleotide divergence detected. In the Kathlaniidae, the ITS sequence data are available in GenBank for M. terdentatum (MN444703–MN444704), Megalobatrachonema hainanensis (MH545567–MH545569), M. wangi (MN245657–MN245659), Falcaustra sinensis (MF061681), Falcaustra sp. (MN727388, MN727391, MN727392) and Cruzia sp. (MT809125). Pairwise comparison of ITS sequences of C. leytensis and these six kathlaniid species displayed 12.1% (Cruzia sp.) to 34.8% (F. sinensis) nucleotide divergence.
Partial 28S region
Two 28S sequence of C. leytensis obtained herein were both 725 bp in length, with no nucleotide divergence detected. In the Kathlaniidae, the 28S sequence data are available in GenBank for M. terdentatum (MN444705–MN444706), M. wangi (MN245660–MN245662), M. hainanensis (MH545569–MH545570), F. sinensis (MF094270), Falcaustra sp. (LC605539–LC605541) and C. americana (U94757). Pairwise comparison of 28S sequences of C. leytensis and these six kathlaniid species displayed 12.5% (C. americana) to 20.0% (M. hainanensis) nucleotide divergence.
Partial cox1 region
Two cox1 sequences of C. leytensis obtained herein were both 384 bp in length, with 0.52% of nucleotide divergence detected. In the Kathlaniidae, the cox1 sequence data are available in GenBank for M. terdentatum (MN444709–MN444710), M. wangi (MN245668–MN245670), F. sinensis (MF113223), Falcaustra sp. (MN729570–MN729572) and C. tentaculata (MN842776–MN842778). Pairwise comparison of cox1 sequences of C. leytensis and these five kathlaniid species displayed 12.3% (Falcaustra sp.) to 53.8% (C. tentaculata) nucleotide divergence.
Partial cox2 region
Two cox2 sequences of C. leytensis obtained herein were both 501 bp in length, with no nucleotide divergence detected. In the Kathlaniidae, the cox2 sequence data are available in GenBank for C. americana (AF179911) and F. sinensis (MF120240). Pairwise comparison of cox2 sequences of C. leytensis and these two kathlaniid species displayed 16.6% (F. sinensis) to 22.0% (C. americana) nucleotide divergence.
Partial 12S region
Two 12S sequences of C. leytensis obtained herein were both 469 bp in length, with no nucleotide divergence detected. In the Kathlaniidae, the 12S sequence data are available in GenBank for M. terdentatum (MN444707–MN444708), M. hainanensis (MN245666–MN245667), M. wangi (MN245663–MN245665) and F. sinensis (MF140337). Pairwise comparison of 12S sequences of C. leytensis and these four kathlaniid species displayed 24.7% (F. sinensis) to 28.6% (M. terdentatum) nucleotide divergence.
Phylogenetic analyses (Figs. 5, 6)
Phylogenetic relationships of representatives of the superfamily Cosmocercoidea using maximum likelihood (a) and Bayesian inference (b) analyses based on the 18S + 28S sequences. Ascaris lumbricoides (Ascaridida: Ascaridoidea) was chosen as the out-group. Bootstrap values exceeding 70% are shown in the phylogenetic trees
Phylogenetic relationships of representatives of the superfamily Cosmocercoidea using maximum likelihood (a) and Bayesian inference (b) analyses based on the ITS sequences. Ascaris lumbricoides (Ascaridida: Ascaridoidea) was chosen as the out-group. Bootstrap values exceeding 70% are shown in the phylogenetic trees
The phylogenetic results of ML and BI trees using 18S + 28S sequence data were more or less identical, with both showing the representatives of the superfamily Cosmocercoidea divided into four large clades (Fig. 5). The species of Cosmocerca + Aplectana + Cosmocercoides formed clade I, which represents the family Cosmocercidae. The species C. americana formed clade II, which represents the subfamily Cruzinae in the Kathlaniidae. The species of Falcaustra + Cissophyllus + Megalobatrachonema formed clade III, which represents the subfamily Kathlaniinae in the Kathlaniidae. The species of Orientatractis + Rondonia formed clade IV, which represents the family Atractidae. Cissophyllus and Falcaustra formed a sister group in the ML tree in clade III, but Cissophyllus clustered together with Falcaustra + Megalobatrachonema in the BI tree (Fig. 5). By contrast, the phylogenetic results of ML and BI trees using ITS sequence data showed the representatives of the superfamily Cosmocercoidea divided into three large clades, due to the lack of available ITS data for atractid species (Fig. 6). The genus Cruzia (clade II) is at the base of the ML and BI trees, and the genus Cissophyllus showed a closer relationship to Megalobatrachonema than Falcaustra with weak support (Fig. 6).
Discussion
Tubangui and Villaamil (1933) [1] described C. leytensis from H. pustulatus in the Philippines. The morphology and measurements of the present specimens are almost identical to the original description of C. leytensis by Tubangui and Villaamil (1933) regarding some important taxonomical features, including the morphology of the lips, the length of the male body and total esophagus, the morphology and length of tail, spicules and gubernaculum, the number and arrangement of caudal papillae, and the absence of the precloacal sucker (see Table 2 for details). It should be noted that the present specimens were collected from the type host H. pustulatus. Therefore, we consider our newly collected specimens to be conspecific with C. leytensis. However, we observed the position of the excretory pore varied from the anterior edge of the isthmus to the level of the middle of the esophageal bulb, and the isthmus slightly inflated (slightly wider than corpus) or nearly as wide as the corpus among different individuals of our specimens. Tubangui and Villaamil (1933) [1] did not mention the intraspecific morphological variation of these characters in their description. The size of eggs and the length of females in the present study are slightly smaller than those of the original description (see Table 2 for details), which were possibly affected by the age/developmental stage or infection intensity of parasites. Some characters important for the specific diagnosis of C. leytensis were reported for the first time: the number of acuminate denticles (lamellae) on each lip, the chitinized pharynx with three flabellate pharyngeal plates, the presence of single medioventral precloacal papilla and the detailed morphology of caudal papillae.
In the genus Cissophyllus, only C. leytensis has been reported from a lizard, with the other three species Cissophyllus laverani, C. roseus and C. penitus all from turtles. Cissophyllus leytensis can be easily distinguished from C. laverani, C. roseus and C. penitus by the absence of a precloacal sucker (vs. the presence of remarkable precloacal sucker). It is very interesting that the species of Cissophyllus parasitic in different hosts (lizard and turtles) showed such distinct morphological differences. However, we do not think that it is reasonable to erect a new genus or subgenus for C. leytensis, because the other generic diagnostic characters of the four species are almost coincident. But the true phylogenetic relationships of the four species should be investigated using phylogenetic analysis based on genetic sequences in the future.
In recent years, some studies have started to expand their morphological descriptions of new species of the superfamily Cosmocercoidea with DNA sequence data [17,18,19,20,21,22,23]. Nevertheless, the vast majority of the c. 410 currently recognized species in the Cosmocercoidea [15] were defined under the traditional morphospecies concept. Within Cissophyllus, none of the four currently recognized species had been characterized using molecular markers since they were originally described. In the present study, the genetic characterization of the partial 18S, ITS, 28S ribosomal DNA, and the partial mitochondrial cox1, cox2 and 12S of C. leytensis are provided for the first time. Based on the molecular analysis of C. leytensis, low levels of intraspecific nucleotide differences were noted only in the cox1 region, but high levels of interspecific genetic variation in all six genetic markers was clear among the genera of Kathlaniidae. These genetic data of C. leytensis obtained herein will be valuable for further investigations on the species identification, population genetics and phylogeny of this poorly known group.
Our phylogenetic analyses based on 18S + 28S and ITS sequence data both showed that the family Kathlaniidae is not a monophyletic group. The present results are consistent with some recent phylogenetic studies [22, 23]. According to the classification by Chabaud (1978) [7], the Kathlaniidae includes three subfamilies, namely Kathlaniinae, Cruziinae and Oxyascaridinae. However, Chabaud’s classification has been challenged by some traditional taxonomical studies and recent molecular phylogenetic studies [22,23,24]. Our phylogenetic results supported the subfamily Cruziinae moved out from the hitherto-defined family Kathlaniidae and elevated to a separate family, which agreed with the proposal by Travassos (1917) and Skrjabin et al. (1960) [33, 34]. Moreover, the present phylogenetic results supported the genus Cissophyllus belonging to the subfamily Kathlaniinae, which is congruent with the traditional classification of Chabaud (1978) [7]. The subfamily Cissophyllinae proposed by Yorke and Maplestone (1926) and Skrjabin et al. (1976) is invalid. The highly specialized structure of the cephalic end of Cissophyllus species can only be treated at the level of a generic diagnostic character.
Conclusions
Molecular phylogenetic results further confirmed that the family Kathlaniidae is not a monophyletic group. The subfamily Cruziinae should be moved from the hitherto-defined family Kathlaniidae and elevated as a separate family Cruziidae. The present phylogeny also negated the validity of the subfamily Cissophyllinae and supported the genus Cissophyllus assigned in the subfamily Kathlaniinae. Molecular analysis indicated that the presence of morphological variation in the isthmus and position of excretory pore among different individuals should be considered as intraspecific variation. Moreover, some characters important for the specific diagnosis of C. leytensis are reported for the first time: the number of acuminate denticles (lamellae) on each lip, the chitinized pharynx with three flabellate pharyngeal plates, the presence of single medioventral precloacal papilla and the detailed morphology of caudal papillae. The present study is only the second record of C. leytensis.
Availability of data and materials
The nuclear and mitochondrial DNA sequences of Cissophyllus leytensis obtained in this study were deposited in GenBank database. Voucher specimens of C. leytensis were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, under the accession numbers HBNU–N-2021R0013L, HBNU–N-2021R0014L, China.
Abbreviations
- am:
-
Amphid
- BI:
-
Bayesian inference
- cox1:
-
Cytochrome c oxidase subunit 1
- cox2:
-
Cytochrome c oxidase subunit 2
- DL:
-
Dorsal lip
- dp:
-
Large double papillae
- ep:
-
Excretory pore
- gu:
-
Gubernaculum
- ITS:
-
Internal transcribed spacer
- lc:
-
Largest cluster of acuminate denticles (lamellae)
- LM:
-
Light microscopy
- mc:
-
Medium cluster of acuminate denticles (lamellae)
- ML:
-
Maximum likelihood
- nr:
-
Nerve ring
- PCR:
-
Polymerase chain reaction
- php:
-
Pharyngeal plates
- pp:
-
Paracloacal papilla
- pvp:
-
Precloacal medioventral papilla
- qp:
-
Single quadrate cuticular plate
- sc:
-
Smallest cluster of acuminate denticles (lamellae)
- SEM:
-
Scanning electron microscopy
- SL:
-
Subventral lip
- sp:
-
Small papilla
- tp:
-
Small triangular cuticular projection
- ttp:
-
Large trilobed tooth plate
- 12S:
-
Small subunit ribosomal RNA gene
- 18S:
-
Small ribosomal subunit
- 28S:
-
Large ribosomal subunit
References
Tubangui MA, Villaamil R. Nematodes in the collection of the Philippine bureau of science. I Oxyuroidea Philipp J Sci. 1933;51:607–15.
Yamaguti S. Systema helminthum. The nematodes of vertebrates. Part I and II, vol. 3. London: Interscience Publisher; 1961. p. 1261.
Purwaningsih E, Mumpuni M. Parasitic nematodes from turtles: new species and new record from Indonesia. J Coast Life Med. 2015;3:607–11.
Railliet A, Henry A. Quelques nématodes parasites des reptiles. Bull Soc Pathol Exol. 1912;5:251–9.
Yorke W, Maplestone PA. The nematode parasites of vertebrates. Trans Amer Microscop Soc. 1926. https://doi.org/10.2307/3221805.
Skrjabin KI, Shikhobalova NP, Lagodovskaya EA. Oxyurata of animals and man. Part 3 Translated from Russian. Jerusalem: Israel Program for Scientific Translations, 1964, 266.
Chabaud AG. Keys to genera of the superfamilies Cosmocercoidea, Seuratoidea, Heterakoidea, and Subuluroidea. In: Anderson RC, Chabaud AG, Willmott S, editors. CIH keys to the nematode parasites of vertebrates. Farnham Royal: Commonwealth Agricultural Bureaux; 1978. p. 71.
Floyd RM, Rogers AD, Lambshead PJD, Smith CR. Nematode-specific PCR primers for the 18S small subunit rRNA gene. Mol Ecol Notes. 2005;5:611–2.
Zhu X, D’Amelio S, Paggi L, Gasser RB. Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitol Res. 2000;86:677–83.
Nadler SA, Hudspeth DSS. Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): implications for morphological evolution and classification. Mol Phylogenet Evol. 1998;10:221–36.
Lazarova SS, Malloch G, Oliveira CMG, Hübschen J, Neilson R. Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. J Nematol. 2006;38:404–10.
Nadler SA, Hudspeth DSS. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: hypotheses of structural and sequence evolution. J Parasitol. 2000;86:380–93.
Li Y, Niu L-L, Wang Q, Zhang Z-H, Chen Z-G, Gu X-B, et al. Molecular characterization and phylogenetic analysis of ascarid nematodes from twenty-one species of captive wild mammals based on mitochondrial and nuclear sequences. Parasitol. 2012;139:1329–38.
Li L, Lü L, Nadler SA, Gibson DI, Zhang L-P, Chen H-X, et al. A terrestrial origin for ascaridoid nematodes in the early carboniferous. Syst Biol. 2018;67:888–900.
Zhang Z-Q. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Zootaxa. 2011;3148:63–95.
Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601.
Tran BT, Sato H, Luc PV. A new Cosmocercoides species (Nematoda: Cosmocercidae), C. tonkinensis n. sp., in the scalebellied tree lizard (Acanthosaura lepidogaster) from Vietnam. Acta Parasitol. 2015;60:407–16.
Chen H-X, Zhang L-P, Minoru N, Li L. Morphological and molecular evidence for a new species of the genus Cosmocercoides Wilkie, 1930 (Ascaridida: Cosmocercoidea) from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura). Parasitol Res. 2018;117:1857–64.
Chen H-X, Zhang L-P, Li L. Morphological and molecular characterization of Megalobatrachonema hainanensis sp. nov. (Nematoda: Ascaridida), with phylogenetic position of Megalobatrachonema in Cosmocercoidea. J Helminthol. 2019;94:1–7.
Chen H-X, Zhang L-P, Feng Y-Y, Li L. Integrated evidence reveals a new species of Cosmocerca (Ascaridomorpha: Cosmocercoidea) from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura). Parasitol Res. 2020;119:1795–802.
Chen H-X, Zhang L-P, Sinsch U, Scheid P, Balczun C, Li L. Molecular phylogeny of Megalobatrachonema (Nematoda: Ascaridida), with description of a new species based on morphological and molecular evidence. Infect Genet Evol. 2020;80:e104172.
Chen H-X, Ni X-F, Gu X-H, Sinsch U, Li L. Morphology, genetic characterization and phylogeny of Aplectana dayaoshanensis n. sp. (Nematoda: Ascaridida) from frogs. Infect Genet Evol. 2021;96:e105123.
Chen H-X, Gu X-H, Ni X-F, Li L. Description of a new species of Aplectana (Nematoda: Ascaridomorpha: Cosmocercidae) using an integrative approach and preliminary phylogenetic study of Cosmocercidae and related taxa. Parasite Vector. 2021;14:e165.
Baker MR, Vaucher C. Parasitic helminths from Paraguay. VII. Systematic position of Oxyascaris Travassos, 1920 (Nematoda: Cosmocercoidea). Rev Suisse Zool. 1985;92:303–10.
Sato A, Hasegawa H, Sekiya K, Tsubouchi T. Is Cosmocerca (Nematoda: Cosmocercidae) parasitic in Japanese amphibians a single species? Japan J Vet Parasitol. 2015;14:7–12.
Liu Y, Yu Q, Shu Y-L, Zhao J-H, Fang J-Y, Wu H-L. A new Cosmocercoides species (Ascaridida: Cosmocercidae), C. wuyiensis n. sp., from the Asiatic frog Amolops wuyiensis (Amphibia: Anura). J Helminthol. 2019;94:1–8.
Hasegawa H, Sato A, Kai M, Uchida A. Helminth parasites of bullfrogs, Lithobates catesbeianus (Shaw, 1802), in Kanto district, Japan, with special reference to those introduced from North America. Japan J Vet Parasitol. 2013;12:1–10.
Sinsch U, Heneberg P, Těšínský M, Balczun C, Scheid P. Helminth endoparasites of the smooth newt Lissotriton vulgaris: linking morphological identification and molecular data. J Helminthol. 2019;93:332–41.
Cavalcante PHO, Silva MT, Santos EGN, Chagas-Moutinho VA, Santos CP. Orientatractis moraveci n. sp. and Rondonia rondoni Travassos, 1920 (Nematoda: Atractidae), parasites of Pimelodus blochii (Osteichthyes, Pimelodidae) from the Acre and Xapuri Rivers, Western Amazon, Brazil. Parasitol. 2016;144:226–36.
Wijová M, Moravec F, Horák A, Lukes J. Evolutionary relationships of Spirurina (Nematoda: Chromadorea: Rhabditida) with special emphasis on dracunculoid nematodes inferred from SSU rRNA gene sequences. Int J Parasitol. 2006;36:1067–75.
Müller E, Neuhaus H, Tobler H, Müller F. The two main rDNA size classes of Ascaris lumbricoides: comparison of transcription termination and spacer organization. Nucleic Acids Res. 1992;20:2977–83.
Sato M, Funayama K, Hoshi R, Takatsuka H, Sato MO. Ascaris lumbricoides found in ashore corpses from Korean peninsula to Japan. Parasitol Int. 2019;70:1–4.
Travassos L. Algunus helminthos de collecção do Instituto Bacteriologico de S. Paulo Brasil Medico. 1917;31:99–100.
Skrjabin KI, Shikhobalova NP, Mozgovoi AA. Key to parasitic nematodes. Oxyurata and Ascaridata. Moscow: Akademiya Nauk SSSR Publishers; 1951. p. 327–8.
Acknowledgements
The authors are grateful to Mr Fang-Wen Zhang for providing the nematode specimens of Cissophyllus leytensis.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 32170442), the Natural Science Foundation of Hebei Province (C2019205094), the Science and Technology Project of Hebei Education Department (SLRC2019033), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB26000000) and the Youth Top Talent Support Program of Hebei Province for Dr. Liang Li.
Author information
Authors and Affiliations
Contributions
XFN and LL contributed to the study design and identification of the nematode specimens. All authors analyzed morphological and genetic data. HXC and LL conducted the phylogenetic analyses. XFN, HXC and LL wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted under the protocol of Hebei Normal University. All applicable national and international guidelines for the protection and use of animals were followed.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ni, XF., Chen, HX., Xu, Z. et al. Morphology, genetic characterization and molecular phylogeny of the poorly known nematode parasite Cissophyllus leytensis Tubangui & Villaamil, 1933 (Nematoda: Ascaridida) from the Philippine sailfin lizard Hydrosaurus pustulatus (Eschscholtz, 1829) (Reptilia: Squamata). Parasites Vectors 15, 116 (2022). https://doi.org/10.1186/s13071-022-05224-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05224-8