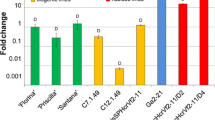
Abstract
Key message
pPPO16, the first Ea-inducible promoter cloned from apple, can be a useful component of intragenic strategies to create fire blight resistant apple genotypes.
Abstract
Intragenesis is an important alternative to transgenesis to produce modified plants containing native DNA only. A key point to develop such a strategy is the availability of regulatory sequences controlling the expression of the gene of interest. With the aim of finding apple gene promoters either inducible by the fire blight pathogen Erwinia amylovora (Ea) or moderately constitutive, we focused on polyphenoloxidase genes (PPO). These genes encode oxidative enzymes involved in many physiological processes and have been previously shown to be upregulated during the Ea infection process. We found ten PPO and two PPO-like sequences in the apple genome and characterized the promoters of MdPPO16 (pPPO16) and MdKFDV02 PPO-like (pKFDV02) for their potential as Ea-inducible and low-constitutive regulatory sequences, respectively. Expression levels of reporter genes fused to these promoters and transiently or stably expressed in apple were quantified after various treatments. Unlike pKFDV02 which displayed a variable activity, pPPO16 allowed a fast and strong expression of transgenes in apple following Ea infection in a Type 3 Secretion System dependent manner. Altogether our results does not confirmed pKFDV02 as a constitutive and weak promoter whereas pPPO16, the first Ea-inducible promoter cloned from apple, can be a useful component of intragenic strategies to create fire blight resistant apple genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Erwinia amylovora (Ea) is a necrogenic enterobacterium causing progressive necrosis on flowers and succulent shoots in members of the Malinae tribe of the Rosaceae family including apple (Malus x domestica Borkh; Vanneste 2000). Rapid invasion of the bacteria into branches and trunks can lead to the death of the trees within a growing season for the most susceptible cultivars. At the cellular level, the bacteria use a Type 3 Secretion System (T3SS) to deliver effectors into the plant cells, to induce membrane disruption and oxidative burst leading to cell death (Vrancken et al. 2013). H2O2 is one of the first detectable ROS (Reactive Oxygen Species) produced during this infection process (Vrancken et al. 2013). Fire blight outbreaks are sporadic, particularly difficult to control and improving host resistance is by far the most effective option to control the disease (Paulin 1996). Breeding for fire blight resistance is therefore an active area of research with the identification of genetic resistance factors including quantitative traits loci (Khan et al. 2012), a “resistance” gene (R gene) implicated in pathogen recognition (FB_MR5; Vogt et al. 2013) or defense mechanisms downstream recognition (Vrancken et al. 2013).
Numerous attempts to create fire blight resistant apple transgenic lines have been performed with various degrees of success. For example, a number of studies were based on the expression of foreign genes encoding insect lytic proteins (Borejsza-Wysocka et al. 2010), a chalcone 3-hydroxylase (Hutabarat et al. 2016), a viral EPS-depolymerase (Flachowsky et al. 2008a) or the Ea HrpN protein (Vergne et al. 2014). Other studies tested the introgression of the fire blight resistance gene FB_MR5 (Broggini et al. 2014; Kost et al. 2015), the overexpression of apple defense genes such as MpNPR1 (Malnoy et al. 2007), MbR4 R gene (Flachowsky et al. 2008b), or the silencing or gene editing of potential apple susceptibility factors such as HIPM (Malnoy et al. 2008; Campa et al. 2019), DIPM (Pompili et al. 2020) or FHT (Flachowsky et al. 2012). To our knowledge, the only cisgenic strategy employed to improve fire blight resistance of apple was performed with the FB_MR5 resistance gene controlled by its native regulatory sequences (Kost et al. 2015).
Intragenesis and cisgenesis are alternatives to transgenesis defined by Rommens et al. (2007) and Schouten et al. (2006) respectively, and are based on the exclusive use of genetic sequences from the same (or a sexually compatible) species. These strategies aim at improving crop breeding while considering the public’s reluctance toward the use of foreign genes usually present in the genetically modified plant varieties. In the case of cisgenesis, coding sequences (CDS) must be in a sense orientation and flanked by their native promoter and terminator sequences, while intragenesis allows a reorganization of both regulatory and coding sequences, as well as the introduction of mutations (e.g., nucleotide substitutions, sequence deletions, duplications and inversions), to fine tune the expression of the CDS of interest (Holme et al. 2013). These techniques are of particular interest for perennial vegetatively propagated crops such as apple for which conventional breeding is very time-consuming (Limera et al. 2017). In addition, the selectable marker gene is eliminated from cisgenic as well as from intragenic plants, thus allowing sequential introduction of a new transgene, using the same selectable marker, in an elite variety (Halpin 2005).
The generation of intragenic/cisgenic apple plants requires the development and combination of different strategies. The selection of transgenic lines can be based on alternative selectable marker genes from apple such as genes implicated in anthocyanin production (Kortstee et al. 2011) or genes of which certain mutation gives resistance to herbicide (acetolactate synthase; Yao et al. 2013). A recombinase-mediated removal of the unwanted selectable marker sequence has also been used (Herzog et al. 2012; Righetti et al. 2014; Kost et al. 2015).
As for the regulatory sequences, so far, only the apple Rubisco promoter has been used to obtain the constitutive and high expression of an intragene, the R gene Rvi6, to control apple scab caused by the fungi Venturia inaequalis (Vi, Joshi et al. 2011). Overexpression of genes downstream R genes in the defense pathways (i.e. regulators and defense genes) can lead to enhanced resistance but with an important energetic cost that might impede primary plant functions or create developmental disorders. For example, constant overexpression of master-switch genes like NPR1 (Pieterse and Van Loon 2004) can lead to lesion mimic phenotypes (Fitzgerald et al. 2004) and be detrimental to plant development (Gurr and Rushton 2005). Overexpression of phytoalexins or other antimicrobial compounds at high level can also damage tissue integrity (Großkinsky et al. 2012). Therefore, the use of pathogen-inducible promoters to drive regulators of defense pathways, PR genes or toxic antimicrobial genes is a necessity (Gurr and Rushton 2005). To create apple intragenic lines resistant to Ea, we were interested in two kinds of regulatory sequences: (i) an inducible promoter with a fast and strong induction after Ea infection and able to trigger the production of defense mechanisms in the right place at the right time against the bacteria and (ii) a constitutive promoter with a moderate expression level. Such a promoter could ensure the permanent presence of immune receptors such as pattern recognition ones or ones encoded by R genes, with minimal negative tradeoff effects. Previous results led us to investigate the family of polyphenol oxidases (PPO) for this purpose. This complex family of enzymes catalyzes the hydroxylation of monophenols and/or the oxidation of di-phenolic compounds into quinones (Pourcel et al. 2007). A high increase of global enzyme activity has been reported in apple after Ea infection (Skłodowska et al. 2011; Gaucher et al. 2013) and preliminary studies on gene expression by RT-qPCR revealed a clear differential induction of PPO genes—or set of genes—after infection (Dugé de Bernonville 2009).
Here, we took advantage of the recent high-quality apple genome (Daccord et al. 2017) to fully describe the apple PPO family and to select individual genes with differential expression after Ea infection. After cloning, promoters of interest were fused to reporter genes and transiently or stably transformed in apple. This allowed the assessment of their activity under various stresses to evaluate their usefulness in future intragenesis strategies for apple resistance to Ea.
Materials and methods
Material, growth and inoculation conditions
Apple
Four Malus x domestica genotypes were used in this work: the ornamental cv. ‘Evereste’, the rootstock ‘MM106’ and the table apples ‘Golden Delicious’ and ‘Gala’. Experiments were performed in greenhouse on actively growing shoots of young grafts (‘Evereste’ and ‘MM106’) grafted on ‘MM106’, or on actively growing plants not grafted (‘Golden Delicious’), and grown under greenhouse conditions (natural photoperiod, temperatures between 17 and 22 °C). Experiments were also performed on in vitro–growing shoots of three to four cm, used 4 weeks after rooting (Online Resource 1). Micropropagation conditions were as described in Righetti et al. (2014) and rooting conditions as previously reported (Faize et al. 2003).
Erwinia amylovora culture, inoculation and experiments
Two Ea strains were used in this study: wild-type Ea CFBP1430 (Ea wt; Paulin and Samson 1973) and PMV6023, a non-pathogenic T3SS-defective mutant of Ea wt, mutated in hrcV (Ea t3ss; Barny 1995). Prior to each experiment, bacteria were subcultured at 26 °C overnight on solid King’s medium B (King et al. 1954) supplemented with chloramphenicol (20 µg/mL) for the mutant. Bacterial inocula were prepared in sterile distilled water to yield a concentration of 107 colony-forming units (CFU)/mL, supplemented with 0.01% (v/v) of wetting agent Silwet (L-77, De Sangosse Ltd, Cambridge, UK). Mock corresponded to sterile water supplemented with the wetting agent Silwet.
For greenhouse growing plants inoculation was performed by vacuum infiltration as described in Pontais et al. (2008). Briefly, the top of growing shoots were submerged in bacterial suspension and the vacuum was applied for 2 min at − 0.09 Mp (Online Resource 1).
In related experiments, leaf samples were immediately frozen in liquid nitrogen and kept at − 80 °C until analysis. Sampling concerned the youngest expanded leaf of each plant labeled the day of the inoculation. Each sample is a pool of leaves from three different plants and two (n = 2; PPO genes expression analysis in ‘Evereste’ and ‘MM106’ genotypes) to three (n = 3; promoters analysis in ‘Golden Delicious’ transgenic lines) biological repeats have been made by condition (genotype/transgenic line x treatment x time).
For in vitro–growing shoots, four weeks after rooting, shoots were separated from their roots, totally submerged in inoculum and vacuum infiltrated for 2 min at − 0.09 Mp. Shoots were then dried on sterile filter paper and placed for 1 day back on micropropagation medium before sampling.
In related experiments, leaf samples were immediately frozen in liquid nitrogen and kept at − 80 °C until analysis. Sampling concerned all the leaves of each shoot. Each sample is a pool of leaves from three different plants and three (n = 3; transient transformation assay on ‘Gala’ genotype) to six (n = 6; in vitro experiments on ‘Golden Delicious’ transgenic lines) biological repeats have been made by condition (genotype/transgenic line x treatment x time).
Venturia inaequalis culture, inoculation and experiment
The apple scab monoconidial isolate used was EU-B04 from the European collection of V. inaequalis from the European project Durable Apple Resistance in Europe (Lespinasse et al. 2000). Inoculum was prepared as described by Parisi and Lespinasse (1996) to obtain a final concentration of 2.5 × 105 conidia/mL. Inoculation was performed as described by Parisi et al. (1993). Briefly, conidial suspension was applied to runoff on leaves with a manual sprayer. Plants were then incubated for two days under plastic tarpaulin and sprayed three times a day to assure constant leaf wetness. The tarpaulin was then removed and plants grew under greenhouse conditions. Mock corresponded to sterile water.
Leaf samples were immediately frozen in liquid nitrogen and kept at − 80 °C until analysis. Sampling concerned the youngest expanded leaf of each plant labeled the day of the inoculation. Each sample is a pool of leaves from three different plants and three biological repeats (n = 3) have been made by condition (transgenic line × treatment × time).
Agrobacterium culture
Agrobacterium tumefaciens EHA105 (Hood et al. 1993) containing binary expression vectors of interest (Online Resource 2) was cultured on LBA (LB Agar, Sigma-Aldrich, St. Louis, MO, USA) supplemented with appropriate antibiotics and incubated at 28 °C for two days.
H2O2 treatment
H2O2 (30% w/v solution, Fisher Scientific, Loughborough, UK) was used at 10 mM concentration on in vitro–growing shoots. Four weeks after rooting, shoots were separated from their roots and either cultured on micropropagation medium supplemented with 10 mM H2O2 during 1 day before sampling or vacuum infiltrated for 2 min at − 0.09 Mp, dried on sterile filter paper and placed for 1 day back on micropropagation medium before sampling.
Leaf samples were immediately frozen in liquid nitrogen and kept at − 80 °C until analysis. Sampling concerned all the leaves of each shoot. Each sample is a pool of leaves from three different plants and six (n = 6; in vitro experiments on ‘Golden Delicious’ transgenic lines) biological repeats have been made by condition (transgenic line x treatment x time).
Transformation of apple
Agroinfiltration of in vitro rooted plants was used for transient transformation experiments. The inoculum for infiltration was a mix of the strain with the T-DNA of interest (Online Resource 2: p35S, pKFDV02 or pPPO16 from MM106) and the strain with the T-DNA carrying the gene coding the p19 protein of tomato bushy stunt virus as a suppresser of gene silencing (Voinnet et al. 2003), respectively at 5 × 108 CFU/mL and 2.5 × 108 CFU/mL. The cultures were re-suspended in induction buffer (10 mM MES pH 5.6, 10 mM MgCl2, 2% (w/v) sucrose and 150 µM acetosyringone) (Santos-Rosa et al. 2008), mixed at the desired concentration, incubated at 28 °C with shaking for 3 h, and then supplemented with 0.002% (v/v) of wetting agent Silwet before use. Four weeks after rooting, shoots were separated from their roots, totally submerged in inoculum and vacuum infiltrated for 2 min at − 0.09 Mp. Shoots were then rinsed in three successive baths of sterile water, dried on sterile filter paper and placed for 6 days back on micropropagation medium without antibiotics before sampling.
Stable transformation experiments were carried out according as previously reported (Righetti et al. 2014). Presence of transgenes and absence of contaminating agrobacteria were monitored by PCR and sequencing of PCR products. Genomic DNA of apple leaves was extracted as described in Fulton et al. (1995). Primers used for the detection of (i) A. tumefaciens presence, (ii) nptII gene, (iii) p35S:GUS straddled amplification (iv) pPPO16:GUS straddled amplification, (v) pKFDV02:GUS straddled amplification and (vi) elongation factor 1α (EF-1α) coding gene as a marker of plant DNA suitability for PCR are available in Online Resource 3. Amplifications were performed using GoTaq® Flexi DNA Polymerase (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. The PCR reaction conditions were identical for the six genes except the hybridization step which was at 55 °C and not 58 °C for A. tumefaciens detection primers: 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 58 °C for 45 s, 72 °C for 1 min and 30 s, with a final extension at 72 °C for 5 min. The PCR products were separated on a 2% agarose gel. Transgenic lines and control plants were then propagated in vitro and acclimatized in a greenhouse as previously reported (Faize et al. 2003). Before acclimatization, the ploidy level of transgenic lines was checked by flow cytometry, as described in Chevreau et al. (2011), and tetraploid lines were eliminated.
Characterization of apple PPO family
The annotated genes of the ‘Golden Delicious’ double haploid 13 genome (Daccord et al. 2017) have been screened for PFAM motifs specific to the PPO family, namely PF12142 and PF12143. The structural annotation of each detected locus was manually evaluated in considering BLASTX results and RNA contig alignments. The integrity of CDS has cautiously been checked to differentiate functional genes from pseudogenes. The twelve protein sequences deduced from complete and short CDS have been analyzed with targetP (Emanuelsson et al. 2007) and Predotar (Small et al. 2004) for the prediction of N-terminal targeting peptide for the plasts. Phylogenetic tree was built from full-length alignment with Neighbor-joining method, excluding gap positions and tested with Bootstrap method (Kumar et al. 2016). The percent identity matrix of CDS and proteins were built with MUSCLE (Edgar 2004) and Clustal Omega (Sievers et al. 2011) respectively.
Cloning of promoters
Sequence of CaMV 35S promoter in pK7WG2D (Karimi et al. 2002) and sequences of about 2 kb upstream MdPPO16 (MD10G1299400) and MdKFDV02 CDS (MD10G1298200) were downloaded. Primers for cloning (Online Resource 3) were designed with primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Genomic DNA of apple ‘MM106’ was used as template for PCR amplification of promoters with a high fidelity DNA polymerase (Phusion Hot Start II DNA Polymerase, ThermoFisher Scientific, MA, USA) used according to the manufacturer instructions. Amplified fragments were then cloned into pGEM-T easy (Promega, Madison,WI, USA) or p-ENTR/D TOPO (Invitrogen, Carlsbad, CA, USA) when subsequent Gateway cloning was planned.
For apple stable transformation, promoters were cloned with the Gateway system via pENTR-D TOPO (Invitrogen, Carlsbad, CA, USA) into the destination vector pKGWFS7 (Karimi et al. 2002). In this vector the sequence under study controls the expression of a GUS-GFP reporter gene. 2219 bp upstream to MdPPO16 and 2030 bp upstream to MdKFDV02 start codons were cloned and the final constructs were transformed in Agrobacterium strain EHA105 with the helper plasmid pBBR-MCS5. As a positive control for stable transformation assays a plasmid pKGWFS7 carrying the CaMV 35S promoter was used (Online Resource 2).
For transient assays we used either the same plasmids as for stable transformation, or the binary vector pGREEN II 0800-LUC (Hellens et al. 2005). This vector is specifically designed to clone the sequence under study upstream to a firefly luciferase coding sequence. A renilla luciferase coding sequence under the control of a constitutive CaMV 35S promoter is also present as an internal control. The presence of the two luciferases in a single T-DNA reduces the intrinsic variability of leaf agroinfiltration and thus allows reproducible promoter activity quantification. Cloning into this vector was achieved by adding specific restriction sites to the primers. KpnI and NcoI sites were added to the 5’ and 3’ ends respectively of MdPPO16 and MdKFDV02 promoters, while KpnI and HindIII were added to primers used for CaMV 35S promoter. After digestion with restriction enzymes of both vectors and inserts, 1177 bp and 2030 bp of the sequences upstream the start codon were cloned for MdPPO16 and MdKFDV02, respectively. As a positive control for transient assays 1027 bp of CaMV 35S promoter amplified from the plasmid pK7WG2D (Karimi et al. 2002) were also cloned. The final constructs were transformed in Agrobacterium strain EHA105 with the helper plasmid pSoup (Online Resource 2).
DNA extraction, RNA extraction, reverse transcription, and gene expression analysis
Genomic DNA of leaves of apple ‘MM106’ was extracted as described in Fulton et al. (1995).
For RNA extraction, frozen leaves were ground to a fine powder in a ball mill (MM301, Retsch, Hann, Germany). RNA from leaves was extracted as described in Venisse et al. (2002). Purity and concentration of the samples were assayed with a Nanodrop spectrophotometer (ThermoScientific, Rockford, IL, USA). Reverse transcription was performed with M-MLV as described by Promega with OligodT 25 ng/µl or specific primers 0.04 µM final concentrations (Online Resource 3). Intron-spanning primers designed on the EF-1α gene were used to check the absence of genomic DNA contamination.
Quantitative PCR was used to quantify cDNA in samples. Briefly, 3.75 µL of the appropriately diluted samples (ranging from 4 to 12.5-fold) were mixed with 7.5 µL of qPCR mastermix (MasterMix Plus for SYBR© Green I with fluorescein, Eurogentec, Liège, Belgium) in a final volume of 15 µL. Primers designed with Primer3Plus were added according to their optimal concentration (determined for reaction efficiency near to 100%; calculated as the slope of a standard dilution curve; Pfaffl 2001). Primer sequences are indicated in Online Resource 3. Reaction was performed on a DNA Engine thermal cycler Chromo4 (Bio-Rad, Hercules, CA, USA) using the following program: 95 °C, 5 min; 35 cycles comprising 95 °C 15 s, 60 °C 45 s and 72 °C 30 s with real-time fluorescence monitoring. Melt curves were performed at the end of each run to check the absence of primer-dimers and non-specific amplification products. Data were acquired and analyzed with MJ Opticon Monitor Software 3.1 (Bio-Rad, Hercules, CA, USA). Expression profiles of endogenous PPO genes were calculated using the 2−∆∆Ct method and were corrected as recommended in Vandesompele et al. (2002), with three internal reference genes (GADPH, TuA and Actin) used for the calculation of a normalization factor. Data were transformed into log2 scale. Expression levels of the GUS and FIRE reporter genes were calculated using the 2−∆∆Ct method and were corrected with the spectinomycin (SPEC) selection gene or the internal control REN respectively. GUS in pKGWFS7 did not possess an intron so in the transient assay this reporter gene actually dosed expression from both the plant and Agrobacterium. SPEC gene expression, specific from the bacteria because present in the plasmid but not in the T-DNA, was used to calibrate samples amongst themselves to eliminate the potential part of expression due to bacteria in the GUS measure differences.
Luciferase and GUS activity assay
Frozen leaves were ground to a fine powder in a ball mill (MM301, Retsch, Hann, Germany). Luciferase activities were measured using the dual luciferase assay system (Promega, Madison, USA) according to the manufacturer’s instructions but with some modifications. 150 µL of Passive Lysis Buffer were added to the resulting powders and samples were placed on ice for 15 min and vortexed several times in the meantime. For luciferase activity measurements (firefly and renilla), 10 µL of each extract were transferred into a 96-well white solid plate (Fisher Scientific ltd., Montreal, Quebec). The luminescence was measured using the FluoStar Optima Luminometer (BMG Lab Technologies, Offenburg, Germany) with the injection of 60 µL of LARII reagent (Firefly luciferase activity) and then 60 µL of the Stop & Glo reagent (Renilla luciferase activity). For Gus activity measurements, 800 µL of extraction buffer (50 mM Na2HPO4, pH 7.0, 1 mM Na2EDTA pH 8.0, 0.1% (v/v) Triton X-100, 0.1% (w/v) sodium lauryl sarcosine) were added to the leaf powders. The homogenates were centrifuged at 4 °C for 1 min at 10,000 rpm. The supernatants were tenfold diluted and 100 µL of each dilution were transferred into a 96-well white solid plate. Quantitative fluorimetric GUS activity assay was performed using the FluoStar Optima Luminometer (BMG Lab Technologies, Offenburg, Germany) with the injection of 4 µL of the substrate 4-methylumbelliferyl β-glucuronide (MUG). Luciferase and GUS activities were standardized to the protein concentration (Bradford 1976) of the extracts and firefly luciferase activity was normalized to renilla luciferase activity.
Statistics analysis
All statistical analyses were performed with R 3.4 (R Development Core Team 2016) using the nonparametric rank-based statistical test Kruskal–Wallis. Treatments with significant influence (p < 0.05) were studied more in depth by Fisher’s least significant difference (LSD) as a post hoc test for pairwise comparisons (α = 0.05). Means with different letters are statistically significant.
Results
PPO encoding genes in the apple genome
Screening the ‘Golden Delicious’ double haploid 13 genome (Daccord et al. 2017) revealed the presence of a PPO gene family encompassing ten members with similar gene structure of one or two exons, encoding proteins ranging from 587 to 610 residues (Table 1). N-terminal signal peptides for chloroplast targeting were predicted for all of them. PPO proteins are characterized by three conserved PFAM domains: the tyrosinase superfamily domain PF00264, the PPOI_DWL domain PF12142 and the PPO1_KFDV domain PF12143. Apple PPO genes are located on two clusters on chromosomes 5 (five genes) and 10 (five genes), two chromosomes known to result from a whole genome duplication (Daccord et al. 2017). Close examination of these two chromosomal regions identified two additional PPO-like encoding genes, one on chromosome 5 and the other on chromosome 10, which conserved only the C-terminal KFDV domain and were also predicted to be addressed to the chloroplast (Table 1). Six pseudogenes were finally found, four on chromosome 5 and two on chromosome 10. Their CDS are disrupted by deletion, transposable element insertion, frameshift and/or stop codons (Table 1). We named PPO genes and pseudogenes MdPPO01 to MdPPO16, and PPO-like genes MdKFDV01 and MdKFDV02. Phylogeny generated from the 30 PPO Rosaceae homologs identified in Genbank database revealed six subfamilies (Fig. 1). Identity matrices obtained using nucleic or protein sequences of the ten apple PPOs showed a very high conservation level between accessions inside each apple PPO sub-family (Online Resource 4).
Phylogenetic tree of PPO homologs in Rosaceae. The tree was built with the neighbor-joining method from the multiple alignment of 30 homologous proteins. Gaps were ignored for tree building and 1000 bootstrap replicates were used to determine the robustness of each node (the bigger the green circle size, the more robust the node). The six PPO subfamilies are highlighted with different colors (white, purple, orange, green, blue and yellow). Except for apple for which gene model ID is used (written in black), each protein is labeled with two letters (species) and its GenBank ID or XP number. Frv, Fragaria vesca (red); Prp, Prunus persica (L.) Batsch (orange); Pyb, Pyrus bretschneideri (green) (colour figure online)
Apple PPO gene expression profiles
To identify PPO promoters differentially responding to Ea infection, we quantified by RT-qPCR the specific expression of PPO genes in apple infected tissues. For this study pseudogenes (MdPPO1, MdPPO4, MdPPO7, MdPPO9, MdPPO11 and MdPPO14) were discarded, as well as MdPPO12, MdPPO13 and MdPPO15 for which the design of specific primers was attempted repeatedly base on SNPs (Single Nucleotide Polymorphism; high level of identity ≥ 97.7%; Online Resource 4), but failed. Primers for the remaining seven PPO and the two PPO-like genes were designed with the aim of quantifying their expression in Ea infected leaves of two apple genotypes with contrasted susceptibilities to fire blight, the susceptible ‘MM106’ and the resistant ‘Evereste’ (Venisse et al. 2002). During the test of primers efficacy performed using as template a cDNA pool from these Ea infected apple genotypes, we obtained very weak amplifications for MdPPO02, MdPPO03, MdPPO05, MdPPO06, MdPPO08 and MdPPO10 contrasting with the substantial ones for MdKFDV01, MdKFDV02 and MdPPO16 (Online Resource 5). Therefore gene expression kinetics are only shown for MdKFDV01, MdKFDV02 and MdPPO16 (Fig. 2). Analyses were performed in untreated leaves and in leaves challenged either with a wild-type strain of Ea (Ea wt) or a T3SS deficient mutant (Ea t3ss) or mock at 6, 24 and 48 h post-treatment (hpt). A higher constitutive expression in untreated leaves of MdPPO16 and MdKFDV02 compared to MdKFDV01 was observed in ‘Evereste’. Ea t3ss and mock treatments triggered similar expression changes in the two genotypes, peaking at 6 hpt especially for MdPPO16 probably due to the stress caused by the infiltration method. A strong increase in MdPPO16 expression was recorded in both genotypes challenged with Ea wt, suggesting a type III effector dependent induction. No noticeable modulation was observed in MdKFDV01 and MdKFDV02 expression levels whatever the treatment, except for Ea wt that seemed to slightly modulate the expression of MdKFDV01 in ‘MM106’ at 24 and 48 hpt in one replicate only. Promoter of MdKFDV02 from ‘MM106’, thereafter named pKFDV02, was selected for further investigation instead of promoter of MdKFDV02 from ‘Evereste’ because expression of MdKFDV02 was more stable throughout the kinetics (Fig. 2). Promoter of MdPPO16 from ‘MM106’, thereafter named pPPO16, was also selected for further investigation instead of promoter of MdPPO16 from ‘Evereste’ because MdPPO16 expression throughout the kinetics was similar for the two genotypes (Fig. 2). We found 95.17% identity between sequences of 2218 bp length upstream MdPPO16 CDS in ‘MM106’ and ‘Evereste’.
Expression profiling of MdPPO16, MdKFDV01 and MdKFDV02 in ‘MM106’ (susceptible to fire blight) and ‘Evereste’ (resistant to fire blight) genotypes. Log2 expression levels were measured in untreated leaves and in mock, Ea t3ss or Ea wt infiltrated-leaves at 6, 24, 48 hpt. Expression levels for each gene were calibrated to the mean expression value of the T0 MM106 samples and normalized with three reference genes (GAPDH, TuA and ACTIN). Bars represent maximum and minimum values from two independent experiments (n = 2)
Promoter activity during transient expression
The regions upstream of MdPPO16 (1177 bp; MK873007 in GenBank repository) and MdKFDV02 (2030 bp; MK873006 in GenBank repository) CDS in ‘MM106’ genotype were cloned, and tested as a first approach in a transient expression assay in apple leaves using GUS (β-glucuronidase) as a reporter to quantify promoter activity in untreated, mock or Ea-infiltrated tissues. Rooted in vitro plants of ‘Gala’ were agroinfiltrated with EHA105 carrying different T-DNAs including pPPO16:GUS, pKFDV02:GUS or p35S:GUS as a control. Five days later, plants were infiltrated with mock or Ea wt and gene expression of GUS measured 24 h later by RT-qPCR and calibrated to eliminate expression differences due to bacteria. GUS gene expression was stable in all samples under the control of pKFDV02 (Fig. 3) and had comparable levels to that observed in Ea-infiltrated leaves under the control of pPPO16. Under the control of pPPO16, a strong induction of the GUS expression was observed in leaves challenged with Ea wt (a fivefold increase approximately, Fig. 3). The same transient expression assay was repeated once in the other genotype ‘Golden Delicious’ with firefly luciferase (FIRE) instead of GUS as a reporter gene (Online Resource 6), to quantify promoter activity both at the transcriptional and enzymatic level. FIRE gene expression and protein activity were stable in all samples under the control of pKFDV02 (Online Resource 6A and 6B respectively) and had comparable levels to that observed in untreated and mock-infiltrated leaves under the control of pPPO16 or p35S. Under the control of pPPO16, a strong induction of the FIRE activity was observed in leaves challenged with Ea wt, both at the transcriptional and enzymatic level (a twofold increase approximately, Online Resource 6A and 6B).
Expression of GUS gene driven by pPPO16 and pKFDV02 in transient assays. Relative expression of GUS reporter gene driven by p35S, pPPO16 and pKFDV02 in untreated (nt, white), mock (light gray) or Ea wt (black)-infiltrated leaves (24 hpt) of transiently transformed ‘Gala’ in vitro plants, 6 days after agroinfiltration. GUS raw expression levels of each sample were calibrated to the mean value of the samples pPPO16:GUS-nt, and normalized with SPEC gene to eliminate expression differences due to bacteria. Bars represent SEM from 3 biological repeats (n = 3). Letters indicate statistical classes (Kruskal–Wallis, p < 0.05)
Promoter activity in stable transgenic clones
The results obtained with the transient assay encouraged us to perform apple stable transformations with two constructs carrying each promoter fused with the GUS gene as marker gene (pPPO16:GUS and pKFDV02:GUS), and to compare these to p35S:GUS transformed control. We respectively obtained one (224C), two (222A and D) and four (217F, O, R, S) transgenic lines of ‘Golden Delicious’ transformed with pPPO16:GUS, pKFDV02:GUS and p35S:GUS. The unique line pPPO16:GUS and the more vigorous line pKFDV02:GUS (222D) were kept for subsequent analyses. For p35S:GUS, subsequent analyses were performed on two lines harboring a moderate GUS expression (lines 217O and S; Online Resource 7). Assessment of transgenic lines selected for the further analyses are displayed in Online Resource 8. After in vitro multiplication, all stable transgenic lines were acclimatized and grown in greenhouse. The expression of the reporter gene was assessed by RT-qPCR in untreated, mock and Ea wt-infiltrated leaves at 24 hpt. In pKFDV02 line, activity was not significantly different from p35S lines in all conditions (nt, mock and Ea wt treatments, Fig. 4). By contrast, GUS expression was very weak in untreated and mock-infiltrated leaves of pPPO16:GUS lines and exhibited a strong and significant tenfold induction in inoculated ones, reaching levels similar to p35S:GUS lines. Altogether these results corroborate those of the transient expression assay and show that pPPO16 but not pKFDV02 is strongly induced by Ea infection. The pPPO16-driven induction of the GUS in leaves challenged with Ea wt was also confirmed at the enzymatic level 40 and 48 hpt (Fig. 5).
pPPO16 and pKFDV02-driven GUS expression in ‘Golden Delicious’ transgenic lines cultivated in greenhouse and challenged with Ea. Relative expression of GUS reporter gene driven by p35S, pPPO16 and pKFDV02 in untreated (nt, white), mock (light gray) or Ea wt (black)-infiltrated leaves (24 hpt) of transgenic lines. GUS raw expression levels of each sample were calibrated to the mean value of the samples pPPO16:GUS-nt, and normalized with ACTIN. 217O and 217S represent independent lines of p35S:GUS, harboring a moderate GUS expression. Bars represent SEM from 3 biological repeats (n = 3). Letters indicate statistical classes (Kruskal–Wallis, p < 0.05)
pPPO16-driven GUS activity in ‘Golden Delicious’ transgenic line challenged with Ea. Enzymatic activities of GUS reporter driven by pPPO16 in untreated (nt, white), mock (light gray) and Ea wt (black)-infiltrated leaves of in vitro plants at 24, 40 and 48 hpt. GUS activity is expressed in nmoles MU (methylumbelliferone)/min/mg of total proteins. Bars represent SEM from 3 biological repeats (n = 3). Letters indicate statistical classes (Kruskal–Wallis, p < 0.05)
To determine which component of the Ea pathogenesis is responsible for the induction of pPPO16, i.e. a functional T3SS of the bacterium and/or the ROS production during the infectious process, GUS expression was recorded in transgenic rooted in vitro plants carrying pPPO16:GUS and pKFDV02:GUS at 24 hpt after the following different treatments: mock, Ea t3ss and Ea wt by leaf infiltration and H2O2 by leaf infiltration or by incorporation in the culture medium (Fig. 6). GUS expression was relatively stable when mediated by the promoter pKFDV02, although a slight but significant decrease of activity was observed after H2O2 treatments (infiltration and culture medium) compared to mock treatment. No change in GUS expression was observed in pPPO16:GUS line treated with mock, Ea t3ss and H2O2, while again a strong and significant tenfold induction was observed when this line was inoculated with Ea wt. Taken together, these results highlight the ability of Ea to strongly and specifically induce pPPO16 (and not pKFDV02), probably as an effect of a functional T3SS rather than H2O2 production.
pPPO16 and pKFDV02-driven GUS expression in ‘Golden Delicious’ transgenic lines challenged with Ea. Relative expression of GUS reporter driven by p35S, pPPO16 and pKFDV02 in untreated (nt), H2O2-medium, mock or H2O2 or Ea t3ss or Ea wt-infiltrated leaves (24 hpt) from in vitro plants of transgenic lines. GUS raw expression levels of each sample were calibrated to the mean value of the samples pPPO16:GUS-nt, and normalized with ACTIN. 217O and 217S represent independent lines of p35S:GUS, harboring a moderate GUS expression. Bars represent SEM from six biological repeats (n = 6). Letters indicate statistical classes (Kruskal–Wallis, p < 0.05)
To check pPPO16 ability to be specifically activated by Ea and to observe pKFDV02 behavior in response to another pathogen, the same transgenic lines were challenged with the pathogenic fungus Vi responsible for apple scab. Transgenic lines were therefore cultivated in greenhouse and GUS expression was assessed in untreated, mock and Vi-sprayed leaves at 1, 3 and 10 days post-treatment (dpt), the development of fungus being slower than that of Ea. Results indicated that up to 3 dpt, the GUS expression mediated by pPPO16 was not affected by Vi in comparison to the corresponding mock controls (Fig. 7). However a strong and significant 15-fold induction was observed at 10 dpt, suggesting that pPPO16 could be activated by another apple pathogen. Regarding pKFDV02, GUS expression was not significantly induced by Vi inoculation in the first 3 days, but considerably raised at 10 dpt in both mock or Vi-sprayed leaves, approximately 20-fold relative to the beginning of the experiment (pKFDV02:GUS-nt). The same phenomenon was also observed at 10 dpt in the youngest leaf of each plant which did not receive any treatment (Online Resource 9), suggesting the presence of a different unknown factor affecting pKFDV02.
pPPO16 and pKFDV02-driven GUS expression in ‘Golden Delicious’ transgenic lines cultivated in greenhouse and challenged with Vi. Relative expression levels of GUS reporter gene driven by p35S, pPPO16 and pKFDV02 in untreated (nt), mock or Vi-sprayed leaves (1, 3, 10 dpt) from transgenic lines. GUS raw expression levels of each sample were calibrated to the mean value of the samples pPPO16:GUS-nt, and normalized with ACTIN. 217O and 217S represent independent lines of p35S:GUS, harboring a moderate GUS expression. Bars represent SEM from 3 biological repeats (n = 3). Letters indicate statistical classes (Kruskal–Wallis, p < 0.05)
Discussion
Our work identified ten potentially functional apple PPO-encoding genes harboring the three known typical domains tyrosinase (PF00264), DWL (PF12142) and KFDV (PF12143), located on two duplicated chromosomes (5 and 10), all being addressed to the chloroplast and distributed in five phylogenetic sub-groups. This result completes the survey that Tran et al. (2012) performed among 25 land plants, describing PPO gene families varying in size (1–13) except in the genus Arabidopsis whose genome does not contain PPO sequences. Clustering of PPO genes at the same chromosomal location has already been observed in other plant species and indicates tandem gene duplications (Tran et al. 2012).
In the same chromosomal regions, we also identified six pseudogenes with similarity to PPO but with discrepancies such as deletions, premature stop codons and/or frameshifts, and two PPO-like genes of unknown function with only the KFDV domain. Doubts can be raised over their function as true polyphenol oxidases considering that they lack the common central domain of tyrosinase responsible of the oxidation process. Despite these doubts, KFDV genes were conserved in our study as PPO-like genes according to the fact that they have homologs in numerous dicot species.
Plant PPO genes are known to be involved in different physiological processes, from stress response to developmental regulation and environmental adaptation, as confirmed by their differential expression patterns in different situations (Thipyapong and Steffens 1997; Constabel and Barbehenn 2008; Tran and Constabel 2011; Thipyapong et al. 2007). This makes regulatory sequences of PPO genes good candidates for diversified strategies of intragenesis. Unfortunately in our experiments, analyses showed that the expression driven by pKFDV02, originally chosen for an expected constitutive activity was modulated by unspecified factors. As only one transgenic line with the pKFDV02 construction was further analyzed and that this modulation is not corroborated by transient expression data, we cannot exclude that it derives from insertion effects, and not from the promoter. But for now, this result invalidated pKFDV02 as a good candidate to drive a constitutive but weak expression for apple intragenesis development. On the other hand the fact that we found differential expression of PPO genes in response to Ea is coherent with previous works in other plant species showing induction in response to biotic stresses only for some PPO genes, in both incompatible and compatible interactions (Tran and Constabel 2011; Rinaldi et al. 2007). In our hands MdPPO16 induction in response to Ea has been recorded in three different genotypes (‘MM106’, ‘Evereste’ and ‘Gala’; Vergne et al. 2014 and this work). MdPPO16 was also shown to be induced by wounding (Boss et al. 1995) and in fruit flesh browning disorder (Di Guardo et al. 2013), suggesting various functions for this gene.
Transient and stable transgenic assays using reporter genes fused to the immediate upstream region from the start codon of MdPPO16 confirmed that this regulatory sequence was efficient to obtain the desired Ea-inducible expression pattern. Only one stable transgenic line was recovered with the pPPO16-GUS construction so we cannot affirm that the observed expression profile in that line is not affected, positively or negatively, by insertion effects. Despite this drawback, pPPO16 promoter in 224C line show a quick and strong induction in leaves challenged with Ea, at the transcriptomic and enzymatic levels, in accordance with results obtained in transient assays with GUS or FIRE reporter genes. Thus we are confident on other results get with this line. In an intragenesis strategy designed to confer resistance to Ea, the use of such a promoter should ensure the precise induction of the intragene from the beginning of the infection process. Because a functional bacterial T3SS was required for this promoter induction, it should also avoid inappropriate activation in response to MAMPs (Microbial Associated Patterns, Choi and Klessig 2016) of Ea or of other bacteria with similar conserved motifs potentially present on or inside the plant.
Induction of pPPO16 seems to be linked to the loss of cellular integrity. Three lines of evidence support this hypothesis: (i) pPPO16 induction requires Ea with a functional T3SS, which enables the injection of the major effector DspA/E into the plant cell, causing cell death (Boureau et al. 2006), (ii) pPPO16 activation in compatible interaction with Vi occurred at 10 dpt in our experiments, which correspond to the beginning of tissue rupture by conidiogenesis (Ortega et al. 1998), and (iii) previous work shows the induction of MdPPO16 after wounding (Boss et al. 1995). A specific induction of pPPO16 linked to cell death is particularly interesting in the objective of controlling fire blight disease. It should ensure the induction of the intragene not only in the case of a real bacterial attack but also as a preventive barrier at wound sites caused by insects or climatic events, both acting as entry points for the bacteria. Despite the strong induction of pPPO16 in response to Vi infection, it seems however unwise to consider this promoter in intragenic strategies for apple scab control, as it is only activated during the late phase of infection, i.e. conidiogenesis. Induction of a PPO gene during urediospore formation was already noticed in hybrid poplar/Melampsora laricipopulina interaction (Tran and Constabel 2011).
We did not observe any response of pPPO16 following exogenous application of H2O2, known as a precocious ROS produced during the oxidative burst accompanying Ea infection process (Vrancken et al. 2013). The concentration of H2O2 used in that work is moderate and known to modulate several defense genes in apple without leading to impaired tissue integrity (Dugé de Bernonville et al. 2014). The non-response of pPPO16 following that moderate treatment should indicate that the expression driven by this promoter will remain stable despite moderate increase of H2O2 concentrations known to occur in various stress conditions (Saxena et al. 2016).
In the search for apple resistance, several cisgenic strategies have already been developed (Kost et al. 2015; Krens et al. 2015), but only one case of intragenic strategy has been tested, against another pathogen than Ea (Vi; Joshi et al. 2011). To create efficient fire blight intragenic resistances in apple, several candidate genes could be placed under the control of the pPPO16 promoter characterized in our study: important regulators of defense pathways like NPR1 (Malnoy et al. 2007), members of calcium-dependant protein kinases family (Kanchiswamy et al. 2013), genes involved in the jasmonic acid pathway (Dugé de Bernonville et al. 2012) or genes that increased oxidation of phenolic compounds (Flachowsky et al. 2010; Gaucher et al. 2013; Hutabarat et al. 2016).
The present work represents the first step towards the development of efficient “all native” solutions for apple fire blight resistance. As far as we know, pPPO16 is the first cloned apple promoter inducible by Ea. Considering the narrowness of the gene pool screened to retrieve it, i.e. the MdPPO family, pPPO16 could not be the best Ea inducible promoter candidate and comprehensive genomic level expression analyses are needed to find such candidates. Further work will be also needed to choose optimal candidate genes combining high efficiency for disease resistance, limited risk of break-down and absence of adverse effects on plant physiology.
Data availability
pKFDV02 (MK873006) and pPPO16 (MK873007) sequences are available in GenBank repository. Accession numbers of other sequences analyzed in this work; from repository GenBank or https://iris.angers.inra.fr/gddh13 (choose “Visit the apple genome”, enter accession number to search, right-click of mouse on the CDS of the “curated CDS” layer and choose view details to access sequence); are in Table 1, Online Resource 2 and 3, or in references given in these tables. Data generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Change history
08 June 2022
A Correction to this paper has been published: https://doi.org/10.1007/s00299-022-02887-6
References
Barny MA (1995) Erwinia amylovora hrpN mutants, blocked in harpin synthesis, express a reduced virulence on host plants and elicit variable hypersensitive reactions on tobacco. Eur J Plant Pathol 101:333–340. https://doi.org/10.1007/BF01874789
Borejsza-Wysocka E, Norelli JL, Aldwinckle HS et al (2010) Stable expression and phenotypic impact of attacin E transgene in orchard grown apple trees over a 12 years period. BMC Biotechnol 10:41. https://doi.org/10.1186/1472-6750-10-41
Boss PK, Gardner RC, Janssen BJ et al (1995) An apple polyphenol oxidase cDNA is up-regulated in wounded tissues. Plant Mol Biol 27:429–433. https://doi.org/10.1007/BF00020197
Boureau T, El Maarouf-Bouteau H, Garnier A et al (2006) DspA/E, a type III effector essential for Erwinia amylovora pathogenicity and growth in planta, induces cell death in host apple and non-host tobacco plants. Mol Plant Microbe Interact 19:16–24. https://doi.org/10.1094/MPMI-19-0016
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Broggini GA, Wöhner T, Fahrentrapp J et al (2014) Engineering fire blight resistance into the apple cultivar “Gala” using the FB_MR5 CC-NBS-LRR resistance gene of Malus × robusta 5. Plant Biotechnol J 12:728–733. https://doi.org/10.1111/pbi.12177
Campa M, Piazza S, Righetti L et al (2019) HIPM is a susceptibility gene of Malus spp.: reduced expression reduces susceptibility to Erwinia amylovora. Mol Plant Microbe Interact 32:167–175. https://doi.org/10.1094/MPMI-05-18-0120-R
Chevreau E, Dupuis F, Taglioni JP et al (2011) Effect of ectopic expression of the eutypine detoxifying gene Vr-ERE in transgenic apple plants. Plant Cell Tissue Org Cult 106:161–168. https://doi.org/10.1007/s11240-010-9904-4
Choi HW, Klessig DF (2016) DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol 16:232. https://doi.org/10.1186/s12870-016-0921-2
Constabel CP, Barbehenn RV (2008) Defensive roles of polyphenol oxidase in plants. In: Schaller A (ed) Induced plant resistance to herbivory. Springer, New York, pp 253–269
Daccord N, Celton JM, Linsmith G et al (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49:1099–1106. https://doi.org/10.1038/ng.3886
Di Guardo M, Tadiello A, Farneti B et al (2013) A multidisciplinary approach providing new insight into fruit flesh browning physiology in apple (Malus x domestica Borkh.). PLoS ONE 8:e78004. https://doi.org/10.1371/journal.pone.0078004
Dugé de Bernonville T (2009) Caractérisations histologique, moléculaire et biochimique des interactions compatible et incompatible entre Erwinia amylovora, agent du feu bactérien, et le pommier (Malus x domestica). PhD thesis Angers University. https://tel.archives-ouvertes.fr/tel-00482385/fr/
Dugé de Bernonville T, Gaucher M, Flors V et al (2012) T3SS-dependent differential modulations of the jasmonic acid pathway in susceptible and resistant genotypes of Malus spp. challenged with Erwinia amylovora. Plant Sci 188–189:1–9. https://doi.org/10.1016/j.plantsci.2012.02.009
Dugé de Bernonville T, Marolleau B, Staub J et al (2014) Using molecular tools to decipher the complex world of plant resistance inducers: an apple case study. J Agric Food Chem 62:11403–11411. https://doi.org/10.1021/jf504221x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Emanuelsson O, Brunak S, von Heijne G et al (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971. https://doi.org/10.1038/nprot.2007.131
Faize M, Malnoy M, Dupuis F et al (2003) Chitinases of Trichoderma atroviride induce scab resistance and some metabolic changes in two cultivars of apple. Phytopathol 93:1496–1504. https://doi.org/10.1094/PHYTO.2003.93.12.1496·
Fitzgerald HA, Chern MS, Navarre R et al (2004) Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol Plant Microbe Interact 17:140–151. https://doi.org/10.1094/MPMI.2004.17.2.140
Flachowsky H, Richter K, Kim WS et al (2008a) Transgenic expression of a viral EPS-depolymerase is potentially useful to induce fire blight resistance in apple. Ann Appl Biol 153:345–355. https://doi.org/10.1111/j.1744-7348.2008.00264.x
Flachowsky H, Peil A, Rollins J et al (2008b) Improved fire blight resistance in transgenic apple lines by constitutive overexpression of the mbr4 gene of Malus x baccata. Acta Hort 793:287–291. https://doi.org/10.17660/ActaHortic.2008.793.42
Flachowsky H, Szankowski I, Fischer TC et al (2010) Transgenic apple plants overexpressing the Lc gene of maize show an altered growth habit and increased resistance to apple scab and fire blight. Planta 231:623–635. https://doi.org/10.1007/s00425-009-1074-4
Flachowsky H, Halbwirth H, Treutter D et al (2012) Silencing of flavanone-3-hydroxylase in apple (Malus x domestica Borkh) leads to accumulation of flavanones, but not to reduced fire blight susceptibility. Plant Physiol Biochem 51:18–25. https://doi.org/10.1016/j.plaphy.2011.10.004
Fulton TM, Chunzoongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209. https://doi.org/10.1007/BF02670897
Gaucher M, Dugé de Bernonville T, Guyot S et al (2013) Same ammo, different weapons: enzymatic extracts from two apple genotypes with contrasted susceptibilities to fire blight (Erwinia amylovora) differentially convert phloridzin and phloretin in vitro. Plant Physiol Biochem 72:178–189. https://doi.org/10.1016/j.plaphy.2013.03.012
Großkinsky DK, van der Graaf E, Roitsch T (2012) Phytoalexin transgenics in crop protection. Plant Sci 195:54–70. https://doi.org/10.1016/j.plantsci.2012.06.008
Gurr SJ, Rushton PJ (2005) Engineering plants with increased disease resistance: what are we going to express? Trends Biotech 23:275–282. https://doi.org/10.1016/j.tibtech.2005.04.007
Halpin C (2005) Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotechnol J 3:141–155. https://doi.org/10.1111/j.1467-7652.2004.00113.x
Hellens RP, Allan AC, Friel EN et al (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:1–13. https://doi.org/10.1186/1746-4811-1-13
Herzog K, Flachowsky H, Deising HB et al (2012) Heat-shock-mediated elimination of the nptII marker gene in transgenic apple (Malus×domestica Borkh.). Gene 498:41–49. https://doi.org/10.1016/j.gene.2012.01.074
Holme IB, Wendt T, Holm PB (2013) Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol J 11:395–407. https://doi.org/10.1111/pbi.12055
Hood EE, Gelvin SB, Melchers LS et al (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218. https://doi.org/10.1007/BF01977351
Hutabarat OS, Flachowsky H, Regos I et al (2016) Transgenic apple plants overexpressing the chalcone 3-hydroxylase gene of Cosmos sulphureus show increased levels of 3-hydroxyphloridzin and reduced susceptibility to apple scab and fire blight. Planta 243:1213–1224. https://doi.org/10.1007/s00425-016-2475-9
Joshi SG, Schaart JG, Groenwold R et al (2011) Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol Biol 75:579–591. https://doi.org/10.1007/s11103-011-9749-1
Kanchiswamy CN, Mohanta TK, Capuzzo A et al (2013) Differential expression of CPKs and cytosolic Ca2+ variation in resistant and susceptible apple cultivars (Malus x domestica) in response to the pathogen Erwinia amylovora and mechanical wounding. BMC Genom 14:760. https://doi.org/10.1186/1471-2164-14-760
Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195. https://doi.org/10.1016/S1360-1385(02)02251-3
Khan MA, Zhao YF, Korban SS (2012) Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol Biol Rep 30:247–260. https://doi.org/10.1007/s11105-011-0334-1
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307
Kortstee AJ, Khan SA, Helderman C et al (2011) Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res 20:1253–1264. https://doi.org/10.1007/s11248-011-9490-1
Kost TD, Gessler C, Jänsch M et al (2015) Development of the first cisgenic apple with increased resistance to fire blight. PLoS ONE 10:e0143980. https://doi.org/10.1371/journal.pone.0143980
Krens FA, Schaart JG, van der Burgh AM et al (2015) Cisgenic apple trees; development, characterization, and performance. Front Plant Sci 6:286. https://doi.org/10.3389/fpls.2015.00286
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lespinasse Y, Durel CE, Parisi L et al (2000) A European project: D.A.R.E. durable apple resistance in Europe. Acta Hortic 538:197–200. https://doi.org/10.17660/ActaHortic.2000.538.32
Limera C, Sabbadini S, Sweet JB et al (2017) New biotechnological tools for the genetic improvement of major woody fruit species. Front Plant Sci 8:1418. https://doi.org/10.3389/fpls.2017.01418
Malnoy M, Jin Q, Borejsza-Wysocka EE et al (2007) Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol Plant Microbe Interact 20:1568–1580. https://doi.org/10.1094/MPMI-20-12-1568
Malnoy M, Borejsza-Wysocka EE, Pascal-Omenaca L et al (2008) Silencing of HIPM, the apple protein that interacts with HrpN of Erwinia amylovora. Acta Hortic 793:261–264. https://doi.org/10.17660/ActaHortic.2008.793.38
Ortega F, Steiner U, Dehne HW (1998) Induced resistance to apple scab: microscopic studies on the infection cycle of Venturia inaequalis (Cke.) Wint. J Phytopathol 146:399–405. https://doi.org/10.1111/j.1439-0434.1998.tb04771.x
Parisi L, Lespinasse Y (1996) Pathogenicity of Venturia inaequalis strains of race 6 on apple clones (Malus sp.). Plant Dis 80:1179–1183. https://doi.org/10.1094/PD-80-1179
Parisi L, Lespinasse Y, Guillaumes J et al (1993) A new race of Venturia inaequalis virulent to apples with resistance due to the Vf gene. Phytopathology 83:533–537. https://doi.org/10.1007/978-94-011-0467-8_16
Paulin JP (1996) Control of fire blight in European pome fruits. Outlook Agric 25:49–55. https://doi.org/10.1177/003072709602500109
Paulin JP, Samson R (1973) Le feu bactérien en France. II. Caractères des souches d’Erwinia amylovora (Burrill) isolées du foyer franco-belge. Annales De Phytopathologie 5:389–397
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Pieterse CM, Van Loon LC (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7:456–464. https://doi.org/10.1016/j.pbi.2004.05.006
Pompili V, Dalla Costa L, Piazza S et al (2020) Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol J 18:845–858. https://doi.org/10.1111/pbi.13253
Pontais I, Treutter D, Paulin JP et al (2008) Erwinia amylovora modifies phenolic profiles of susceptible and resistant apple through its type III secretion system. Physiol Plant 132:262–271. https://doi.org/10.1111/j.1399-3054.2007.01004.x
Pourcel L, Routaboul JM, Cheynier V et al (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 12:29–36. https://doi.org/10.1016/j.tplants.2006.11.006
R Development Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Righetti L, Djennane S, Berthelot P et al (2014) Elimination of the nptII marker gene in transgenic apple and pear with a chemically inducible R/Rs recombinase. Plant Cell Tissue Org Cult 117:335–348. https://doi.org/10.1016/j.gene.2012.01.074
Rinaldi C, Kohler A, Frey P et al (2007) Transcript prowling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora laricipopulina. Plant Physiol 144:347–366. https://doi.org/10.1104/pp.106.094987
Rommens CM, Haring MA, Swords K et al (2007) The intragenic approach as a new extension to traditional plant breeding. Trends Plant Sci 12:397–403. https://doi.org/10.1016/j.tplants.2007.08.001
Santos-Rosa M, Poutaraud A, Merdinoglu D et al (2008) Development of a transient expression system in grapevine via agro-infiltration. Plant Cell Rep 27:1053. https://doi.org/10.1007/s00299-008-0531-z
Saxena I, Srikanth S, Chen Z (2016) Talk between H2O2 and interacting signal molecules under plant stress response. Front Plant Sci 7:570. https://doi.org/10.3389/fpls.2016.00570
Schouten HJ, Krens FA, Jacobsen E (2006) Do cisgenic plants warrant less stringent oversight? Nat Biotechnol 24:753. https://doi.org/10.1038/nbt0706-753
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Skłodowska M, Gajewska E, Kuźniak E et al (2011) Antioxidant profile and polyphenol oxidase activities in apple leaves after Erwinia amylovora infection and pretreatment with a benzothiadiazole-type resistance inducer (BTH). J Phytopathol 159:495–504. https://doi.org/10.1111/j.1439-0434.2011.01793.x
Small I, Peeters N, Legeai F et al (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590. https://doi.org/10.1002/pmic.200300776
Thipyapong P, Steffens JC (1997) Tomato polyphenol oxidase (differential response of the polyphenol oxidase F promoter to injuries and wound signals). Plant Physiol 115:409–418. https://doi.org/10.1104/pp.115.2.409·
Thipyapong P, Stout MJ, Attajarusit J (2007) Functional analysis of polyphenol oxidases by antisense/sense technology. Molecules 12:1569–1595. https://doi.org/10.3390/12081569·
Tran LT, Constabel CP (2011) The polyphenol oxidase gene family in poplar: phylogeny, differential expression and identification of a novel, vacuolar isoform. Planta 234:799–813. https://doi.org/10.1007/s00425-011-1441-9
Tran LT, Taylor JS, Constabel CP (2012) The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC Genom 13:395. https://doi.org/10.1186/1471-2164-13-395
Vandesompele J, De Preter K, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034
Vanneste JL (2000) Fire blight: the disease and its causative agent, Erwinia amylovora. CABI, Wallingford
Venisse JS, Malnoy M, Faize M et al (2002) Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol Plant Microbe Interact 15:1204–1212. https://doi.org/10.1094/MPMI.2002.15.12.1204
Vergne E, Dugé de Bernonville T, Dupuis F et al (2014) Membrane targeted HrpNEa can modulate apple defense gene expression. Mol Plant Microbe Interact 27:125–135. https://doi.org/10.1094/MPMI-10-13-0305-R
Vogt I, Wöhner T, Richter K et al (2013) Gene-for-gene relationship in the host-pathogen system Malus x robusta 5-Erwinia amylovora. New Phytol 197:1262–1275. https://doi.org/10.1111/nph.12094
Voinnet O, Rivas S, Mestre P et al (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein oftomato bushy stunt virus. Plant J 33:949–956. https://doi.org/10.1046/j.1365-313X.2003.01676.x
Vrancken K, Holtappels M, Schoofs H et al (2013) Pathogenicity and infection strategies of the fire blight pathogen Erwinia amylovora in Rosaceae: state of the art. Microbiol 159:823–832. https://doi.org/10.1099/mic.0.064881-0
Yao JL, Tomes S, Gleave AP (2013) Transformation of apple (Malus × domestica) using mutants of apple acetolactate synthase as a selectable marker and analysis of the T-DNA integration sites. Plant Cell Rep 32:703–714. https://doi.org/10.1007/s00299-013-1404-7
Acknowledgements
The plasmid pGREEN II 0800-LUC was kindly provided by Dr. A. Allan (PFR, New Zealand). The plasmid pKGWFS7-35S was kindly provided by J. Jeauffre (IRHS, France). The authors gratefully acknowledge the IRHS-ImHorPhen team and the experimental unit HORTI of INRAE Angers for technical assistance in plant maintenance, B. Billy (SNES-GEVES) for technical assistance in flow cytometry and the technical platform ANAN. Technical contributions from M. Jacq, J.G. Bertault and P. Berthelot are also gratefully acknowledged. The authors wish to thank their collaborator Alexandre Degrave for his careful and critical reading of the manuscript.
Funding
This project was funded by the INTRAPOM Project (INRAE BAP division) and post-doctoral grants from Region Pays de la Loire and Angers Agglomération (E. Vergne, L. Righetti and M. Gaucher).
Author information
Authors and Affiliations
Contributions
MG and LR were the main investigators in this study. They performed most of the experiments, analyzed and interpreted data, drafted the manuscript and revised it. EV designed the study, performed part of the experiments, analyzed and interpreted data, drafted the manuscript and revised it. SA, TDB, MNB and EC actively contributed to the analysis and interpretation of data and revised the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Roger Thilmony.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gaucher, M., Righetti, L., Aubourg, S. et al. An Erwinia amylovora inducible promoter for improvement of apple fire blight resistance. Plant Cell Rep 41, 1499–1513 (2022). https://doi.org/10.1007/s00299-022-02869-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02869-8