Abstract
Key message
Expression of Cre recombinase by AtRps5apro or AtDD45pro enabled Cre/lox-mediated recombination at an early embryonic developmental stage upon crossing, activating transgenes in the hybrid cowpea and tobacco.
Abstract
Genetic engineering ideally results in precise spatiotemporal control of transgene expression. To activate transgenes exclusively in a hybrid upon fertilization, we evaluated a Cre/lox-mediated gene activation system with the Cre recombinase expressed by either AtRps5a or AtDD45 promoters that showed activity in egg cells and young embryos. In crosses between Cre recombinase lines and transgenic lines harboring a lox-excision reporter cassette with ZsGreen driven by the AtUbq3 promoter after Cre/lox-mediated recombination, we observed complete excision of the lox-flanked intervening DNA sequence between the AtUbq3pro and the ZsGreen coding sequence in F1 progeny upon genotyping but no ZsGreen expression in F1 seeds or seedlings. The incapability to observe ZsGreen fluorescence was attributed to the activity of the AtUbq3pro. Strong ZsGreen expression in F1 seeds was observed after recombination when ZsGreen was driven by the AtUbq10 promoter. Using the AtDD45pro to express Cre resulted in more variation in recombination frequencies between transgenic lines and crosses. Regardless of the promoter used to regulate Cre, mosaic F1 progeny were rare, suggesting gene activation at an early embryo-developmental stage. Observation of ZsGreen-expressing tobacco embryos at the globular stage from crosses with the AtRps5aproCre lines pollinated by the AtUbq3prolox line supported the early activation mode.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Precise gene regulation is critical in genetic engineering. Conditional transgene activation usually is preferred since ectopic expression of transgenes may result in undesired phenotypes. The application of tissue-specific promoters or inducible promoters is a common approach to achieve conditional transgene activation. Using two-component systems for gene activation wherein the transgene remains inactive until both the components converge in the same cell can offer an alternative switch perspective. With the “activator” regulated by a tissue-specific or inducible promoter, a two-component system potentially can lead to tighter control of spatiotemporal transgene expression.
The Cre/lox system is one of the most well-characterized two-component systems and has been broadly used in eukaryotic organisms, including plants (Branda and Dymecki 2004; Gilbertson 2003; Srivastava and Thomson 2016). In the Cre/lox system, a 38-kDa recombinase Cre from bacteriophage P1 can recognize a 34-bp lox (locus of crossover) site by binding to the inverted repeats and effecting recombination at the 8-bp spacer inside the lox site (Van Duyne 2001). Depending on the numbers of the lox sites employed and their orientations, Cre/lox-mediated recombination can result in excision, inversion, integration, or sequence exchange between two nonhomologous chromosomes (Branda and Dymecki 2004; Srivastava and Thomson 2016). The excision reaction has been the basis of gene activation (Odell et al. 1994; Zhang et al. 2003; Heidstra et al. 2004) and inactivation (Russell et al. 1992; Kopertekh et al. 2010; Lowe et al. 2016) through Cre/lox-mediated recombination. For gene activation, an intervening DNA sequence flanked by two lox sites in the same orientation is placed between the promoter and coding sequence (CDS) of the gene of interest (GOI) so that the GOI remains inactive until the Cre recombinase excises the lox-flanked intervening sequence. The recombination results in a circular DNA molecule of the intervening sequence and leaves one lox site as a “scar” between the promoter and CDS. Gene activation by Cre/lox-mediated recombination has been evaluated in plants with the Cre gene regulated by a constitutive promoter (Bayley et al. 1992; Hoa et al. 2002; Zhang et al. 2003). Conditional gene activation in lox-containing plants also was achieved using inducible promoters (Hoff et al. 2001; Zuo et al. 2001) or tissue-specific promoters (Odell et al. 1994; Chen et al. 2017) to express Cre. To activate the GOI in crosses between Cre- and lox-containing lines, promoters active in reproductive tissues are preferable for Cre expression, as the presence of Cre recombinase in gametes presumably enables excision of the lox-flanked intervening sequence upon fertilization and activates the GOI as early as possible. Delayed expression of the Cre gene likely results in mosaic embryos as observed previously when the Cre gene was regulated by seed-specific promoters that became active at the heart stage of embryo development (Odell et al. 1994).
Careful selection of promoters that express Cre in gametes or zygotes presumably will be able to activate the GOI at the zygote stage or at least an early embryo-developmental stage. Several reproduction-specific promoters, such as flower (Bai et al. 2008; Verweire et al. 2007), pollen (Luo et al. 2007; Mlynárová et al. 2006; Verweire et al. 2007), and pollen-and-embryo promoters (Polóniová et al. 2015) have been evaluated to express Cre for marker-gene excision but never for gene activation. Moreover, the promoters tested were predominantly male gamete-specific, probably because marker-gene excision generally intends to prevent unwanted gene flow to non-transgenic plants via pollen and seeds. The use of female gamete-specific promoters to express Cre remains unreported. The promoter of Arabidopsis RIBOSOMAL PROTEIN S5A (AtRps5a) can bring about a visible expression of marker genes in the egg cells and sperm cell nuclei before fertilization (Maruyama et al. 2013) and strong expression was found in young embryos from the zygote stage up until the globular stage (Weijers et al. 2001; Maruyama et al. 2013; Gooh et al. 2015). The activity in egg cells and young embryos makes the AtRps5apro a candidate for expressing Cre to activate a GOI upon fertilization. However, the AtRps5apro also showed high activity in the floral meristem, shoot/root apical meristem, and leaf primordia (Weijers et al. 2001). It is unknown whether continuous high expression of the DNA-binding protein Cre in these tissues could cause abnormal phenotypes as suggested in some species (Coppoolse et al. 2003). As a precaution, additional tissue-specific promoters, such as the promoter of Arabidopsis DOWN REGULATED IN DETERMINANT INFERTILE (DD) 45/EGG CELL-SECRETED (EC) 1.2 (AtDD45/EC1.2), can be tested for providing an alternative to the AtRps5apro. The AtDD45/EC1.2pro (referred to as AtDD45pro for short in the remaining text) can drive gene expression specifically in egg cells before fertilization (Steffen et al. 2007; Sprunck et al. 2012) and express the transgene in early developing embryos no later than the eight-cell stage after fertilization (Lawit et al. 2013). It is unknown yet whether shortening the expression time of Cre with the AtDD45pro will lead to lower recombination efficiency and more mosaic F1 embryos than with the AtRps5apro.
Cowpea (Vigna unguiculata [L.] Walp.) is one of several important crop plants for food security in sub-Saharan Africa, given its resilience to drought and high temperature (Singh 2014; Carvalho et al. 2017). As a legume, cowpea can also tolerate low-fertility soil thanks to its capability to establish a symbiotic association with nitrogen-fixing bacteria and vesicular-arbuscular mycorrhizal fungi. Cowpea also is an ideal niche crop for cereal-based multiple cropping systems considering the availability of early-maturing and pest-resistant varieties (Singh 2014), and its importance may increase around the globe under changing climate conditions. Currently, cowpea is mostly grown in tropical and subtropical regions and consumed in the human diet or used as fodder in West and Central Africa, Europe, Asia, and America. The increase in cowpea production and consumption has been considerable (Singh 2014; FAOSTAT 2018) and improvement in cowpea varieties has been made in the last several decades (Boukar et al. 2020; Singh 2014). Cowpea improvement likely will accelerate thanks to the availability of new genomic and transcriptomic resources (Huynh et al. 2018; Lonardi et al. 2019; Muñoz-Amatriaín et al. 2017; Spriggs et al. 2018; Yao et al. 2016) as well as an efficient transformation method (Che et al. 2021) that enables transgene introduction and genome editing. The novelty of the current study is in demonstrating the efficacy of the Cre/lox system for gene activation in early embryo development through the use of early embryo and female gametophyte promoters AtRps5apro and AtDD45pro. The results will pave the way for utilizing the Cre/lox system to activate transgenes conditionally in cowpea.
Materials and methods
Cre and lox-reporter gene constructs
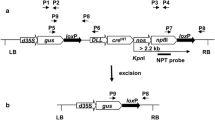
To test the efficacy of gene activation via Cre/lox-mediated excision in tobacco and cowpea, we first constructed two cassettes AtRps5apro:Cre:phaseolinterm (AtRps5aproCre), and AtUbq3pro:lox-PINIIterm-lox:ZsGreen:NOSterm (AtUbq3prolox). In the cassette AtRps5aproCre (Fig. 1a), the Cre gene was controlled by the promoter (1684 bp) from Arabidopsis thaliana ribosomal protein subunit 5a (AtRps5a, AT3G11940; Weijers et al. 2001) and the phaseolin 3’ terminator. In the cassette AtUbq3prolox (Fig. 1b), the ZsGreen gene was controlled by the promoter (1721 bp) from A. thaliana polyubiquitin 3 (AtUbq3, AT5G03240, including its 5′ UTR and leading intron) and the nopaline synthase (NOS) terminator. A potato Proteinase Inhibitor II (PINII) terminator (Keil et al. 1986) flanked by two modified lox sites was inserted between the AtUbq3pro and ZsGreen coding sequence (CDS). The cassette AtRps5aproCre, together with a fluorescent-marker cassette GmEF1apro:DsRed:NOSterm (GmEF1aproDsRed) and a selectable-marker cassette, was transferred into the T-DNA region of a binary vector pPZP201BK (Covert et al. 2001) (Table S1). In the cassette GmEF1aproDsRed, a DsRed gene was controlled by the promoter (2166 bp) from soybean (Glycine max) elongation factor 1a (GmEF1a, Glyma.17G186600, including its 5′ UTR and leading intron; Li et al. 2015) and the NOS terminator. In the selectable-marker cassette, the selectable marker gene was controlled by the promoter (930 bp) from potato (Solanum tuberosum) polyubiquitin (StUbq, GenBank: L22576; Garbarino and Belknap 1994) and the NOS terminator. Flanked by the same promoter and terminator, three selectable-marker CDS, neomycin phosphotransferase II (nptII), hygromycin phosphotransferase (hpt), and Bialaphos resistance (bar), were evaluated side by side for their effect on the efficiency of cowpea transformation. The cassette AtUbq3prolox was transferred into the T-DNA region of the binary vector pORE (Coutu et al. 2007), together with a kanamycin resistance cassette (S1pronptII) wherein the nptII gene containing a Ricinus communis catalase 1 (CAT1) intron was controlled by the S1 promoter and S3 terminator from subterranean clover stunt virus (SCSV) (Schünmann et al. 2003) (Table S1). The cassettes AtUbq3prolox and S1pronptII were also transferred into the T-DNA region of the binary vector pPZP201BK, together with the cassette GmEF1aproDsRed as a fluorescent marker (Table S1).
Four cassettes used to evaluate Cre/lox-mediated recombination. a Cassettes AtRps5aproCre and AtDD45proCre, dash lines in the AtDD45pro indicate a “T” to “A” mutation and a “G” to “T” mutation created respectively at −264 bp and −979 bp upstream from the start site of Cre during synthesis to facilitate the cassette assembly using Golden Gate Cloning (Engler et al. 2008); b, c diagrams of two inactive lox-reporter cassettes and their activated versions after Cre/lox-mediated recombination; b cassette AtUbq3prolox, the 8-bp spacer in the lox sites was oriented as its reverse complement counterpart (Albert et al. 1995) so that the two ATGs in the spacer were in the reverse orientation with respect to the promoter; c cassette AtUbq10prolox, ZsGreen-ER and tdTomato-ER are ZsGreen and tdTomato containing an endoplasmic reticulum signal peptide (ERSP) at the 5′ end and a KDEL-ER retention motif at the 3′ end, respectively; the sizes shown are the fragment amplified by PCR with the specific primers (see Table S3).
To further evaluate Cre/lox-mediated gene activation in cowpea, two additional cassettes were constructed as AtDD45pro:Cre:OCSterm (AtDD45proCre) and AtUbq10pro:loxP-GmUbq3pro:tdTomatoER:OCS-loxP:ZsGreen:NOSterm (AtUbq10prolox). In the cassette AtDD45proCre (Fig. 1a), the synthetic codon-optimized Cre gene containing a modified CAT1 intron was controlled by the promoter (1002 bp) from A. thaliana DOWN REGULATED IN DETERMINANT INFERTILE (DD) 45/Egg Cell-secreted protein 1.2 (AtDD45/EC1.2, AT2G21740; Steffen et al. 2007; Sprunck et al. 2012) and the octopine synthase (OCS) terminator. In the cassette AtUbq10prolox (Fig. 1c), the synthetic codon-optimized ZsGreen gene was controlled by the promoter (1500 bp) from A. thaliana polyubiquitin 10 (AtUbq10, AT4G05320, including its 5’UTR and leading intron) and the NOS terminator. Between the AtUbq10pro and the ZsGreen CDS, a cassette GmUbq3pro:tdTomatoER:OCSterm (GmUbq3protdTomato) flanked by two loxP (the original lox from bacteriophage P1) sites was inserted in a reverse direction for the AtUbq10 promoter. The tdTomato gene (a variant of DsRed) (Shaner et al. 2004) was controlled by the promoter (919 bp) from soybean (G. max) ubiquitin 3 (GmUbq3, Glyma.20G141600, including its 5’UTR and leading intron) and the OCS terminator (Fig. 1c). The cassettes AtDD45proCre and AtUbq10prolox were transferred respectively into the T-DNA region of the binary vector pAGM4673 (Weber et al. 2011), together with a spectinomycin resistance cassette (Che et al. 2021) (Table S1). Considering that the 35S enhancer in the spectinomycin resistance cassette might interact with the AtDD45pro in the cassette AtDD45proCre and affect its tissue-specificity, a 2 kb transformation booster sequence (TBS) (Singer et al. 2011) was placed between the cassette AtDD45proCre and the spectinomycin resistance cassette (Table S1).
Transient assay by microprojectile bombardment
Mature cowpea seeds were surface sterilized with 10% commercial bleach (6% (w/v) sodium hypochlorite; Clorox, Oakland, CA, USA) for 30 min with agitation at 150 rpm, followed by rinsing with sterilized reverse osmosis deionized (RODI) water at least five times. After sterilization, seeds were immersed in sterilized RODI water overnight. Cotyledons were excised from the imbibed seeds and the embryo axes were used for microprojectile bombardment. With one side touching the medium, ten embryo axes were laid within a circle of 2 cm in diameter at the center of a Petri dish containing 0 MS medium composed of 1× Murashige and Skoog (MS) salts and vitamins, 3% (w/v) sucrose (Research Products International, Mt Prospect, IL, USA), and 8 mg/l agarose (pH 5.7). Microprojectile bombardment was conducted using a PDS-1000/HeTM system (Bio-Rad Laboratories, Hercules, CA, USA) under 1550 psi helium in a vacuum of 23 in Hg, with the Petri dish carrying embryo axes positioned on the sample platform 5 cm below the launch assembly. Each bombardment delivered approximately 156 ng DNA composed of an equal molar amount of the Cre cassette and lox-reporter gene cassette and 50 µg gold microcarriers 0.6 µm in diameter (Bio-Rad Laboratories). Each sample was bombarded twice. Upon evaluating the cassettes AtRps5aproCre and AtUbq3prolox harbored in the plasmid pBlueScript KS(+), a trace amount of the cassette GmEF1aproDsRed harbored in the plasmid pBlueScript KS(+) also was added so that we could check the quality of each bombardment. With the lox-flanked GmUbq3protdTomato in the lox-reporter cassette, the cassette GmEF1aproDsRed was omitted upon evaluating the cassette AtDD45proCre in vector RC2677 and the cassette AtUbq10prolox in vector RC2717 (Table S1). Cowpea embryo axes were observed for ZsGreen expression 24 h after bombardment.
Agrobacterium inoculum and plant transformation
All recombinant binary vectors (Table S1) were introduced into competent cells of Agrobacterium strain AGL1 using the freeze-thaw method (Chen et al. 1994). Agrobacterium carrying the recombinant vectors was stored in 15% glycerol at −80 °C. For tobacco transformation and cowpea transformation using Method 1 and 2 (see below), Agrobacterium inoculum was prepared as follows: Agrobacterium from the glycerol stocks was cultured in 2-ml liquid Lysogeny broth (LB) medium (ThermoFisher Scientific, Waltham, MA, USA) containing 100 mg/l kanamycin, 50 mg/l rifampicin, and 50 mg/l carbenicillin, with incubation at 150 rpm and 28 °C for 24 h. The bacterial culture was diluted 1:100 with liquid LB medium containing the same antibiotics and incubated overnight at 150 rpm, 28 °C. The culture was centrifuged for 10 min at 6000 rpm using a J2-21M centrifuge (Beckman Coulter, Brea, CA, USA), and re-suspended in liquid co-culture medium, with OD600 adjusted to 0.5–0.6. For cowpea transformation using Method 3, Agrobacterium inoculum was prepared according to Che et al. (2021).
The AtRps5aproCre and AtUbq3prolox were introduced into tobacco PI 552484 (seeds purchased from Lehle Seeds, Round Rock, TX, USA) following the protocol of Clemente (2006) with some modifications (Zhang et al. 2020). Transgenic plants containing either the cassette AtRps5aproCre or AtUbq3prolox were recovered with selection on either 20 mg/l hygromycin or 200 mg/l kanamycin, according to the selectable marker in the plasmids used for transformation (Table S1). Plants derived from different leaf disc explants were independent, while those from the same explants were potentially the same lines.
Different Cre or lox-reporter cassettes were introduced into cowpea IT86D-1010 following three distinct methods. The cassettes AtRps5aproCre and AtUbq3prolox were introduced by either Method 1 described by Popelka et al. (2006) with some modification or Method 2 wherein explants were cotyledonary nodes from seedlings pre-conditioned with 5 mg/l 6-benzylaminopurine (BA). Both methods started with mature cowpea seeds surface sterilized as previously described (Sect. “Transient assay by microprojectile bombardment”). In Method 1, sterilized seeds were immersed in sterilized RODI water overnight. The cotyledonary nodes from the imbibed seeds were used as explants for Agrobacterium-mediated transformation after removing the seed coat, shoot tips, cotyledons, and radicals. Every ten explants were immersed in 2-ml inoculum consisting of Agrobacterium suspended in a liquid co-culture medium (CCM, Table S2) supplemented with 0.02% (v/v) Silwet-77 (Lehle Seeds, Round Rock, TX, USA) in a 10-ml borosilicate glass test tube (Cat# 14-961-27, ThermoFisher Scientific, Waltham, MA, USA), then sonicated for 20 s using an FS30H sonicator (ThermoFisher Scientific). After 30 min incubation at room temperature, explants were blotted dry with sterile filter paper and transferred onto CCM (Table S2) with a piece of filter paper on top of the medium to prevent direct contact between explants and the medium, 20–25 explants each plate. In Method 2, sterilized mature seeds were cultured on a germination medium (GM, Table S2) for 4 d. Seedlings with a greening cotyledonary node were selected, followed by excision of cotyledons, shoot tips, and roots. The remaining cotyledonary nodes were used as explants for transformation. Every five explants were immersed in 2-ml inoculum consisting of Agrobacterium resuspended in a liquid modified co-culture medium (CCM’, Table S2) supplemented with 0.02% (v/v) Silwet-77 in a 10-ml borosilicate glass test tube then sonicated for 4 min. After 30-min incubation at room temperature, explants were blotted dry and placed on CCM’ plates (Table S2), 12 explants each, with a piece of filter paper between explants and the medium. In both methods, after a 5-day co-culture, explants were washed in a liquid shoot induction medium (SIM, Table S2) then blotted dry. Explants were transferred to SIM (Table S2) supplemented with 20 mg/l hygromycin, 5 mg/l phosphinothricin (PPT), or 200 mg/l kanamycin according to the selectable marker gene used in the vectors (Table S1). Explants forming shoots were transferred to fresh SIM every other week and monitored for DsRed expression. Explants with transgenic shoots were transferred to a shoot elongation medium (SEM, Table S2) supplemented with the same selective agent used in SIM for shoot development and rooting.
Transgenic shoots that failed to form roots were recovered by in vitro grafting adapted from a method for sunflower (Zhang and Finer 2016). Sterilized mature seeds were germinated under a sterilized folded paper towel saturated with sterile RODI water in MagentaTM GA7 vessels (Sigma-Aldrich, St Louis, MO, USA), with ten seeds in each box. Rootstocks were made from 5-day-old seedlings by removing the shoot tips and cotyledons. As a scion, a developed transgenic shoot (> 0.5 cm) with its base shaped into a wedge was inserted into the longitudinal incision (0.5–1 cm) made in the side of the rootstock’s hypocotyl. The hypocotyl tissue from both sides held the scion in place. The grafted plants were grown in GA7 vessels containing 0MS medium supplemented with 30 mg/l meropenem (ABBLIS Chemicals, Houston, TX, USA) for 2–3 weeks. Elongation of the scions indicated graft success. The successfully rooted or grafted plants were transferred to soil, acclimatized in plastic containers with the lids gradually opened over a 3-week period. Plants were transferred to the greenhouse to set seeds when they were sufficiently hardened and growing vigorously.
The cassettes AtDD45proCre and AtUbq10prolox were introduced into cowpea using Method 3 (Che et al. 2021) given its higher efficiency in recovering transgenic plants. In brief, mature seeds were surface-sterilized overnight using chlorine gas, then soaked overnight in a bean germination medium (Che et al. 2021). Embryo axes were isolated by removing seed coats, cotyledons, and plumules without damaging the meristematic dome and collected in sterile RODI water. After removing water, embryo axes (100–200 pieces) were infected with 15-ml Agrobacterium inoculum plus 50-μl sterile Poloxamer 188 10% solution in a 100 × 25 mm Petri dish (ThermoFisher Scientific). After sonication for 3 s, 10 ml of inoculum was added to each Petri dish and incubated at 60 rpm, room temperature, for 1.5 h in the dark. Explants were removed from inoculum and transferred to filter paper (VWR Cat# 28320-020) wetted with 700 μl of infection medium (Che et al. 2021) in a 100 × 25 mm Petri dish, with every 30 explants in a pile. After 2-day co-culture at 21 °C under dim light with a 16/8 h light cycle, embryo axes were inserted into SIM (Table S2) supplemented with 50 mg/l spectinomycin with shoot apex and cotyledonary node above the medium. The shoot apex was removed after 5-day incubation on SIM to facilitate the formation of axillary shoots. After 4 weeks, transgenic shoots were either transferred to a rooting medium (Che et al. 2021) for rooting or a shoot elongation medium (Che et al. 2021) for further growth before transfer to the rooting medium. Plantlets derived from different cotyledonary-node explants were independent, while those from the same explants were considered as potentially the same lines.
All plant tissues were incubated at 25 °C, with a 16/8 h light cycle, except for the cowpea transformation using Method 3 wherein plant tissues were incubated at 25 °C and 24-h light after co-culture. MS salts, MS vitamins, and acetosyringone (AS) were from PhytoTechnology Laboratories (Lenexa, KS, USA). Unless otherwise noted, all chemicals were from Millipore Sigma (St. Louis, MO, USA).
Genotyping
Genomic DNA of tobacco and cowpea was extracted from leaf tissue using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). The presence of the transgene was verified by polymerase chain reaction (PCR) using corresponding primer combinations (Table S3). We conducted PCR using PrimeStar GXL DNA polymerase (Takara Bio, Kusatsu, Japan) in a 25 µl reaction for amplicons larger than 1 kb and using GoTaq® Master Mix (M7123, Promega, Madison, WI, USA) in a 20 µl reaction for amplicons smaller than 1 kb. The PCR setup followed the manufacturers' instructions with 20–100 ng genomic DNA as a template. PCR products were visualized under UV light after electrophoresis in 1% agarose gels and staining with ethidium bromide.
Evaluation of transgene segregation in transgenic lines
In tobacco, after surface sterilization according to Zhang et al. (2020), T1 seeds of the lines harboring the cassette AtRps5aproCre were germinated on 0MS medium containing either 200 mg/l kanamycin or 20 mg/l hygromycin depending on the selectable marker used in the vectors (Table S1). After 2 weeks, seedlings were observed for DsRed expression and counted. Surface-sterilized T1 seeds of tobacco lines harboring the cassette AtUbq3prolox (from vector pZZ017, Table S1) were germinated on 0MS medium containing 600 mg/l kanamycin for a stricter selection to avoid possible false positives. After 2 weeks, the green seedlings were counted as transgenic while the seedlings showing chlorosis at cotyledons or the shoot apex were counted as non-transgenic.
In cowpea, mature T1 seeds of the lines harboring the cassette AtRps5aproCre, AtUbq3prolox (from vector pZZ031B, Table S1), or AtUbq10prolox were observed for DsRed/tdTomato expression and counted. If fluorescence was not evident in dry seeds, a proportion of T1 seeds from those lines was germinated between sheets of filter paper saturated with sterile RODI water and observed for DsRed/tdTomato fluorescence after 5 days. For the cowpea lines harboring the cassette AtDD45proCre, the presence or absence of the transgene in T1 progeny was determined by PCR using genomic DNA extracted from leaf tissues of 2-week-old T1 seedlings grown in the greenhouse. The segregation ratios of the transgene in tobacco and cowpea transgenic lines were calculated and tested by the Chi-square test.
Estimation of copy number by quantitative PCR (qPCR)
Genomic DNA from transgenic plants harboring the cassette AtRps5aproCre or AtUbq3prolox was digested by restriction endonuclease HindIII-HF® (New England Biolabs, Ipswich, MA, USA) while genomic DNA from transgenic plants harboring the cassette AtDD45proCre or AtUbq10prolox was digested by NcoI-HF® (New England Biolabs). After purification with the DNA Clean and Concentration-5 Kit (Zymo Research, Irvine, CA, USA), digested DNAs in 20-fold dilution served as the qPCR templates. The qPCR reactions were conducted with the SYBR green I/HRM dye program in a LightCycler® 480 system (Roche, Basel, Switzerland) following the manufacturer’s protocol of LightCycler 480 SYBR Green I Master V13 (Roche, Mannheim, Germany) in a 20 µl volume. The amplification efficiency of each primer combination was estimated based on qPCR data of a 5-log serial dilution (0.0016, 0.008, 0.04, 0.2, 1×) of a DNA mixture that contained an equal amount of DNA from each tested sample using the absolute quantification method in the LightCycler 480 software (Roche, Release 1.5.0). The qPCR results with two technical replicates for each sample were analyzed using the advanced relative quantification method in the LightCycler 480 software. Primers for qPCR were designed using either Geneious R10 (Auckland, New Zealand) or Realtime PCR Tool from Integrated DNA Technology (San Jose, CA, USA) (Table S4). The tobacco tubulin α-chain gene (NCBI accession # XM_016623993) served as a reference gene (Głowacka et al. 2016) for the assays in tobacco. To identify single-copy genes used as reference genes for cowpea, the Plants Datasets (Embryophyta odb9) from Benchmarking Universal Single-Copy Orthologs (BUSCO) was aligned to the whole-genome shot-gun (WGS) assembly (MATU00000000 at NCBI) of cowpea IT97K-499-35 (Muñoz-Amatriaín et al. 2017) using the BUSCO v3 software (Simão et al. 2015; Waterhouse et al. 2018). The amino acid sequences of those orthologs classified as “complete” BUSCO genes were aligned to the reference protein database at NCBI using the BLASTP program to acquire the annotation of the genes. After removing mitochondrial and chloroplastic genes, the cowpea nucleotide sequences which encode the amino acid sequences were aligned to the cowpea WGS assembly to verify whether the gene sequence was unique in the assembly. The genes without any duplication in the genome were considered as single-copy genes in the cowpea genome and potentially used as reference genes for qPCR. After testing the amplification efficiency and specificity of the first six genes from the list, a predicted cowpea F-box protein (VuFbox, Phytozome12 transcript name Vigun07g146600) was selected and used as the reference gene for the assays in cowpea. The copy numbers of the transgene were calculated based on the ratios of the transgene to the reference gene.
Southern blot
Genomic DNA of 10 µg from the cowpea lines of the cassettes AtRps5aproCre and AtUbq3prolox was digested overnight by SpeI-HF® and MluI-HF® (New England Biolabs), respectively. Restriction fragments were separated by gel electrophoresis and then transferred to GeneScreen Plus® hybridization transfer membrane (Cat# NEF1017001PK, PerkinElmer, Waltham, MA, USA). Hybridization was conducted using a digoxigenin (DIG)-labeled probe targeting either the Cre gene for the cassette AtRps5aproCre or the ZsGreen gene for the cassette AtUbq3prolox and using DIG Easy HybTM as hybridization buffer following the Roche DIG application manual for filter hybridization (Eisel et al. 2008). The probes were generated and labeled by PCR with primers p3769/p3770 and p3700/p3701 (Table S3) for Cre and ZsGreen, respectively using the Roche PCR DIG labeling mix following the manufacturer’s protocol. Detection was conducted using the chromogenic assay with NBT/BCIP according to the Roche DIG application manual for filter hybridization (Eisel et al. 2008). All chemicals used for making buffers, as well as blocking reagent (Cat# 11096176001), anti-DIG-AP (Cat# 11093274910), and NBT/BCIP (Cat# 11681451001), were from Millipore Sigma.
Identification of cowpea plants homozygous for the transgenes
The ideal plant materials for evaluating Cre/lox-mediated gene activation upon crossing were homozygous plants derived from lines where the transgene segregated as a single locus. For the cassette AtRps5aproCre, at least six transgenic T1 seedlings from the single-locus lines were grown in the greenhouse to set seeds, and their T2 seeds were observed for DsRed expression. If all T2 seeds harvested displayed DsRed fluorescence, the T1 plants were homozygous for the transgene. For the cassettes AtUbq3prolox, AtDD45proCre and AtUbq10prolox, the homozygotes were identified using qPCR (Sect. “Estimation of copy number by quantitative PCR (qPCR)”) from the single-locus lines. If the ratio of the transgene to the reference gene in the T1 progeny doubled compared to the T0 mother plants, the T1 progeny would be homozygous for the transgene. Expression of DsRed/tdTomato in T2 seeds from the homozygous T1 of the AtUbq3prolox and AtUbq10prolox lines also was observed to verify their homozygosity. The homozygous plants were grown in the greenhouse to increase seeds and make crosses.
We also attempted to identify homozygous progeny that inherited a single locus from the AtRps5aproCre or AtUbq3prolox lines with multiple copies of the transgene. The numbers of cowpea transgenic lines for the cassettes AtRps5aproCre and AtUbq3prolox were low due to the low efficiency of the transformation methods applied. To include additional lines harboring those cassettes in the analysis, it was necessary to utilize high copy-number lines. Since the transgenes segregated in the T1 generation, progeny inheriting only one of the transgene loci from those lines could be obtained if enough progeny were screened by qPCR, T1 progeny that likely carried two copies of the transgene were identified and grown in the greenhouse to set seeds. If all the T2 seeds from a T1 displayed DsRed fluorescence, the T1 progeny had become homozygous for the single locus inherited from the T0 parents. If the fluorescence trait segregated among T2 seeds from a T1 progeny, the segregation ratio would indicate the number of transgene loci inherited by the T1 progeny. The T1 progeny with an approximately 3:1 segregation ratio were further advanced to obtain homozygous plants using the same approach as previously described.
Tobacco crossing and F1 embryo isolation
Flowers were emasculated at stage 10 (Koltunow et al. 1990) and covered with a tailored pollination envelope. After 2 days, the emasculated flowers on the AtRps5aproCre lines were pollinated with pollen from the AtUbq3prolox line. Meanwhile, the AtRps5aproCre lines and the AtUbq3prolox line were self-pollinated as controls. The pollinated flowers were tagged and covered with tailored pollination envelopes. Ovules were harvested 6–8 days after pollination (DAP), and embryos were isolated using the method described by Fu et al. (1996) with some modifications (Zhang et al. 2020). Isolated embryos were observed for ZsGreen expression.
Cowpea crossing and F1 progeny analysis
Cowpea flowers were emasculated in the evening before anthesis, followed by covering the peduncles of emasculated flowers with moist tailored pollination envelopes. The next morning, emasculated flowers were pollinated and tagged, and the peduncles of the pollinated flowers were covered by moist tailored pollination envelopes. After harvest, mature F1 seeds were observed for ZsGreen expression. Genomic DNA was extracted from leaf tissues of F1 seedlings. F1 progeny were genotyped by PCR to verify whether they inherited both the Cre and lox-reporter genes. With the use of p3785/p3786 and p4825/p4826, PCR could also confirm the Cre/lox-mediated excision in F1 progeny inheriting both the Cre and lox-reporter genes, as the excision would result in a smaller amplicon due to the removal of the intervening DNA sequence flanked by two lox sites (Fig. 1b, c).
Observation of fluorescence
Expression of fluorescent marker genes (i.e., ZsGreen, DsRed, and tdTomato) in plant tissues was observed using a Stemi SVII stereomicroscope equipped with an HBO illuminator (Zeiss, Thornwood, NY, USA), a FITC filter set (λexcitation = 480 nm, and λemission = 515 nm; Chroma Technology, Bellows Falls, VT, USA), and a DsRED filter (λexcitation = 545/25 nm, dichroic 565LP, λemission = 605/70 nm; Chroma Technology). Expression of ZsGreen in isolated embryos was observed under a Zeiss Axioskop 2 plus fluorescence microscope equipped with an 89 North® PhotoFluor LM-75 illumination system (Chroma Technology) and a FITC filter set. Images were taken using an AxioCam camera (Carl Zeiss, Oberkochen, Germany) and the AxioVision LE64 software.
Nomenclature of transgenic cowpea
We gave a unique code to every T0 transgenic cowpea plant recovered from tissue culture. For the AtRps5aproCre and AtUbq3prolox, each plant was named by a six-digit code. The first two digits were the last two digits of the vector used; the middle two digits indicated the independent explant giving rise to transgenic shoots (i.e., independent lines); and the last two digits indicated the plants recovered from the particular line. In the code “120200”, for example, ‘12’ indicated the use of vector pZZ012; ‘02’ indicated the independent line 2; and ‘00’ indicated the first plant recovered from line 1202. For the AtDD45proCre and AtUbq10prolox, each plant was named by a code starting with “R” followed by six digits. In the code “R771402”, for example, ‘77’ indicated the use of vector RC2677; ‘14’ indicated the independent line 14; and ‘02’ indicated the second plant recovered from line R7714. Regardless of the last two digits, as long as the first four digits were the same, the plants were potentially clones from the same line unless additional evidence showed that they were not.
Results
Transient Cre/lox-mediated gene activation
To validate whether the Cre and lox-reporter cassettes were functional in cowpea before creating stable transformants, the Cre and lox-reporter cassettes were co-introduced into cowpea embryo axes by microprojectile bombardment. Upon co-bombardment with the cassettes AtRps5aproCre and AtUbq3prolox, cowpea embryo axes displayed ZsGreen expression 24 h after bombardment (Fig. 2), though the intensity of ZsGreen expression was weaker than expected. In preliminary experiments, we observed a stronger expression of ZsGreen when cowpea tissues were bombarded by a cassette with the AtUbq3pro directly followed by the ZsGreen CDS and a cassette with a modified lox site (see Sect. “Cre and lox‑reporter gene constructs”, Fig. 1b) present between the AtUbq3pro and the ZsGreen CDS (i.e., the product of treating the cassette AtUbq3prolox with Cre recombinase in vitro) (Fig. S1). Upon co-bombardment with the cassettes AtDD45proCre and AtUbq10prolox, ZsGreen expression was also observed in cowpea embryo axes in 24 h (Fig. 2h). The tissue specificity of the AtDD45pro might relax under the condition of bombardment. Compared with the cassette AtUbq3prolox, the reporter gene ZsGreen in the cassette AtUbq10prolox appeared to have leaky expression, but the bright ZsGreen fluorescence after activation was distinguishable from the leaky expression (Fig. 2).
Gene activation via Cre/lox-mediated recombination by microprojectile bombardment. Each column displays primary leaves of a cowpea embryo axis 24 h after bombardment under bright field (top), FITC filter (middle), and DsRED filter (bottom). e–h ZsGreen expression indicates activation of the reporter gene; i, j, DsRed expression as an internal control indicates the success of the bombardment; k, l, tdTomato expression from the lox-flanked cassette; bar = 1 mm
Transgenic tobacco
Recovery of transgenic tobacco
We introduced the cassettes AtRps5aproCre and AtUbq3prolox into tobacco and first evaluated their efficiency for Cre/lox-mediated gene activation by making crosses in tobacco prior to recovery of transgenic cowpea. Seven independent lines of the cassette AtRps5aproCre and three independent lines of the cassette AtUbq3prolox were recovered, showing no visual abnormality in plant development compared with non-transgenic. Upon genotyping with primers p3753/p3768, all AtRps5aproCre lines carried the full-length CDS of the Cre gene and the phaseolin terminator. Expression of Cre was detected in ovules of unpollinated flowers (data not shown). Upon genotyping with primers p3785/p3908, only one AtUbq3prolox line harbored the full-length CDS of the ZsGreen gene and the NOS terminator without truncation. Based on the DsRed expression among T1 progeny from the AtRps5aproCre T0 lines, the transgene segregated in a 3:1 ratio as a single locus in two lines (pZZ010_1.1 and pZZ010_202.3), in a 15:1 ratio as two independent loci in four lines (pZZ010_2.2, pZZ010_201.2, pZZ012_1.4, and pZZ012_2.1), and likely as four independent loci or more in one line (pZZ010_1.3) (Table S5). Based on the response to kanamycin selection among T1 progeny from the AtUbq3prolox line (pZZ017_1.1) carrying the complete cassette, the transgene likely segregated as four independent loci (Table S5). Based on the phenotype of T2 progeny, we identified homozygous T1 progeny from two single-locus AtRps5aproCre lines where all T2 progeny displayed DsRed fluorescence and resistance to hygromycin selection (Table S6).
Cre/lox-mediated gene activation in tobacco by crossing
With six AtRps5aproCre T0 lines pollinated by the AtUbq3prolox T0 line (line pZZ017_1.1), ZsGreen expression was observed in isolated F1 embryos at an early developmental stage (Fig. 3). Activation of ZsGreen occurred in F1 embryos from crosses with five AtRps5aproCre lines at a frequency ranging from 12.1% to 82.6% (Table 1). In contrast, self-pollination of the AtUbq3prolox T0 line or AtRps5aproCre T0 lines resulted in no ZsGreen-expressing embryos. The expected ratios of ZsGreen-positive to ZsGreen-negative embryos were calculated based on the assumption that the transgenes in all loci were functional and excision occurred at 100% (Table 1). Expression of ZsGreen segregated as expected in F1 embryos of three lines, while the number of ZsGreen-positive F1 embryos was lower than expected in line pZZ010_1.1 and pZZ010_201.2 (Table 1). With the homozygous T1 progeny of a single-locus AtRps5aproCre line (pZZ010_1.1) pollinated by T1 progeny of the AtUbq3prolox line pZZ017_1.1, the frequency of isolated ZsGreen-expressing F1 embryos varied from 7.1% to 70.0% (Table S7). Without identifying progeny of the AtUbq3prolox line homozygous for all transgene loci, we could not know whether an isolated embryo inherited both the Cre and lox-reporter cassette. Therefore, we decided to estimate the efficiency of Cre/lox-mediated gene activation based on the ZsGreen expression in F1 seedlings rather than isolated F1 embryos, since the F1 seedlings could be tested for both transgenes by genotyping. When F1 seeds from crosses made between the homozygous T2 plants of line pZZ010_1.1 and the T2 plants of the AtUbq3prolox T0 line pZZ017_1.1 germinated, strong ZsGreen expression was not observed as expected in any F1 seedlings and dim ZsGreen fluorescence was detected only in root tissues (Fig. 4). However, genotyping by PCR indicated removal of the PINIIterm in F1 seedlings inheriting both the Cre and the lox-reporter cassette given the amplification of a 333-bp fragment rather than a 699-bp one with p3785/p3786 (Fig. 4e). Upon searching for the expression profile of AtUbq3 (AT5G03240) visualized on the Arabidopsis eFP Browser (Schmid et al. 2005; Winter et al. 2007), we found that the AtUbq3pro drives consistently low gene expression across many tissues, except for dry seeds, the second internode, and mature pollen with a relatively higher expression (Fig. S2). The difficulty to detect ZsGreen fluorescence in F1 seedlings could be attributable to the low expression of ZsGreen driven by the AtUbq3pro. Genotyping results on a larger scale showed that all F1 seedlings inheriting both transgenes yielded only the 333 bp amplicon, indicating complete removal of PINIIterm by Cre/lox-mediated recombination at 100% across samples and the unlikeliness of mosaic F1 progeny (Table 2).
Cre/lox-mediated recombination in F1 progeny of tobacco from a cross between the AtRps5aproCre line and the AtUbq3prolox line. a, b F1 seedlings from a cross between a homozygous plant of the AtRps5aproCre line and a T1 plant of the AtUbq3prolox line; c, d T2 seedlings of the AtUbq3prolox line; a, c bright field; b, d FITC filter; e electrophoresis of PCR products with p3785/p3786 (top panel, the lox-reporter cassette) and p3753/p3769 (bottom panel, Cre); 1–18, F1 seedlings randomly sampled; lox+, the AtUbq3prolox line; Cre+, the AtRps5aproCre line; WT wild type; PCR-, no-template control; M, DNA ladder; bar = 5 mm
Transgenic cowpea
Recovery of transgenic cowpea
When the cassettes AtRps5aproCre and AtUbq3prolox were introduced into cowpea using Method 1 and 2, transgenic plant recovery was very inefficient. For the cassette AtRps5aproCre, three independent lines (out of 464 explants) and one transgenic line (out of 489 explants) were recovered through kanamycin or PPT selection, respectively. No transgenic plant (out of 481 explants) was recovered through hygromycin selection. All transgenic lines harbored the full-length CDS of Cre and the phaseolin terminator as determined by genotyping with primers p3753/p3768. According to qPCR, one line (1201) had a single copy of the transgene, one line (1101) likely had two copies, one line (1202) had four copies, and one line (1203) likely had 6-8 copies of the transgene (Table S8). Southern blot analysis verified that line 1201 had one transgene copy, line 1101 had two copies, and lines 1202 and 1203 had multiple copies that were too complex to separate by gel electrophoresis (data not shown). The consistency between qPCR and Southern blot suggested that the designed qPCR assay could effectively estimate the copy number of the transgene, at least for the low copy-number lines; therefore, qPCR was used for the rest of the study. Based on the expression of DsRed among T1 seeds, the transgene segregated in a 3:1 ratio as a single locus in the AtRps5aproCre lines 1101 and 1201 (Table S9). For the cassette AtUbq3prolox, 2 independent lines (out of 456 explants) were recovered through kanamycin selection, and both harbored the full-length CDS of the ZsGreen and the terminator as shown by genotyping (primers p3785/p3908). According to qPCR, one line (3102) contained a single copy of the transgene while the other line (3101) likely contained six copies.
When the cassettes AtDD45proCre and AtUbq10prolox were introduced into cowpea using Method 3 with spectinomycin selection, recovery of transgenic plants was much more efficient. Nine lines (out of 271 explants) and 13 lines (out of 271 explants) were recovered for these 2 cassettes, respectively, (Table S8) without the use of in vitro micrografting. All transgenic lines harbored the full-length CDS of the transgene and the terminator based on PCR genotyping. According to qPCR for the AtDD45proCre lines, one line (R7719) had a single copy of the transgene, one line (R7743) had 1-2 copies, one line (R7715) had 2-3 copies, and the other six lines had at least three copies (Table S8). As line R7719 senesced quickly after entering the reproductive stage and produced only one pod with one non-transgenic seed capable of germination, we could not maintain this single-copy AtDD45proCre line for further analysis. The transgene segregated as a single locus for several lines, although they harbored more than one copy of the transgene. When T1 seedlings of lines R7743, R7714, and R7715 were genotyped with primers p4261/p4262, transgene segregation fit a 3:1 ratio indicating a single locus in these three lines (Table S10). According to qPCR for the AtUbq10prolox lines, eight lines (R170102, R170103, R1702, R1713, R1714, R1716, R1720, and R1739) had 1-2 copies of the transgene, three lines (R1704, R1719, and R1744) had 2-3 copies and the other two lines (R1723 and R1725) had at least three copies (Table S8). Twelve AtUbq10prolox lines were capable of producing T1 seeds except for line R1744. Since the expression of tdTomato in mature T1 seeds of the AtUbq10prolox lines was relatively weak except for line R1702, the transgene segregation ratio of these lines based on tdTomato fluorescence was calculated from seedlings instead of dry seeds. Based on the expression of tdTomato in T1 progeny of seven lines, the transgene segregated in a 3:1 ratio in lines R1702, R1719, and R1739 that carried at least two copies of the transgene (Table S10). The transgene segregated in a 15:1 ratio as two independent loci in lines R170103 and R1714. Segregation distortion was found in lines R170102, R1713, and R1720 (Table S10), and they likely carried 1-2 copies of the transgene according to the qPCR results (Table S8).
Identifying homozygous transgenic cowpea plants for making genetic crosses
Homozygous progeny of the lines with a simple transgene integration are preferable to evaluate Cre/lox-mediated gene activation upon crossing. Homozygous T1 progeny of the two single-locus AtRps5aproCre lines (1101 and 1201) were identified based on the expression of DsRed in T2 seeds (Table S11). By qPCR, we identified homozygous T1 progeny of the AtUbq3prolox line 3102 (Table S12), three single-locus AtDD45proCre lines (R7714, R7715, and R7743), and three single-locus AtUbq10prolox lines (R1702, R1719, and R1719) (Table S13).
Since only a few transgenic lines were recovered for the cassette AtRps5aproCre, T1 progeny that inherited a single transgene locus from the high-copy transgenic lines were identified to evaluate Cre/lox-mediated gene activation with the Cre expressed by the AtRps5apro. T1 transgenic progeny of lines 1202 and 1203 that likely inherited two copies of the transgene AtRps5aproCre were identified based on qPCR, and a homozygous plant of line 1203 was identified based on expression of DsRed in T2 seeds (Table S14).
Cre/lox-mediated gene activation in cowpea by crossing
Gene activation by Cre/lox-mediated recombination was first evaluated in the crosses between the AtRps5aproCre lines and the AtUbq3prolox lines. Similar to the observation in tobacco, ZsGreen fluorescence was probably too weak to detect in the F1 mature seeds or any tissues of the seedlings. The F1 progeny inheriting both transgenes showed excision of the PINIIterm, as evidenced by the predicted smaller amplicon (Fig. 5). ZsGreen expression was observed in pods during the maturation process as well as the immature F2 seeds from the F1 plants with the PINIIterm excised (Fig. S3). The ZsGreen fluorescence was undetected in younger pods, dry pods, or dry seeds (data not shown). ZsGreen expression was not observed in any other tissues except for the peduncle tip when it whitened (Fig. S4). These observations suggested that the difficulty to observe ZsGreen fluorescence in dry seeds and seedlings was possibly due to the low activity of the AtUbq3pro in these tissues.
An example of electrophoresis of PCR amplicons with p3785/p3786 (top, the lox-reporter cassette) and p3753/p3796 (bottom, Cre) for cowpea F1 progeny of crosses between homozygous plants of the AtRps5aproCre lines and an AtUbq3proloxP line. C18-11 was the 11th seedling of cross 18 (line 3102 × line 1201); C19-1 to C19-9 were the nine seedlings of cross 19 (line 3102 × line 1201); C21-1 to C21-3 were the three seedlings of cross 21 (line 3102 × line 1203); C22-1 to C22-9 were nine seedlings of cross 22 (line 3102 × line 1203); C23-1 to C23-2 were two seedlings of cross 23 (line 3102 × line 1203); C24-1 was the only seedling of cross 24 (line 3102 × line 1203); C31-1 to C31-11 were ten seedlings of cross 31 (line 1203 × line 3102); lox+, the AtUbq3prolox line; cre+, the AtRps5aproCre line; gen−, non-transgenic IT86D-1010; PCR−, no-template control for PCR; M, DNA ladder
Although we were unable to evaluate gene activation by screening for ZsGreen expression in F1 seeds or seedlings from the crosses with the AtUbq3prolox lines, the efficiency of Cre/lox-mediated excision could be determined by genotyping. The efficiency of excision should theoretically be the same as the efficiency of activating the lox-reporter gene if it were visible. Based on genotyping, Cre/lox-mediated excision occurred in all F1 progeny from crosses between homozygous plants of AtRps5aproCre lines 1101, 1201, and 1203 pollinated by homozygous plants of AtUbq3prolox line 3102 (Table 3). In their reciprocal crosses, excision also took place in all F1 progeny (Table 3), except for a cross between line 3102 and line 1203 where three F1 progeny showed the 699-bp instead of the 333 bp amplicon (Fig. 5). With line 3101 pollinated by four AtRps5aproCre lines, excision occurred in all F1 progeny inheriting both transgenes except for one cross (line 3101 pollinated by line 1202) (Table 3) wherein F1 progeny (2/6) displayed both the 333-bp and the 699-bp amplicons together. Among five crosses between homozygous progeny of line 3102 and an AtDD45proCre line (R7743) at the T0 generation (i.e., hemizygous), Cre/lox-mediated excision seemed to take place in fewer F1 progeny inheriting both transgenes, compared with the crosses between line 3102 and the AtRps5aproCre lines (Table 3). Although the AtDD45pro presumably expressed Cre in a much shorter time course and more tissue-specifically than the AtRps5apro, no F1 progeny showed both 333-bp and 699-bp amplicons together upon genotyping with p3785/p3786, indicating that a tighter expression of Cre did not result in higher mosaicism.
To improve the ability to observe fluorescent reporter gene activation in F1 seeds or seedlings, we generated another lox-reporter cassette (i.e., AtUbq10prolox) with the ZsGreen regulated by the AtUbq10pro after Cre/lox-mediated recombination. A pilot experiment with homozygous plants from an AtRps5aproCre line (1101) pollinated by two AtUbq10prolox lines (R1701 and R1714) at the T0 generation showed that all F1 seeds displayed ZsGreen fluorescence if they inherited both transgenes, indicating that gene activation could be visualized in F1 seeds with the new lox-reporter cassette. Genotyping by PCR with primers p4797/p4798 verified the excision of the lox-flanked cassette GmUbq3protdTomato located in F1 progeny, showing an amplicon of 2589 bp rather than 5822 bp (data not shown).
Among crosses between homozygous progeny of the single-locus AtUbq10prolox lines and the single-locus AtRps5aproCre or AtDD45proCre lines, gene activation generally occurred at a comparable frequency. Except for a few crosses, gene activation occurred in all F1 progeny from the crosses between the AtUbq10prolox line R1719 and the Cre lines (Table 4) based on the ZsGreen expression in mature F1 seeds (Fig. 6). For three AtRps5aproCre lines pollinated by line R1719, ZsGreen was expressed in over 98% of F1 progeny (Table 4). For three AtDD45proCre lines pollinated by line R1719, ZsGreen was expressed in all F1 progeny for both lines R7714 and R7715 while less than a half of the F1 progeny for line R7743 (Table 4). In the reciprocal crosses with line R1719 pollinated by the Cre lines, gene activation occurred in over 83% of F1 progeny for two AtRps5aproCre lines (1201 and 1203) and two AtDD45proCre lines (R7715 and R7743) (Table 4). With line R1719 pollinated by line R7714, gene activation occurred in 56% of F1 progeny in one cross and was undetected in the other one. Strong ZsGreen-expression was observed in F1 seedlings (Fig. 7a). Upon PCR genotyping with primers p4825/p4826, all ZsGreen-expressing seedlings showed 1049 bp amplicon, indicating excision of the lox-flanked cassette (Fig. 7c). A weak amplicon of about 4 kb showed in some progeny tested (Fig. 7c) suggesting the presence of the intact AtUbq10prolox cassette without the lox-flanked DNA sequence excised. Considering that line 1719 carried 2–3 copies of the transgene (Table S8), Cre/lox-mediated excision could fail to occur in every copy of the transgene in those F1 progeny but may have taken place in every cell, as none of the F1 seedlings appeared to be mosaic by showing a sector without ZsGreen fluorescence (Fig. 7a). All F1 progeny from crosses of two AtRps5aproCre lines (1203 and 1101) with another AtUbq10prolox line (R1702) displayed ZsGreen fluorescence (Table 4), indicating gene activation at a frequency of 100%.
Cre/lox-mediated recombination in cowpea F1 progeny from a cross between an AtRps5aproCre line (1101) and an AtUbq10prolox line (R1719). a, b F1 seedlings from a cross between homozygous plants of line 1101 and line R1719; a FITC filter; b bright field; c electrophoresis of PCR products for five F1 progeny (C1-1 to C1-5) from the same cross with p4825/p4826 and p3753/p3769; PCR−, no-template control; M, DNA ladder; bar = 5 mm
Discussion
This study demonstrates the efficacy of Cre/lox-mediated gene activation through genetic crosses in cowpea. Gene activation through Cre/lox-mediated excision occurred at a high frequency and probably at an early developmental stage of embryos upon expression of Cre by the AtRps5apro or AtDD45pro. We first evaluated Cre/lox-mediated recombination by sexual hybridization with Cre driven by the AtRps5apro and the “reporter” gene driven by the AtUbq3pro after excision of the lox-flanked DNA sequence. Because the process of recovering transgenic cowpea was inefficient at the time we introduced these two vectors into cowpea, we also evaluated gene activation with these two vectors in tobacco. The function of Cre/lox-mediated recombination has been reported in tobacco with Cre expressed by either the CaMV35Spro (Odell et al. 1990; Bayley et al. 1992; Medberry et al. 1995) or seed-specific promoters (Odell et al. 1994). Compared to those reports with Cre expressed by either the CaMV35Spro or seed-specific promoters, the current study showed that recombination could occur at an earlier developmental stage and higher frequency with Cre controlled by the AtRps5apro. Although we did not obtain direct evidence of gene activation at the zygote stage, gene activation via Cre/lox-mediated recombination occurred early in embryo development based on suspensor and globular embryo fluorescence. If the activity of the AtRps5apro in tobacco mirrored the expression profile observed in Arabidopsis (Weijers et al. 2001; Maruyama et al. 2013; Gooh et al. 2015), Cre was expressed in egg cells and young embryos which activated ZsGreen early after fertilization. Such results were not achieved by expressing Cre with the CaMV35Spro or seed-specific promoters (Odell et al. 1994). The results confirmed that gene activation via Cre/lox-mediated recombination can occur earlier than previously reported if Cre is expressed by a promoter having activity in egg cells and young embryos.
Using the AtRps5apro to express Cre also contributed to the rarity of mosaic F1 progeny both in tobacco and cowpea based on the genotyping results of the crosses with the AtUbq3prolox lines. Amplification of both fragments in the F1 progeny in a few instances may not necessarily indicate mosaicism. Alternatively, it might reflect that excision did not happen in every copy of the transgene inherited from a parent line that harbored multiple copies of the AtUbq3prolox cassette (Table S8). The absence of mosaicism in most F1 progeny was likely ascribed to the expression of Cre by the AtRps5apro in the egg cells and young embryos. The presence of Cre recombinase in the egg cells ideally would enable the excision of the lox-flanked DNA sequence and activation of ZsGreen in the zygote that could pass the activated ZsGreen to all of its daughter cells. Even if gene activation did not occur in the zygote stage, a strong expression of Cre by the AtRps5apro during early cell divisions could complete the activation process quickly in the majority of cells when the embryos only consisted of a few cells. Although the AtRps5apro activity would be inclined to early activation of the lox-reporter gene, we never observed ZsGreen expression in all isolated tobacco embryos from any crosses with the AtRps5aproCre lines pollinated by the AtUbq3prolox line (Table 1, S7). Without identifying homozygous plants of the AtUbq3prolox line carrying multiple transgene loci, we could not determine whether the absence of ZsGreen expression in the isolated F1 embryos was attributable to inactivation of the reporter gene or the lack of the lox-reporter cassette. Considering that the AtRps5apro remains active in embryos after the globular stage and even in the meristematic regions during later plant development (Weijers et al. 2001), we cannot exclude the possibility that gene activation might occur at later developmental stages. Such a pattern was observed in gene editing by CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated 9) in A. thaliana with the Cas9 expressed by the AtRps5apro (Tsutsui and Higashiyama 2017). The activity of the AtRps5apro at later developmental stages likely caused heterogeneity in mutations, which appeared to be line-dependent (Tsutsui and Higashiyama 2017). Unlike mutagenesis induced by CRISPR/Cas9, the outcome of Cre/lox-mediated recombination is unique, thus early events cannot be genetically distinguished from later events.
Further characterization of Cre/lox-mediated recombination in cowpea with Cre expressed by the AtDD45pro verified that expressing Cre in the egg cells and young embryos was sufficient to complete excision in most if not all cells at a very early embryonic developmental stage. Compared with the AtRps5apro, the AtDD45pro has a more restricted expression profile limited to egg cells and young embryos before the eight-cell stage (Steffen et al. 2007; Lawit et al. 2013). Hence, Cre/lox-mediated recombination should end soon after the eight-cell stage if the AtDD45pro maintained the same tissue-specificity in cowpea as in Arabidopsis. This would exclude the possibility of activating the reporter gene at later developmental stages. Considering that the genotyping results with p3785/p3786 indicated the absence of mosaic F1 progeny from the crosses with an AtDD45proCre line (R7743) pollinated by an AtUbq3prolox line (3102), Cre/lox-mediated recombination should have completed in all cells at an early developmental stage despite expression of Cre over a shorter period. Even without mosaic F1 progeny identified in those crosses between line R7743 and line 3102, excision failure tended to occur more frequently than in the crosses between the AtRps5aproCre lines (1101, 1201, and 1203) and the same AtUbq3prolox line (Table 3). Nevertheless, in the crosses between another AtDD45proCre line (R7715) and an AtUbq10prolox line (R1719), excision occurred at a frequency as high as in the crosses between the AtRps5aproCre lines and the same AtUbq10prolox line (Table 4), suggesting that differences in the recombination efficiency could be line-dependent and ascribed to other factors (e.g., integration and copy number) rather than the promoter activity per se. Differences in the pattern and efficiency of gene activation mediated by Cre/lox between lines have also been reported in tobacco (Odell et al. 1994) and maize (Zhang et al. 2003). The efficiency of gene activation with Cre expressed by the AtDD45pro tended to vary more between lines and crosses compared with the AtRps5apro (Table 4), suggesting that an extended Cre expression could be important to complete gene activation for some lines with a lower recombination efficiency at the early developmental stage. Activation of the lox-reporter gene at later developmental stages by a prolonged expression of Cre in embryos and meristematic regions might make recombination failure at the early developmental stage undetectable in seedlings. As a result, the final recombination frequency in the AtRps5aproCre lines might have shown less variation than the AtDD45proCre lines in which such compensation would be absent. In addition, given that the coding sequence of Cre used in the AtDD45proCre cassette was codon-optimized, we cannot exclude the possibility that differences in the recombination efficiency between two constructs could also stem from differences between the coding sequences of the Cre genes which was not examined. Overall, shortening the expressing period of Cre did not necessarily decrease the efficiency of Cre/lox-mediated recombination, similar to the observation in Arabidopsis that the use of AtRps5apro and AtDD45pro to express Cas9 ended in no significant difference in the efficiency of gene editing (Ordon et al. 2020).
The efficiency of Cre/lox-mediated recombination changed little in cowpea when the Cre lines served as a pollen donor. Among the crosses with the lox-reporter lines pollinated by the Cre lines, Cre/lox-mediated recombination mostly came about at a frequency comparable to their corresponding reciprocal crosses (Tables 3, 4), suggesting that the expression of Cre in the egg cells might not be essential for activating the reporter gene in every cell of F1 embryos as long as Cre was expressed after fertilization. Given that both AtRps5apro (Weijers et al. 2001; Maruyama et al. 2013) and AtDD45pro (Steffen et al. 2007; Lawit et al. 2013) showed activity in zygotes and young embryos, expression of Cre during this post-fertilization period probably accounted for the gene activation in F1 embryos from crosses with the Cre lines as the paternal parent.
The original plan to use the AtUbq3pro in the lox-reporter cassette to evaluate Cre/lox-mediated gene activation was unsuccessful because we could not observe ZsGreen expression in F1 seeds or seedlings. We thus designed and generated another lox-reporter cassette with the AtUbq10pro expressing the reporter gene to evaluate gene activation in cowpea. The initial selection of the AtUbq3pro was based on the strong ZsGreen expression in cowpea embryo axis tissue after microprojectile bombardment (Fig. S1c). Activation of the ZsGreen after co-bombardment with the cassettes AtRps5aproCre and AtUbq3prolox also indicated that these two cassettes functioned in cowpea (Fig. S1d). However, ZsGreen activation was invisible in F1 mature seeds and seedlings but visible in some specific tissues (Figs. S3, S4) on F1 plants with the lox-flanked DNA sequence excised, suggesting either tissue-specificity for the AtUbq3pro or some mechanisms of gene silencing in those lines that we did not explore. Although both the AtUbq3pro and AtUbq10pro were considered as “constitutive” promoters (Norris et al. 1993), expression from AtUbq3 was shown to vary more than AtUbq10 during plant development (Sun and Callis 1997; Schmid et al. 2005) and could even be inducible by dark (Sun and Callis 1997). The AtUbq10 and β-glucuronidase controlled by the AtUbq10pro were expressed much stronger than AtUbq3 and β-glucuronidase controlled by the AtUbq3pro, respectively, across various tissue types analyzed (Schmid et al. 2005; Sun and Callis 1997), supporting our observation with the lox-reporter cassettes. Using the AtUbq10pro to drive the reporter gene in the lox-reporter cassette made the outcome of Cre/lox-mediated recombination easy to screen in F1 seeds or seedlings. As a result, we could gauge the recombination efficiency by phenotyping F1 progeny based on ZsGreen expression rather than genotyping them to determine the excision of the lox-flanked intervening DNA sequence. The cassette AtUbq10prolox contained a larger intervening DNA sequence between the lox sites than AtUbq3prolox; however, the size of the intervening sequence between lox sites did not appear to affect recombination efficiency.
In summary, we characterized the Cre/lox gene activation system in both cowpea and tobacco and found that Cre/lox-mediated recombination could occur early and efficiently when Cre was controlled by AtRps5apro and AtDD45pro. The two-component gene activation system via Cre/lox-mediated recombination described here provides a useful tool for genetic engineering in cowpea and beyond.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. Novel biological materials described in this publication may be available to the academic community and other not-for-profit institutions solely for non-commercial research purposes upon acceptance and signing of a material transfer agreement between the author's institution and the requestor. In some cases, such materials may originally contain genetic elements described in the manuscript that were obtained from a third party(s), and the authors may not be able to provide materials including third party genetic elements to the requestor because of certain third-party contractual restrictions placed on the author's institution. In such cases, the requester will be required to obtain such materials directly from the third party. The authors and authors' institution do not make any express or implied permission(s) to the requester to make, use, sell, offer for sale, or import third party proprietary materials. Obtaining any such permission(s) will be the sole responsibility of the requestor. Plant germplasm and transgenic material will not be made available except at the discretion of the owner and then only in accordance with all applicable governmental regulations.
Code availability
Not applicable.
References
Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7(4):649–659
Bai X, Wang Q, Chu C (2008) Excision of a selective marker in transgenic rice using a novel Cre/loxP system controlled by a floral specific promoter. Transgenic Res 17(6):1035–1043
Bayley CC, Morgan M, Dale EC, Ow DW (1992) Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol Biol 18(2):353–361
Boukar O, Abberton M, Oyatomi O, Togola A, Tripathi L, Fatokun C (2020) Introgression breeding in cowpea [Vigna unguiculata (L.) Walp.]. Front Plant Sci 11:567425
Branda CS, Dymecki SM (2004) Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6(1):7–28
Carvalho M, Lino-Neto T, Rosa E, Carnide V (2017) Cowpea: a legume crop for a challenging environment. J Sci Food Agric 97(13):4273–4284
Che P, Chang S, Simon MK, Zhang Z, Shaharyar A, Ourada J, O’Neil D, Torres-Mendoza M, Guo Y, Marasigan KM, Vielle-Calzada J, Ozias-Akins P, Albertsen MC, Jones TJ (2021) Developing a rapid and highly efficient cowpea regeneration, transformation and genome editing system using embryonic axis explants. Plant J 106:817–830
Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16(4):664
Chen H, Luo J, Zheng P, Zhang X, Zhang C, Li X, Wang M, Huang Y, Liu X, Jan M, Liu Y, Hu P, Tu J (2017) Application of Cre-lox gene switch to limit the Cry expression in rice green tissues. Sci Rep 7(1):1–14
Clemente T (2006) Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). In: Wang K (ed) Agrobacterium protocols, vol 1. Humana Press, New Jersey, pp 143–154
Coppoolse ER, de Vroomen MJ, Roelofs D, Smit J, van Gennip F, Hersmus BJM, Nijkamp HJJ, van Haaren MJ (2003) Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol 51(2):263–279
Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16(6):771–781
Covert SF, Kapoor P, Lee MH, Briley A, Nairn CJ (2001) Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res 105(3):259–264
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(13):39–40
Eisel D, Seth O, Grünewald-Janho S, Kruchen B, Rüger B (2008) DIG application manual for filter hybridization. Editorial Roche Diagnostics GmbH 204
Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3(11):e3647
FAOSTAT (2018) Food and Agriculture Organization of the United Nations (FAO). FAOSTAT Database. http://www.fao.org/faostat/en/#data/QC. Assessed 15 December, 2020
Fu C, Sun M, Zhou C, Yang H (1996) Isolation of fertilized embryo sacs and zygotes and triggering of zygote division in Nicotiana tabacum. Acta Bot Sin 38(4):262–267
Garbarino JE, Belknap WR (1994) Isolation of a ubiquitin-ribosomal protein gene (ubi3) from potato and expression of its promoter in transgenic plants. Plant Mol Biol 24(1):119–127
Gilbertson L (2003) Cre–lox recombination: creative tools for plant biotechnology. Trends Biotechnol 21(12):550–555
Głowacka K, Kromdijk J, Leonelli L, Niyogi KK, Clemente TE, Long SP (2016) An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ 39(4):908–917
Gooh K, Ueda M, Aruga K, Park J, Arata H, Higashiyama T, Kurihara D (2015) Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev Cell 34(2):242–251
Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18(16):1964–1969
Hoa TTC, Bong BB, Huq E, Hodges TK (2002) Cre/lox site-specific recombination controls the excision of a transgene from the rice genome. Theor Appl Genet 104(4):518–525
Hoff T, Schnorr KM, Mundy J (2001) A recombinase-mediated transcriptional induction system in transgenic plants. Plant Mol Biol 45(1):41–49
Huynh BL, Ehlers JD, Huang BE, Muñoz-Amatriaín M, Lonardi S, Santos JR, Ndeve A, Batieno BJ, Boukar O, Cisse N, Drabo I, Fatokun C, Kusi F, Agyare RY, Guo Y, Herniter I, Lo S, Wanamaker SI, Xu S, Close TJ, Roberts PA (2018) A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J 93(6):1129–1142
Keil M, Sanchez-Serrano J, Schell J, Willmitzer L (1986) Primary structure of a proteinase inhibitor II gene from potato (Solanum tuberosum). Nucleic Acids Res 14(14):5641–5650
Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2(12):1201–1224
Kopertekh L, Schulze K, Frolov A, Strack D, Broer I, Schiemann J (2010) Cre-mediated seed-specific transgene excision in tobacco. Plant Mol Biol 72(6):597–605
Lawit SJ, Chamberlin MA, Agee A, Caswell ES, Albertsen MC (2013) Transgenic manipulation of plant embryo sacs tracked through cell-type-specific fluorescent markers: cell labeling, cell ablation, and adventitious embryos. Plant Reprod 26(2):125–137
Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward T, Clifton E, Falco SC, Cigan AM (2015) Cas9-guide RNA directed genome editing in soybean. Plant Physiol 169(2):960–970
Lonardi S, Muñoz-Amatriaín M, Liang Q, Shu S, Wanamaker SI, Lo S, Tanskanen J, Schulman AH, Zhu T, Luo M, Alhakami H, Ounit R, Hasan AM, Verdier J, Roberts PA, Santos JRP, Ndeve A, Doležel J, Vrána J, Hokin SA, Farmer AD, Cannon SB, Close TJ (2019) The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98(5):767–782
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao Z, Xu D, Jones T, Gordon-Kamm W (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28(9):1998–2015
Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN Jr, McAvoy R, Jiang X, Wu Y, He A, Pei Y, Li Y (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5(2):263–374
Maruyama D, Hamamura Y, Takeuchi H, Susaki D, Nishimaki M, Kurihara D, Kasahara RD, Higashiyama T (2013) Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev Cell 25(3):317–323
Medberry SL, Dale E, Qin M, Ow DW (1995) Intra-chromosomal rearrangements generated by Cre-lox site-specific recombination. Nucleic Acids Res 23(3):485–490
Mlynárová L, Conner AJ, Nap JP (2006) Directed microspore-specific recombination of transgenic alleles to prevent pollen-mediated transmission of transgenes. Plant Biotechnol J 4(4):445–452
Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo M, Alhakami H, Alpert M, Batieno BJ, Boukar O, Bozdag S, Cisse N, Drabo I, Ehlers JD, Farmer A, Fatokun C, Gu YQ, Guo Y, Huynh B, Jackson SA, Kusi F, Lawley CT, Lucas MR, Ma Y, Timko MP, Wu J, You F, Barkley NA, Roberts PA, Lonardi S, Close TJ (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J 89(5):1042–1054
Norris SR, Meyer SE, Callis J (1993) The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol 21(5):895–906
Odell J, Caimi P, Sauer B, Russell S (1990) Site-directed recombination in the genome of transgenic tobacco. Mol Gen Genet 223(3):369–378
Odell JT, Hoopes JL, Vermerris W (1994) Seed-specific gene activation mediated by the Cre/lox site-specific recombination system. Plant Physiol 106(2):447–458
Ordon J, Bressan M, Kretschmer C, Dall’Osto L, Marillonnet S, Bassi R, Stuttmann J (2020) Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct Integr Genomics 20(1):151–162
Polóniová Z, Jopčík M, Matušíková I, Libantová J, Moravčíková J (2015) The pollen-and embryo-specific Arabidopsis DLL promoter bears good potential for application in marker-free Cre/loxP self-excision strategy. Plant Cell Rep 34(3):469–481
Popelka JC, Gollasch S, Moore A, Molvig L, Higgins TJ (2006) Genetic transformation of cowpea (Vigna unguiculata L.) and stable transmission of the transgenes to progeny. Plant Cell Rep 25(4):304–312
Russell SH, Hoopes JL, Odell JT (1992) Directed excision of a transgene from the plant genome. Mol Genet Genomics 234(1):49–59
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37(5):501–506
Schünmann PH, Llewellyn DJ, Surin B, Boevink P, De Feyter RC, Waterhouse PM (2003) A suite of novel promoters and terminators for plant biotechnology. Funct Plant Biol 30(4):443–452
Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22(12):1567–1572
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212
Singer SD, Hily JM, Cox KD (2011) Analysis of the enhancer-blocking function of the TBS element from Petunia hybrida in transgenic Arabidopsis thaliana and Nicotiana tabacum. Plant Cell Rep 30(11):2013–2025
Singh BB (2014) Cowpea: the food legume of the 21st century. Crop Science Society of America
Spriggs A, Henderson ST, Hand ML, Johnson SD, Taylor JM, Koltunow A (2018) Assembled genomic and tissue-specific transcriptomic data resources for two genetically distinct lines of Cowpea (Vigna unguiculata (L.) Walp). Gates Open Res 2
Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell–secreted EC1 triggers sperm cell activation during double fertilization. Science 338(6110):1093–1097
Srivastava V, Thomson J (2016) Gene stacking by recombinases. Plant Biotechnol J 14(2):471–482
Steffen JG, Kang IH, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51(2):281–292
Sun CW, Callis J (1997) Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J 11(5):1017–1027
Tsutsui H, Higashiyama T (2017) pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol 58(1):46–56
Van Duyne GD (2001) A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct 30(1):87–104
Verweire D, Verleyen K, De Buck S, Claeys M, Angenon G (2007) Marker-free transgenic plants through genetically programmed auto-excision. Plant Physiol 145(4):1220–1231
Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM (2018) BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35(3):543–548
Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6(2):e16765
Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128(21):4289–4299
Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2(8):e718
Yao S, Jiang C, Huang Z, Torres-Jerez I, Chang J, Zhang H, Udvardi M, Liu R, Verdier J (2016) The Vigna unguiculata Gene Expression Atlas (VuGEA) from de novo assembly and quantification of RNA-seq data provides insights into seed maturation mechanisms. Plant J 88(2):318–327
Zhang Z, Conner J, Guo Y, Ozias-Akins P (2020) Haploidy in tobacco induced by PsASGR-BBML transgenes via parthenogenesis. Genes 11(9):1072
Zhang Z, Finer JJ (2016) Use of cytokinin pulse treatments and micrografting to improve sunflower (Helianthus annuus L.) plant recovery from cotyledonary tissues of mature seeds. In Vitro Cell Dev Biol-Plant 52(4):391–399
Zhang W, Subbarao S, Addae P, Shen A, Armstrong C, Peschke V, Gilbertson L (2003) Cre/lox-mediated marker gene excision in transgenic maize (Zea mays L.) plants. Theor Appl Genet 107(7):1157–1168
Zuo J, Niu QW, Møller SG, Chua NH (2001) Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19(2):157–161
Acknowledgements
We thank Commonwealth Scientific and Industrial Research Organisation (CSIRO) and Corteva for providing the clones of AtRps5apro, AtUbq3pro, GmEF1apro, Cre, DsRed, ZsGreen, phaseolinterm, and the cassette of nptII controlled by the S1 promoter and S3 terminator from subterranean clover stunt virus (SCSV). We thank Dr. Ray Collier at the Wisconsin Crop Innovation Center for generating the vectors RC2677 and RC2717, and Dr. Joe Nairn (School of Forest Resources, University of Georgia) for the clone of StUbqpro. We thank Gregory B. Thomas for taking care of the greenhouse facilities and the growing plants. We acknowledge the Capturing Heterosis team, in particular Anna Koltunow and Marc Albertsen, for their vision and support for this research direction.
Funding
This research was supported through a sub-award from the Commonwealth Scientific and Industrial Research Organisation (CSIRO) under the Capturing Heterosis for Smallholder Farmers grant from the Bill and Melinda Gates Foundation (OPP1076280). Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission.
Author information
Authors and Affiliations
Contributions
Conceptualization, Z.Z., J.C., and P.O-A.; methodology, Z.Z., J.C., and P.O-A.; formal analysis, Z.Z., Y.G., K.M., J.C.; investigation, Z.Z., Y.G., K.M., J.C.; writing−original draft preparation, Z.Z.; writing−review and editing, J.C., P.O-A.; supervision, P.O-A.; funding acquisition, P.O-A.. All authors have read and approved the published version of the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no financial or non-financial interests in any material discussed in this article. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Baochun Li.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1
Expression of ZsGreen in cowpea embryo axes 24 h after microprojectile bombardment with two AtUbq3pro expression cassettes; left column, PHP43648, AtUbq3pro:ZsGreen:NOSterm; right column, pZZ006, AtUbq3pro:lox:ZsGreen:NOSterm; a, b, bright field; c, d, FITC filter; bar = 1 mm. Supplementary file1 (TIFF 17886 KB)
Fig. S2
Expression of AtUbq3 (AT5G03240) and AtUbq10 (AT4G05320) in Arabidopsis across 47 tissue types obtained from the Arabidopsis eFP Browser (Schmid et al. 2005; Winter et al. 2007). Supplementary file2 (TIFF 24905 KB)
Fig. S3
Expression of ZsGreen in F2 seeds of cowpea from F1 plants derived from a cross between an AtRps5aproCre line (1101) and an AtUbq3prolox line (3102). a, b, F2 from an F1 progeny with the lox-flanked PINIIterm excised; c, d, F2 from an F1 progeny without the lox-flanked PINIIterm excised due to the absence of the Cre; a, c, bright field; b, d, FITC filter; bar = 5 mm. Supplementary file3 (TIFF 22527 KB)
Fig. S4
Expression of ZsGreen at the tip of peduncles from F1 plants of cowpea derived from a cross between an AtRps5aproCre line (1101) and an AtUbq3prolox line (3102). a, b, F1 progeny with the lox-flanked PINIIterm excised; c, d, F1 progeny without the lox-flanked PINIIterm excised due to the absence of the Cre; a, c, bright field; b, d, FITC filter; bar = 5 mm. Supplementary file4 (TIFF 20094 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Z., Guo, Y., Marasigan, K.M. et al. Gene activation via Cre/lox-mediated excision in cowpea (Vigna unguiculata). Plant Cell Rep 41, 119–138 (2022). https://doi.org/10.1007/s00299-021-02789-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-021-02789-z