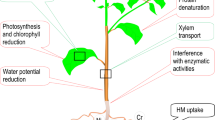
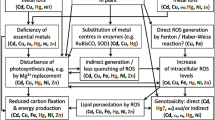
Abstract
Arsenic contaminations, often adversely influencing the living organisms, including plants, animals, and the microbial communities, are of grave apprehension. Many physical, chemical, and biological techniques are now being explored to minimize the adverse affects of arsenic toxicity. Bioremediation of arsenic species using arsenic loving bacteria has drawn much attention. Arsenate and arsenite are mostly uptaken by bacteria through aquaglycoporins and phosphate transporters. After entering arsenic inside bacterial cell arsenic get metabolized (e.g., reduction, oxidation, methylation, etc.) into different forms. Arsenite is sequentially methylated into monomethyl arsenic acid (MMA) and dimethyl arsenic acid (DMA), followed by a transformation of less toxic, volatile trimethyl arsenic acid (TMA). Passive remediation techniques, including adsorption, biomineralization, bioaccumulation, bioleaching, and so on are exploited by bacteria. Rhizospheric bacterial association with some specific plants enhances phytoextraction process. Arsenic-resistant rhizospheric bacteria have immense role in enhancement of crop plant growth and development, but their applications are not well studied till date. Emerging techniques like phytosuction separation (PS-S) have a promising future, but still light to be focused on these techniques. Plant-associated bioremediation processes like phytoextraction and phytosuction separation (PS-S) techniques might be modified by treating with potent bacteria for furtherance.
Graphical Abstract



Similar content being viewed by others
Data Availability
Open to all at any time.
References
Chen QY, Costa M (2021) Arsenic: a global environmental challenge. Annu Rev Pharmacol Toxicol 61:47–63. https://doi.org/10.1146/annurev-pharmtox-030220-013418
Zhang Y, Xu B, Guo Z, Han J, Li H, Jin L, Chen F, Xiong Y (2019) Human health risk assessment of groundwater arsenic contamination in Jinghui irrigation district, China. J Environ Manage 237:163–169. https://doi.org/10.1016/j.jenvman.2019.02.067
Mateos LM, Ordóñez E, Letek M, Gil JA (2006) Corynebacterium glutamicum as a model bacterium for the bioremediation of arsenic. Int Microbiol 9:207–215
Villaescusa I, Bollinger JC (2008) Arsenic in drinking water: sources, occurrence and health effects (a review). Rev Environ Sci 7:307–323. https://doi.org/10.1007/s11157-008-9138-7
Tariq A, Ullah U, Asif M, Sadiq I (2019) Biosorption of arsenic through bacteria isolated from Pakistan. Int Microbiol 22:59–68. https://doi.org/10.1007/s10123-018-0028-8
Zacarías-Estrada OL, Ballinas-Casarrubias L, Montero-Cabrera ME, Loredo-Portales R, Orrantia-Borunda E, Luna-Velasco A (2020) Arsenic removal and activity of a sulfate reducing bacteria-enriched anaerobic sludge using zero valent iron as electron donor. J Hazard Mater 384:121392. https://doi.org/10.1016/j.jhazmat.2019.121392
Smith AH, Lingas EO, Rahman M (2000) Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull World Health Organ 78:1093–1103
Berg M, Tran HC, Nguyen TC, Pham HV, Schertenleib R, Giger W (2001) Arsenic contamination of groundwater and drinking water in Vietnam: a human health threat. Environ Sci Technol 35:2621–2626. https://doi.org/10.1021/es010027y
Mukhopadhyay R, Rosen BP, Phung LT, Silver S (2002) Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26:311–325. https://doi.org/10.1111/j.1574-6976.2002.tb00617.x
Lindberg AL, Goessler W, Gurzau E, Koppova K, Rudnai P, Kumar R, Fletcher T, Leonardi G, Slotova K, Gheorghiu E, Vahter M (2006) Arsenic exposure in Hungary, Romania and Slovakia. J Environ Monitor 8:203–208
Naito S, Matsumoto E, Shindoh K, Nishimura T (2015) Effects of polishing, cooking, and storing on total arsenic and arsenic species concentrations in rice cultivated in Japan. Food Chem 168:294–301. https://doi.org/10.1016/j.foodchem.2014.07.060
Moens M, Branco R, Morais PV (2020) Arsenic accumulation by a rhizosphere bacterial strain Ochrobactrum tritici reduces rice plant arsenic levels. World J Microbiol Biotechnol 36:23. https://doi.org/10.1007/s11274-020-2800-0
Smedley PL, Zhang M, Zhang G, Luo Z (2003) Mobilisation of arsenic and other trace elements in fluviolacustrine aquifers of the Huhhot Basin, Inner Mongolia. Appl Geochem 18:1453–1477. https://doi.org/10.1016/S0883-2927(03)00062-3
Uppal JS, Zheng Q, Le XC (2019) Arsenic in drinking water—recent examples and updates from Southeast Asia. Curr Opin Environ Sci Health 7:126–135. https://doi.org/10.1016/j.coesh.2019.01.004
Aryal M, Ziagova M, Liakopoulou-Kyriakides M (2010) Study on arsenic biosorption using Fe(III)-treated biomass of Staphylococcus xylosus. Chem Eng J 162:178–185. https://doi.org/10.1016/j.cej.2010.05.026
Cavalca L, Corsini A, Zaccheo P, Andreoni V, Muyzer G (2013) Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol 8:753–768. https://doi.org/10.2217/fmb.13.38
Yang Q, Tu S, Wang G, Liao X, Yan X (2012) Effectiveness of applying arsenate reducing bacteria to enhance arsenic removal from polluted soils by Pteris vittata L. Int J phytoremediat 14:89–99. https://doi.org/10.1080/15226510903567471
Kumar M, Yadav A, Ramanathan AL (2020) Arsenic contamination in environment, ecotoxicological and health effects, and bioremediation strategies for its detoxification. In: Saxena G, Bharagava RN (eds) Bioremediation of industrial waste for environmental safety. Springer, Singapore, pp 245–264
Jackson CR, Dugas SL, Harrison KG (2005) Enumeration and characterization of arsenate-resistant bacteria in arsenic free soils. Soil Biol Biochem 37:2319–2322. https://doi.org/10.1016/j.soilbio.2005.04.010
Irshad S, Xie Z, Wang J, Nawaz A, Luo Y, Wang Y, Mehmood S (2020) Indigenous strain Bacillus XZM assisted phytoremediation and detoxification of arsenic in Vallisneria denseserrulata. J Hazard Mater 381:120903. https://doi.org/10.1016/j.jhazmat.2019.120903
Kruger MC, Bertin PN, Heipieper HJ, Arsène-Ploetze F (2013) Bacterial metabolism of environmental arsenic—mechanisms and biotechnological applications. Appl Microbiol 97:3827–3841. https://doi.org/10.1007/s00253-013-4838-5
Chen C, Li L, Huang K, Zhang J, Xie WY, Lu Y, Dong S, Zhao FJ (2019) Sulfate-reducing bacteria and methanogens are involved in arsenic methylation and demethylation in paddy soils. ISME J. https://doi.org/10.1038/s41396-019-0451-7
Tsai SL, Singh S, Chen W (2009) Arsenic metabolism by microbes in nature and the impact on arsenic remediation. Curr Opin Biotechnol 20:659–667. https://doi.org/10.1016/j.copbio.2009.09.013
Pandey S, Rai R, Rai LC (2015) Biochemical and molecular basis of arsenic toxicity and tolerance in microbes and plants. In: Flora SJS (ed) Handbook of arsenic toxicology. Academic Press, New York, pp 627–674
Rosen BP, Tamás MJ (2010) Arsenic transport in prokaryotes and eukaryotic microbes. In: Jahn TP, Bienert GP (eds) MIPs and their role in the exchange of metalloids. Springer, New York, pp 47–55
Kumari N, Jagadevan S (2016) Genetic identification of arsenate reductase and arsenite oxidase in redox transformations carried out by arsenic metabolising prokaryotes—a comprehensive review. Chemosphere 163:400–412. https://doi.org/10.1016/j.chemosphere.2016.08.044
Nies DH, Silver S (1995) Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol 14:186–199. https://doi.org/10.1007/BF01569902
Rosen BP, Liu Z (2009) Transport pathways for arsenic and selenium: a minireview. Environ Int 35:512–515. https://doi.org/10.1016/j.envint.2008.07.023
Yan L, Yin H, Zhang S, Leng F, Nan W, Li H (2010) Biosorption of inorganic and organic arsenic from aqueous solution by Acidithiobacillus ferrooxidans BY-3. J Hazard Mater 178:209–217. https://doi.org/10.1016/j.jhazmat.2010.01.065
Sanders OI, Rensing C, Kuroda M, Mitra B, Rosen BP (1997) Antimonite is accumulated by the glycerol facilitator GlpF in Escherichia coli. J Bacteriol 179:3365–3367. https://doi.org/10.1128/jb.179.10.3365-3367.1997
Anderson CR, Cook GM (2004) Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr Microbiol 48:341–347. https://doi.org/10.1007/s00284-003-4205-3
Sunita MSL, Prashant S, Chari PB, Rao SN, Balaravi P, Kishor PK (2012) Molecular identification of arsenic-resistant estuarine bacteria and characterization of their ars genotype. Ecotoxicology 21:202–212. https://doi.org/10.1007/s10646-011-0779-x
Satyapal GK, Rani S, Kumar M, Kumar N (2016) Potential role of arsenic resistant bacteria in bioremediation: current status and future prospects. J Microb Biochem Technol 8:256–258. https://doi.org/10.4172/1948-5948.1000294
Zhao C, Zhang Y, Chan Z, Chen S, Yang S (2015) Insights into arsenic multi-operons expression and resistance mechanisms in Rhodopseudomonas palustris CGA009. Front Microbiol 6:986. https://doi.org/10.3389/fmicb.2015.00986
Fekih IB, Zhang C, Li YP, Zhao Y, Alwathnani HA, Saquib Q, Rensing C, Cervantes C (2018) Distribution of arsenic resistance genes in prokaryotes. Front Microbiol 9:2473. https://doi.org/10.3389/fmicb.2018.02473
Liao VHC, Chu YJ, Su YC, Hsiao SY, Wei CC, Liu CW, Liao CM, Shen WC, Chang FJ (2011) Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J Contam Hydrol 123:20–29. https://doi.org/10.1016/j.jconhyd.2010.12.003
Crognale S, Amalfitano S, Casentini B, Fazi S, Petruccioli M, Rossetti S (2017) Arsenic-related microorganisms in groundwater: a review on distribution, metabolic activities and potential use in arsenic removal processes. Rev Environ Sci 16:647–665. https://doi.org/10.1007/s11157-017-9448-8
Vidal FV, Vidal VMV (1980) Arsenic metabolism in marine bacteria and yeast. Mar Biol 60:1–7. https://doi.org/10.1007/BF00395600
Rowland IR, Davies MJ (1981) In vitro metabolism of inorganic arsenic by the gastro-intestinal microflora of the rat. J Appl Toxicol 1:278–283. https://doi.org/10.1002/jat.2550010508
Silver S, Phung LT (2005) Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71:599–608
Rosen BP (2002) Biochemistry of arsenic detoxification. FEBS Lett 529:86–92. https://doi.org/10.1016/S0014-5793(02)03186-1
Ji G, Silver S (1992) Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci USA 89:9474–9478. https://doi.org/10.1073/pnas.89.20.9474
Glasser NR, Oyala PH, Osborne TH, Santini JM, Newman DK (2018) Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations. Proc Natl Acad Sci USA 115:E8614–E8623. https://doi.org/10.1073/pnas.1807984115
Song B, Chyun E, Jaffé PR, Ward BB (2009) Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microbiol Ecol 68:108–117. https://doi.org/10.1111/j.1574-6941.2009.00657.x
Saleem H, Rehman Y (2019) Arsenic respiration and detoxification by purple non-sulphur bacteria under anaerobic conditions. C R Biol 342:101–107. https://doi.org/10.1016/j.crvi.2019.02.001
Anderson GL, Williams J, Hille R (1992) The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem 267:23674–23682
Ellis PJ, Conrads T, Hille R, Kuhn P (2001) Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9:125–132. https://doi.org/10.1016/S0969-2126(01)00566-4
Kalimuthu P, Heath MD, Santini JM, Kappler U, Bernhardt PV (2014) Electrochemically driven catalysis of Rhizobium sp. NT-26 arsenite oxidase with its native electron acceptor cytochrome c552. Biochim Biophys Acta 1837:112–120. https://doi.org/10.1016/j.bbabio.2013.07.010
Chen J, Qin J, Zhu YG, de Lorenzo V, Rosen BP (2013) Engineering the soil bacterium Pseudomonas putida for arsenic methylation. Appl Environ Microbiol 79:4493–4495
Ngegla JV, Zhou X, Chen X, Zhu X, Liu Z, Feng J, Zeng XC (2020) Unique diversity and functions of the arsenic-methylating microorganisms from the tailings of Shimen Realgar Mine. Ecotoxicology 29:86–96. https://doi.org/10.1007/s10646-019-02144-9
Wang PP, Bao P, Sun GX (2015) Identification and catalytic residues of the arsenite methyltransferase from a sulfate-reducing bacterium, Clostridium sp. BXM FEMS Microbiol Lett 362:1–8
Ahsan N, Faruque K, Shamma F, Islam N, Akhand AA (2011) Arsenic adsorption by bacterial extracellular polymeric substances. Banglad J Microbiol 28:80–83. https://doi.org/10.3329/bjm.v28i2.11821
Kostal J, Yang R, Wu CH, Mulchandani A, Chen W (2004) Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Appl Environ Microbiol 70:4582–4587
Liu S, Zhang F, Chen J, Sun G (2011) Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J Environ Sci 23:1544–1550. https://doi.org/10.1016/S1001-0742(10)60570-0
Huang K, Chen C, Shen Q, Rosen BP, Zhao FJ (2015) Genetically engineering Bacillus subtilis with a heat-resistant arsenite methyltransferase for bioremediation of arsenic-contaminated organic waste. Appl Environ Microbiol 81:6718–6724
Singh S, Mulchandani A, Chen W (2008) Highly selective and rapid arsenic removal by metabolically engineered Escherichia coli cells expressing Fucus vesiculosus metallothionein. Appl Environ Microbiol 74:2924–2927
Crameri A, Dawes G, Rodriguez E Jr, Silver S, Stemmer WP (1997) Molecular evolution of an arsenate detoxification pathway by DNA shuffling. Nat Biotechnol 15:436. https://doi.org/10.1038/nbt0597-436
Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP (2014) Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci 42:443–467. https://doi.org/10.1146/annurev-earth-060313-054942
Yamamura S, Ike M, Fujita M (2003) Dissimilatory arsenate reduction by a facultative anaerobe, Bacillus sp. strain SF-1. J Biosci Bioeng 96:454–460. https://doi.org/10.1016/S1389-1723(03)70131-5
Uhrynowski W, Radlinska M, Drewniak L (2019) Genomic analysis of Shewanella sp. O23S—the natural host of the psheb plasmid carrying genes for arsenic resistance and dissimilatory reduction. Int J Mol Sci 20:1018. https://doi.org/10.3390/ijms20051018
Stolz JF, Perera E, Kilonzo B, Kail B, Crable B, Fisher E, Ranganatanm M, Wormer L, Basu P (2007) Biotransformation of 3-nitro-4-hydroxybenzene arsonic acid (roxarsone) and release of inorganic arsenic by Clostridium species. Environ Sci Technol 41:818–823. https://doi.org/10.1021/es061802i
Huang K, Peng H, Gao F, Liu Q, Lu X, Shen Q, Le XC, Zhao FJ (2019) Biotransformation of arsenic-containing roxarsone by an aerobic soil bacterium Enterobacter sp. CZ-1. Environ Pollut 247:482–487. https://doi.org/10.1016/j.envpol.2019.01.076
Das S, Jean JS, Kar S, Chou ML, Chen CY (2014) Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from agricultural soil and their potential implication for arsenic bioremediation. J Hazard Mater 272:112–120. https://doi.org/10.1016/j.jhazmat.2014.03.012
Nguyen VK, Lee MH, Park HJ, Lee JU (2015) Bioleaching of arsenic and heavy metals from mine tailings by pure and mixed cultures of Acidithiobacillus spp. J Ind Eng Chem 21:451–458. https://doi.org/10.1016/j.jiec.2014.03.004
Zhang J, Zhang X, Ni Y, Yang X, Li H (2007) Bioleaching of arsenic from medicinal realgar by pure and mixed cultures. Process Biochem 42:1265–1271. https://doi.org/10.1016/j.procbio.2007.05.021
Cheng C, Nie ZW, He LY, Sheng XF (2020) Rice-derived facultative endophytic Serratia liquefaciens F2 decreases rice grain arsenic accumulation in arsenic-polluted soil. Environ Pollut 259:113832. https://doi.org/10.1016/j.envpol.2019.113832
Banerjee S, Majumdar J, Samal AC, Bhattachariya P, Santra SC (2013) Biotransformation and bioaccumulation of arsenic by Brevibacillus brevis isolated from arsenic contaminated region of West Bengal. IOSR J Environ Sci Toxicol Food Technol 3:1–10
Irshad S, Xie Z, Mehmood S, Nawaz A, Ditta A, Mahmood Q (2021) Insights into conventional and recent technologies for arsenic bioremediation: a systematic review. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-021-12487-8
Podder MS, Majumder CB (2017) Simultaneous biosorption and bioaccumulation: a novel technique for the efficient removal of arsenic. Sustain Water Resour Manag 3:357–389. https://doi.org/10.1007/s40899-017-0103-x
Tazaki K, Rafiqul IA, Nagai K, Kurihara T (2003) FeAs2 biomineralization on encrusted bacteria in hot springs: an ecological role of symbiotic bacteria. Can J Earth Sci 40:1725–1738. https://doi.org/10.1139/e03-081
Roh Y, Chon CM, Moon JW (2007) Metal reduction and biomineralization by an alkaliphilic metal-reducing bacterium, Alkaliphilus metalliredigens (QYMF). Geosci J 11:415–423. https://doi.org/10.1007/BF02857056
Rodriguez-Freire L, Sierra-Alvarez R, Root R, Chorover J, Field JA (2014) Biomineralization of arsenate to arsenic sulfides is greatly enhanced at mildly acidic conditions. Water Res 66:242–253. https://doi.org/10.1016/j.watres.2014.08.016
Antenozio ML, Giannelli G, Marabottini R, Brunetti P, Allevato E, Marzi D et al (2021) Phytoextraction efficiency of Pteris vittata grown on a naturally As-rich soil and characterization of As-resistant rhizosphere bacteria. Sci Rep 11:1–11. https://doi.org/10.1038/s41598-021-86076-7
Lampis S, Santi C, Ciurli A, Andreolli M, Vallini G (2015) Promotion of arsenic phytoextraction efficiency in the fern Pteris vittata by the inoculation of As-resistant bacteria: a soil bioremediation perspective. Front Plant Sci 6:80. https://doi.org/10.3389/fpls.2015.00080
Franchi E, Cosmina P, Pedron F, Rosellini I, Barbafieri M, Petruzzelli G, Vocciante M (2019) Improved arsenic phytoextraction by combined use of mobilizing chemicals and autochthonous soil bacteria. Sci Total Environ 655:328–336. https://doi.org/10.1016/j.scitotenv.2018.11.242
Mesa V, Navazas A, González-Gil R, González A, Weyens N, Lauga B et al (2017) Use of endophytic and rhizosphere bacteria to improve phytoremediation of arsenic-contaminated industrial soils by autochthonous Betula celtiberica. Appl Environ Microbiol 83:e03411-e3416. https://doi.org/10.1128/AEM.03411-16
Sessitsch A, Kuffner M, Kidd P, Vangronsveld J, Wenzel WW, Fallmann K, Puschenreiter M (2013) The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol Biochem 60:182–194. https://doi.org/10.1016/j.soilbio.2013.01.012
Katoh M, Hashimoto K, Sato T (2016) Lead and antimony removal from contaminated soil by phytoremediation combined with an immobilization material. Clean: Soil, Air, Water 44:1717–1724. https://doi.org/10.1002/clen.201500162
Arita S, Katoh M (2018) Arsenic removal from contaminated soil by phytoremediation combined with chemical immobilization. The international congress on environmental geotechnics. Springer, Singapore, pp 280–285
Yan M, Zeng X, Wang J, Meharg AA, Meharg C, Tang X et al (2020) Dissolved organic matter differentially influences arsenic methylation and volatilization in paddy soils. J Hazard Mater 388:121795. https://doi.org/10.1016/j.jhazmat.2019.121795
Glick BR (2003) Phytoremediation: synergistic use of plants and bacteria to clean up the environment. Biotechnol Adv 21:383–393. https://doi.org/10.1016/S0734-9750(03)00055-7
Kabiraj A, Majhi K, Halder U, Let M, Bandopadhyay R (2020) Role of plant growth promoting rhizobacteria (PGPR) for crop stress management. In: Roychowdhury R, Choudhury S, Hasanuzzaman M, Srivastava S (eds) Sustainable agriculture in the era of climate change. Springer, Cham, pp 367–389
Cavalca L, Zanchi R, Corsini A, Colombo M, Romagnoli C, Canzi E, Andreoni V (2010) Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst Appl Microbiol 33:154–164. https://doi.org/10.1016/j.syapm.2010.02.004
Gihring TM, Druschel GK, McCleskey RB, Hamers RJ, Banfield JF (2001) Rapid arsenite oxidation by Thermus aquaticus and Thermus thermophilus: field and laboratory investigations. Environ Sci Technol 35:3857–3862. https://doi.org/10.1021/es010816f
Ghosh PK, Maiti TK, Pramanik K, Ghosh SK, Mitra S, De TK (2018) The role of arsenic resistant Bacillus aryabhattai MCC3374 in promotion of rice seedlings growth and alleviation of arsenic phytotoxicity. Chemosphere 211:407–419. https://doi.org/10.1016/j.chemosphere.2018.07.148
Banerjee A, Hazra A, Das S, Sengupta C (2020) Groundwater inhabited Bacillus and Paenibacillus strains alleviate arsenic-induced phytotoxicity of rice plant. Int J Phytoremediat. https://doi.org/10.1080/15226514.2020.1725871
Sarkar A, Kazy SK, Sar P (2013) Characterization of arsenic resistant bacteria from arsenic rich groundwater of West Bengal, India. Ecotoxicology 22:363–376. https://doi.org/10.1007/s10646-012-1031-z
Bhakta JN, Munekage Y, Ohnishi K, Jana BB, Balcazar JL (2014) Isolation and characterization of cadmium-and arsenic-absorbing bacteria for bioremediation. Water Air Soil Pollut 225:2151
Pepi M, Volterrani M, Renzi M, Marvasi M, Gasperini S, Franchi E, Focardi SE (2007) Arsenic-resistant bacteria isolated from contaminated sediments of the Orbetello Lagoon, Italy, and their characterization. J Appl Microbiol 103:2299–2308. https://doi.org/10.1111/j.1365-2672.2007.03471.x
Bachate SP, Cavalca L, Andreoni V (2009) Arsenic-resistant bacteria isolated from agricultural soils of Bangladesh and characterization of arsenate-reducing strains. J Appl Microbiol 107:145–156. https://doi.org/10.1111/j.1365-2672.2009.04188.x
Ghodsi H, Hoodaji M, Tahmourespour A, Gheisari MM (2011) Investigation of bioremediation of arsenic by bacteria isolated from contaminated soil. Afr J Microbiol Res 5:5889–5895
Shakoori FR, Aziz I, Rehman A, Shakoori AR (2010) Isolation and characterization of arsenic reducing bacteria from industrial effluents and their potential use in bioremediation of wastewater. Pak J Zool 42:331–338
Chitpirom K, Akaracharanya A, Tanasupawat S, Leepipatpibooim N, Kim KW (2009) Isolation and characterization of arsenic resistant bacteria from tannery wastes and agricultural soils in Thailand. Ann Microbiol 59:649–656. https://doi.org/10.1007/BF03179204
Del Giudice I, Limauro D, Pedone E, Bartolucci S, Fiorentino G (2013) A novel arsenate reductase from the bacterium Thermus thermophilus HB27: its role in arsenic detoxification. BBA-Proteins Proteom 1834:2071–2079. https://doi.org/10.1016/j.bbapap.2013.06.007
Collinet MN, Morin D (1990) Characterization of arsenopyrite oxidizing Thiobacillus. tolerance to arsenite, arsenate, ferrous and ferric iron. Anton Leeuw Int J G 57:237–244. https://doi.org/10.1007/BF00400155
Banerjee A, Sarkar S, Gorai S, Kabiraj A, Bandopadhyay R (2021) High arsenic tolerance in Brevundimonas aurantiaca PFAB1 from an arsenic-rich Indian hot spring. Electron J Biotechnol 53:1–7. https://doi.org/10.1016/j.ejbt.2021.05.006
Li Q, Luo J, Xu R, Yang Y, Xu B, Jiang T, Yin H (2021) Synergistic enhancement effect of Ag+ and organic ligands on the bioleaching of arsenic-bearing gold concentrate. Hydrometallurgy 204:105723. https://doi.org/10.1016/j.hydromet.2021.105723
Luo X, Jiang X, Xue S, Tang X, Zhou C, Wu C et al (2021) Arsenic biomineralization by iron oxidizing strain (Ochrobactrum sp.) isolated from a paddy soil in Hunan, China. Land Degrad Dev 32:2082–2093. https://doi.org/10.1002/ldr.3842
Yang C, Ho YN, Makita R, Inoue C, Chien MF (2020) Cupriavidus basilensis strain r507, a toxic arsenic phytoextraction facilitator, potentiates the arsenic accumulation by Pteris vittata. Ecotoxicol Environ Saf 190:110075. https://doi.org/10.1016/j.ecoenv.2019.110075
Parsania S, Mohammadi P, Soudi MR (2021) Biotransformation and removal of arsenic oxyanions by Alishewanella agri PMS5 in biofilm and planktonic states. Chemosphere 284:131336. https://doi.org/10.1016/j.chemosphere.2021.131336
Acknowledgements
We are thankful to UGC-Centre for Advanced Study, Department of Botany and DST-FIST, The University of Burdwan, Burdwan, for pursuing all the research facilities. AK is thankful to DHESTBT (WB-DBT) [Memo. No. 30 (Sanc.)-BT/ST/P/S&T/2G-48/2017] and UH is thankful to SRF (State Fund) for the financial assistance [Fc (Sc.)/RS/SF/BOT./2017-18/22].
Funding
No funding is available for this work.
Author information
Authors and Affiliations
Contributions
RB1 and AK adopted the idea and wrote the manuscript. RB2 and UH collected the information. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
Authors declare that this manuscript is a review work, and therefore, it does not include any studies using animal and human beings.
Consent to Publication
All authors read and approved the final manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kabiraj, A., Biswas, R., Halder, U. et al. Bacterial Arsenic Metabolism and Its Role in Arsenic Bioremediation. Curr Microbiol 79, 131 (2022). https://doi.org/10.1007/s00284-022-02810-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02810-y