Abstract
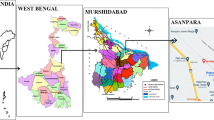
Sixty-four arsenic (As) resistant bacteria isolated from an arsenic rich groundwater sample of West Bengal were characterized to investigate their potential role in subsurface arsenic mobilization. Among the isolated strains predominance of genera Agrobacterium/Rhizobium, Ochrobactrum and Achromobacter which could grow chemolitrophically and utilize arsenic as electron donor were detected. Higher tolerance to As3+ [maximum tolerable concentration (MTC): ≥10 mM], As5+ (MTC: ≥100 mM) and other heavy metals like Cu2+, Cr2+, Ni2+ etc. (MTC: ≥10 mM), presence of arsenate reductase and siderophore was frequently observed among the isolates. Ability to produce arsenite oxidase and phosphatase enzyme was detected in 50 and 34 % of the isolates, respectively. Although no direct correlation among taxonomic identity of bacterial strains and their metabolic abilities as mentioned above was apparent, several isolates affiliated to genera Ochrobactrum, Achromobacter and unclassified Rhizobiaceae members were found to be highly resistant to As3+ and As5+ and positive for all the test properties. Arsenate reductase activity was found to be conferred by arsC gene, which in many strains was coupled with arsenite efflux gene arsB as well. Phylogenetic incongruence between the 16S rRNA and ars genes lineages indicated possible incidence of horizontal gene transfer for ars genes. Based on the results we propose that under the prevailing low nutrient condition inhabitant bacteria capable of using inorganic electron donors play a synergistic role wherein siderophores and phosphatase activities facilitate the release of sediment bound As5+, which is subsequently reduced by arsenate reductase resulting into the mobilization of As3+ in groundwater.






Similar content being viewed by others
References
Achour-Rokbani A, Bauda P, Billard P (2007) Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res Microbiol 158:128–137
Achour-Rokbani A, Cordi A, Poupin P, Bauda P, Billard P (2010) Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. Strain A33. Appl Microbiol 76:948–955
Bachate SP, Khapare RM, Kodam KM (2012) Oxidation of arsenite by two β-proteobacteria isolated from soil. Appl Microbiol Biotechnol 93:2135–2145
Bertics VJ, Ziebis W (2009) Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME 3:1269–1285
Cai L, Liu G, Rensing C, Wang G (2009) Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol 9:4
Cavalca L, Zanchi R, Corsini A, Colombo M, Romagnoli C, Canzi E, Andreoni V (2010) Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst Appl Microbiol 33:154–164
Cheah SM, Kraemer J, Cervini S, Sposito G (2003) Steady-state dissolution kinetics of goethite in the presence of desferrioxamine B and oxalate ligands: implications for the microbial acquisition of iron. Chem Geol 198:63–75
Coombs JM, Barkay T (2004) Molecular evidence for the evolution of metal homeostasis genes by lateral gene transfer in bacteria from the deep terrestrial subsurface. Appl Environ Microbiol 70:1698–1707
Drewniak L, Styczek A, Lopatka MM, Sklodowska A (2008) Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potential role in dissemination of arsenic pollution. Environ Pollut 156:1069–1074
Drewniak L, Maryan N, Lewandowski W, Kaczanowski S, Sklodowska A (2012) The contribution of microbial mats to the arsenic geochemistry of an ancient gold mine. Environ Pollut 162:190–201
Ellis PJ, Conrads T, Hille R, Kuhn P (2001) Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 angstrom and 2.03 angstrom. Structure 9:125–132
Fan H et al (2008) Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, northwestern China. J Appl Microbiol 105:529–539
Fendorf S, Michael HA, van Geen A (2010) Spatial and temporal variations of groundwater arsenic in south and southeast Asia. Science 328:1123–1127
Gihring TM, Banfield JF (2001) Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol Lett 204:335–340
Gorra R, Webster G, Martin M, Celi L, Mapelli F, Weightman AJ (2012) Dynamic microbial community associated with iron–arsenic co-precipitation products from a groundwater storage system in Bangladesh. Microb Ecol 64:171–186
Hery M, Van Dongen BE, Gill F, Mondal D, Vaughan DJ, Pancost RD, Polya DA, Lloyd JR (2010) Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology 8:155–168
Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR (2004) Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68–71
Jackson CR, Dugas SL (2003) Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol Biol 3:18–27
Jackson CR, Jackson EF, Dugas SL, Gamble K, Williams SE (2003) Microbial transformations of arsenite and arsenate in natural environments. Recent Res Dev Microbiol 7:103–118
Kazy SK, Sar P, Asthana RK, Singh SP (1999) Copper uptake and its compartmentalization in Pseudomonas aeruginosa strains: chemical nature of cellular metal. World J Microbiol Biotechnol 15:599–605
Krumholz LR (1998) Microbial ecosystems in the earth’s subsurface. ASM News 64:4
Liao VH-C, Chu YJ, Su YC, Hsiao SY, Wei CC, Liu CW, Liao CM, Shen WC, Chang FJ (2011) Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J Contam Hydrol 123:20–29
Lillis TO, Bissonnette GK (2001) Detection and characterization of filterable hetero trophic bacteria from rural groundwater supplies. Lett Appl Microbiol 32:268–272
Luli GW, Talnagi JW, Strohl WR, Pfister RM (1983) Hexavalent chromium resistant bacteria isolated from river sediments. Appl Environ Microbiol 46:846–854
Mailloux BJ, Alexandrova E, Alison R et al (2009) Microbial mineral weathering for nutrient acquisition releases arsenic. Appl Environ Microbiol 75:2558–2565
Maria FVT, Oscar CBR, Camilo S, Martha JVF, Jenny D (2011) Horizontal arsC gene transfer among microorganisms isolated from arsenic polluted soil. Int Biodeterior Biodegradation 65:147–152
Mukhopadhyay R, Rosen BP, Phung LT, Silver S (2002) Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26:311–325
Muller D, Medigue C, Koechler S et al (2007) A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet 3:53
Mumford AC, Barringer JL, Benzel WM, Reilly PA, Young LY (2012) Microbial transformations of arsenic: mobilization from glauconitic sediments to water. Water Res 46:2859–2868
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nair A, Juwarkar AA, Singh SK (2007) Production and characterization of siderophores and its application in arsenic removal from contaminated soil. Water Air Soil Pollut 180:199–212
Oremland RS, Kulp TR, Switzer BJ, Hoeft SE, Baesman S, Miller G, Stolz JF (2005) A microbial arsenic cycle in a salt saturated, extreme environment. Science 308:1305–1308
Osborne FH, Ehrlich HL (1976) Oxidation of arsenite by a soil isolate of Alcaligenes. J Appl Bacteriol 41:295–304
Pal T, Mukherjee PK (2009) Study of subsurface geology in locating arsenic-free groundwater in Bengal delta, West Bengal, India. Environ Geol 56:1211–1225
Piotrowska SZ, Kozdroj J (2008) Changes in culturable bacterial community of soil treated with high dosages of Cu or Cd. Plant Soil Environ 54:520–528
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Riccio ML, Rossolini GM, Lombardi G, Chiesurin A, Satta G (1997) Expression cloning of different bacterial phosphatase-encoding genes by histochemical screening of genomic libraries onto an indicator medium containing phenolphthalein diphosphate and methyl green. J Appl Microbiol 82:177–185
Saltikov CW, Olson BH (2002) Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl Environ Microbiol 68:280–288
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Silver S, Phung LT (2005) Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol 71:599–608
Simeonova DD, Lievremont D, Lagarde F, Muller DA, Groudeva VI, Lett MC (2004) Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol Lett 237:249–253
Slyemi D, Bonnefoy V (2011) How prokaryotes deal with arsenic. Environ Microbiol Rep 1:16
Sriyosachati S, Cox CD (1986) Siderophore-mediated iron acquisition from transferrin by Pseudomonas aeruginosa. Infect Immun 52:885–891
Sun Y, Polischuk EA, Radoja U, Cullen WR (2004) Identification and quantification of arsC genes in environmental samples by using real-time PCR. J Microbiol Methods 58:335–349
Sutton NB, van der Kraan GM, van Loosdrecht MC, Muyzer G, Bruining J, Schotting RJ (2009) Characterization of geochemical constituents and bacterial populations associated with As mobilization in deep and shallow tube wells in Bangladesh. Water Res 43:1720–1730
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vala AK, Dave BP, Dube HC (2006) Chemical characterization and quantification of siderophores produced by marine and terrestrial aspergilli. Can J Microbiol 52:603–607
vanden Hoven RN, Santini GM (2004) Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: the arsenite oxidase and its physiological electron acceptor. Biochim Biophys Acta 1656:148–155
Wang SW, Liu CW, Lu KL, Lin LH (2012) Biogeochemical cycling of ferric oxyhydroxide affecting As partition in groundwater aquitard. Environ Geochem Health 34(4):467–479
Acknowledgments
Angana Sarkar acknowledges Indian Institute of Technology, Kharagpur for financial support. Pinaki Sar and Sufia K Kazy acknowledge support from Department of Biotechnology, Government of India (Rapid Grant for Young Investigator scheme). Generous help from Mr. Buddhadev Banerjee, Barasat, West Bengal in sample collection is gratefully acknowledged.
Ethical standards
The authors declare that all the experiments comply with the current laws of the India.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Plot of PCA-scores on arsenic and others heavy metals resistances of arsenic resistant bacteria isolated from As-contaminated ground water. Each bacterial strain is represented by its ID number (DOC 49 kb)
Rights and permissions
About this article
Cite this article
Sarkar, A., Kazy, S.K. & Sar, P. Characterization of arsenic resistant bacteria from arsenic rich groundwater of West Bengal, India. Ecotoxicology 22, 363–376 (2013). https://doi.org/10.1007/s10646-012-1031-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-012-1031-z