Abstract
Haemoglobin (Hb) H-constant spring (CS) alpha thalassaemia (- -/-αCS) is the most common type of nondeletional Hb H disease in southern China. The CRISPR/Cas9-based gene correction of patient-specific induced pluripotent stem cells (iPSCs) and cell transplantation now represent a therapeutic solution for this genetic disease. We designed primers for the target sites using CRISPR/Cas9 to specifically edit the HBA2 gene with an Hb-CS mutation. After applying a correction-specific PCR assay to purify the corrected clones followed by sequencing to confirm the mutation correction, we verified that the purified clones retained full pluripotency and exhibited a normal karyotype. This strategy may be promising in the future, although it is far from representing a solution for the treatment of HbH-CS thalassemia now.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haemoglobin-Constant Spring (Hb-CS, α142, Term→Gln, TAA>CAA (α2), αcsα/) is a nondeletional form of α-thalassemia (α-Thal) with a nucleotide substitution at the termination codon CD142 (UAA>CAA) of the α2-globin gene [1]. Although the heterozygote of Hb-CS is clinically normal, when associated with a0-thalassemia, it can result in HbH-CS disease (--/aCSa), which is the most common type of nondeletional HbH disease (β4) in southern China [2]. In addition, HbH-CS disease-affected individuals are usually more anaemic, more symptomatic, and more prone to significant hepatosplenomegaly, especially compared to an individual with a triple gene deletion involving the a-globin gene (--/-a), who is more likely to require transfusions [3]. Normally, haematopoietic stem cell (HSC) transplantation is an efficient means and the only way to cure severe thalassemia, while a lack of HLA-matched healthy donors, immune complications, and viral vector safety concerns has limited its use [4,5,6].
The generation of patient-specific α-Thal-induced pluripotent stem cells (iPSCs) from patient somatic cells, the correction of disease-causing mutations in those cells, and differentiation into haematopoietic stem cells (HSCs) offer a new therapeutic strategy for this monogenic disease [7,8,9]. Recently, single-strand oligodeoxynucleotides (ssODNs), high-fidelity CRISPR/Cas9 nuclease and small molecules were used to achieve a seamless correction of the β-41/42 (TCTT) deletion mutation in β-thalassemia patient-specific iPSCs with remarkable efficiency [10]. However, there has been no similar attempt in HbH-CS disease.
In this study, we successfully corrected the HBA2 gene of the Hb-CS mutation in Hb-CS-CS-Thal iPSCs by the combination of ssODNs and CRISPR/Cas9 gRNA, providing a potential method for the treatment of HbH-CS thalassemia.
Materials and methods
Ethics statement
All participants in this study signed a written informed consent for donating peripheral blood for stem cell generation, and all animal care and experiments were performed in accordance with the institutional ethical guidelines for animal experiments (the IRB at the Third Affiliated Hospital of Guangzhou Medical University (No. 2016-001)). All methods were performed in accordance with the approved guidelines.
Cell culture and iPSC generation
Five-millilitre peripheral blood was obtained from a Hb-CS thalassemia 6.3-month-old boy patient (ααcs/--SEA) and a normal male adult without thalassemia trait or alpha globin chain defect (control). And peripheral blood lymphocyte (SU-PBMC, control-PBMC) were isolated and cultured in DMEM (Gibco, USA, CAT#11995) supplemented with 10% FBS (Gibco, 26140079) and 1% GlutaMAX (Gibco, 35050061). The iPS-HbH-CS cell line (SU-iPS-1) and control cell line (control-iPS) were generated with Sendai virus using a CytoTune-iPS 2.0 Sendai Reprogramming Kit (Invitrogen, A16518), which included the four Yamanaka factors, including Oct4, Sox2, Klf4, and c-Myc. Cells were subsequently counted and infected with Sendai virus following the manufacturer’s instructions.
Gene editing
Construction of primers and repair of the mutated template
According to the HBA2 gene sequences in GenBank (NC_000016.10 (172847...173710), we designed a gene-targeting gRNA on the CRISPR design website (http://crispr.mit.edu/). The homologous oligo sequence was 5′- CTGGACAAGTTCCTGGCTTCTGTGAGCACCGTGCTGACCTCCAAATACCGTTAAGCTTGAGCCTCGGTAGCCGTTCCTCCTGCCCGCTGGGCCTCCCAAC-3′. The HBA2 gene mutation primers and the upstream and downstream loci after repair of the region are presented in Fig. 2a. The primers and homologous oligos were synthesized by Jie Rui Biological Technology Co., Ltd. (China).
Plasmid structure of the targeted HBA2 gene gRNA
The sequence of procedures was followed as previously reported [10]. Briefly, the spCas9-HF1 expression vector was constructed using the wild-type spCas9 expression vector pX458, and the coding sequence of spCas9-HF1 (N497A, 374 R661A, Q695A, and Q926A) was generated via standard PCR mutagenesis of wild-type spCas9 as previously reported. Next, spCas9 was replaced by spCas9-HF1 in the pX458 vector. gRNA two-direction primers were synthesized to contain sticky ends for annealing, form a double-stranded molecule, and contain a BsmbI linearized carrier of the skeleton (SpCas9-HF1). The primers were incubated at 16 °C for annealing and transformed into DH5-alpha competent cells, and monoclonal bacteria in LB medium containing 100 mu (g/mL) benzylamine with temperature resistance at 37 °C were picked and centrifuged at 200 r/min for 16 h. The plasmid was extracted and, finally, the positive cloning sequence was identified.
Gene editing based on the electrorevolution technique
The antiapoptotic factor Y-27632 (Sigma) was added at a 10 μm concentration to cells with a growth density of 60 to 70%. Two hours after the addition of EDTA or pancreatic enzyme for cell digestion, cells were washed in PBS at 37 °C for 4 min and collected by centrifugation at 200×g for 5 min, and the washing was repeated. The mixed plasmid, template oligo, and transfer solution were fully resuspended for cell precipitation and transferred to a rotor cup after the single cells were in a uniform state, and an electric shock was subsequently administered to the LONZA point rotor. After the electric shock was completed, the cells were distributed evenly onto a petri dish coated with Matrigel, antiapoptotic factors were added, and the cells were returned to the incubator. Daily routine exchange of fluid was performed for obvious clone formation and for positive clone screening.
DNA extraction and HBA2 gene sequencing
Portions of each clone were chosen to isolate DNA for PCR screening. Colonies were lysed in 20 μL of lysis buffer as previously described. A portion of each sample was used for PCR (20 μL total volume) to detect modifications. The modified sequences were verified by Sanger sequencing. After the collection of cell deposits, the cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and precipitated. The DNA was extracted using a commercial kit (Tiangen Cat. No. DP304-03) after pretreatment of the collected samples at 37 °C for 30 min. A Nanodrop 2000 was used to test the purity and concentration of the DNA, and samples were stored at − 20 °C. PCR amplification was performed using human HBA2-specific primers (Table 1), and several subclones were selected for sequencing and identification.
Validation of pluripotent-repaired HbH-CS iPS cell lines
Immunofluorescence (IF) staining was performed using primary antibodies (all at 1:200 dilution) to detect OCT4, SOX2, and SSEA-4. Next, nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) at a final concentration of 0.01 mg/mL for 10 min. Chromosome analysis was performed as previously reported [11]. At least 20 cells were analysed in each group of chromosomes at a resolution of 400 bands. Teratoma formation was examined according to a previously reported method. For DNA fingerprinting, genomic DNA was extracted from the corrected C-SU-iPS-1 cells and the patient’s SU-iPS-1 cells. The extracted DNA was amplified for 15 different genetic loci using a Promega PowerPlex 16 System Kit (Promega, USA). Capillary electrophoresis was performed using an ABI 3100 Genetic Analyser (Applied Biosystems, USA).
Whole-exome capture sequencing and gene-corrected off-target analysis
We aimed to perform whole-genome sequencing at × 100 coverage. Paired-end sequencing was performed using Illumina GAIIx/HiSeq 2000 instruments (Illumina, San Diego, CA, USA), and exon capture was performed using Agilent SureSelect Technology (Agilent, Santa Clara, CA, USA). For sequence alignment, variant calling, and annotation, these sequences were mapped to their locations with the human genome reference sequence (hg19; NCBI Build 37.1) using a Burrows-Wheeler Aligner (BWA) (v.0.5.9-r16). Local realignment of the potential insertion/deletion sites was carried out with a genome analysis tool (GATK). Single-nucleotide variations (SNVs) and indel variants were assessed against the reference dbSNP138. All variants were annotated with reference to the consensus coding sequences (CCDS) (NCBI release: 20090902) and RefSeq (UCSC distribution: 20101004). The novel variants were checked using the Integrative Genomics Viewer (IGV) [12, 13].
Extracorporeal induction of iPSC haematopoietic differentiation
OP9 cells were cultured in a 100-mm culture dish, and iPSCs were transferred to OP9 cells. The stem cells were washed twice with PBS, 2 ml of DMEM/F12 was added, and a feeder layer cell coculture of iPSCs in 1 ml was also used. One millilitre of Essential 8 Medium-cultivated iPSCs were scraped off the plate and transferred to a 15 ml centrifuge tube for gravity sedimentation for 3 min. The supernatant was discarded, and 1 ml of fresh mTeSR medium was added, evenly and gently covering the OP9 cells in the dish. Cells were cultured at 37 °C and 5% CO2 for 1 day, and dead cells (cells that did not stick to the wall) were removed on the second day. The following day, half of the liquid medium was changed to improve differentiation efficiency. The cells were collected on the 14th day for follow-up experiments.
Flow cytometric analysis of haematopoietic differentiation
After 14 days in culture, cells were washed twice with PBS, digested with 1 mg/ml of collagenase IV for 20 min followed by 0.25% trypsin for 15–20 min, after which the digest was resuspended. Blast cells were a single-cell suspension. Cells were collected to a 15 ml volume by centrifugation at 1000 rpm for 5 min, reduced to 1 ml after discarding the supernatant containing 2% serum, and again passed through a 100-μm sterile cell screen. A CD34-PE-CY7 antibody was added, and the cells were cleaned by streaming buffer at 4 °C for 30 min. Finally, the cells were brought to a density of 5 × 106 suspension cells/ml, injected into an Aria III flow cytometer, and analysed with FACS software to determine differentiation efficiency.
Colony-forming assay
A clone-forming experiment was used to evaluate the in vitro differentiation ability of haematopoietic cells (HSCs). The CD34+ HSCs were grown in heavy suspension with 2 ml of methyl fibrin to 5–10 × 104 cells with inoculation in low-adsorption 6-well panels, and the cells were cultured at 37 °C and 5% CO2 for 21 days. One millilitre of fresh methylcellulose-based feed medium was added every 3 days. Methylcellulose-based feed medium containing Iscove’s Modified Dulbecco’s Medium (IMDM), 30% H4434, 0.75 mg/mL bovine serum albumin, 20% FBS, 2 mM GlutaMAX, 0.5 mM β-ME, insulin-transferrin-sodium (ITS), 50 ng/ml SCF, 10 ng/ml IL-3, 10 ng/ml IL-6, 10 ng/mL granulocyte-macrophage colony-stimulating factor (GMCSF), 6 U/ml EPO, and 1% penicillin/streptomycin was added every 3 days. HSCs can be differentiated into erythroid (E), macrophage, granulocyte (G), and granulocyte macrophage (granulocyte/macrophage, GM) lineages that exhibit granulocyte/red blood cell and macrophage/megakaryocyte pluripotency (granulocyte/erythrocyte/macrophage/megakaryocyte, GEMM), i.e., into the direction of the five different terminal differentiations. Fourteen days after cultivation, typical clones had formed. Clones were imaged and scored according to their morphological criteria. Representative images were recorded using a microscope (Zeiss, Axio Vert, Germany) .
Real-time quantitative PCR was used to detect haematopoietic correlation genes
After collecting the cells of normal-person-iPS (control), SU-iPS-1, and C-SU-iPS-1, RNA was extracted using the TRIZOL method. After collecting normal-person-iPS (control), SU-iPS-1, and C-SU-iPS-1 cells, RNA was extracted using the TRIZOL method. Normal-person iPS cells were induced from a normal male adult without thalassemia or alpha globin chain defects. The RNA concentration was measured, and 1 μg of cDNA was synthesized and diluted 10-fold. Quantitative fluorescence PCR was performed using a SYBR Green qPCR SuperMix-UDG with ROX Kit according to the manufacturer’s instructions. The expression levels of LD2, CD34, CD71, HBB, EPOR, HBA, and HBG were detected with beta-actin as an internal reference. The reaction mixture proceeded as follows: with the diluted cDNA, 2 mu/l sample was adsorbed, 10 mu of the upstream and downstream primers at 10 mu (mol/l) were added, and later, 10 mu/l SYBR Green Mix and water were added to 20 mu/l. The thermal cycling conditions included an initial denaturation at 95 °C for 10 min, 40 cycles of 15 s at 95 °C and 60 °C modified annealing for 1 min, and fluorescence signal collection in each cycle’s annealing stage. Data analysis was conducted using ABI StepOne™ v2.2.2. The sequence information of the primers is shown in Table 2.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 software to detect significant differences in measured variables among groups. A value of P < 0.05 was considered to indicate a statistically significant difference.
Results
Validation of iPSCs
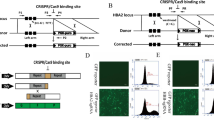
Karyotyping of the patient’s PBMCs (SU-PBMCs) and iPSCs (SU-iPSc-1 and SU-iPSc-2) was performed with no abnormality (46, XY). The results of pluripotent validation included immunocytochemistry for the detection of TRA-1-81, SSEA4, SOX2, and OCT4 (Fig. 1a) and quantitative PCR for the expression of OCT4, SOX2, and NANOG (Fig. 1d). The SU-iPS-1 cells spontaneously differentiated into cells that highly expressed AFP, SMA, and NESTIN (Fig. 1b). Teratoma formation (Fig. 1c) was also observed. The cell lines (46, XY) were maintained in our laboratory and cultured in irradiated mTeSR (STEM CELL, Inc., Rockville, Maryland).
Validation of iPSCs (SU-iPS-1). a Immunocytochemistry of SU-iPS-1 cells for the detection of TRA-1-81, SSEA4, SOX2, and OCT4. b The iPSCs spontaneously differentiated into cells that highly expressed AFP, SMA, and NESTIN. c Teratoma formation. d The results of pluripotent validation included quantitative PCR analysis of the expression of OCT4, SOX2, and NANOG
Specific RNA-guided Cas9 nuclease-mediated HBA2 gene correction
We sought to correct these disease-causing mutations in α-Thal iPSCs (SU-iPS-1) through in situ gene targeting using the CRISPR/Cas9 system (Fig. 2a). We obtained the corrected clone (C-SU-iPS-1) using confirmation by direct sequencing analysis of the PCR fragment clones, which were randomly selected from 126 clones. The sequencing results showed that 11 clones were correctly repaired. As a result, the correction efficiency was 8.7%. Representative sequencing results are shown in Fig. 2b.
Pluripotency of gene-corrected α-Thal iPSCs (C-SU-iPS-1)
To characterize the pluripotency of the gene-corrected α-Thal iPSC clones, immunostaining was performed to detect pluripotent gene expression. The results showed that typical pluripotent markers, such as OCT4, SSEA-4, and TRA-1-81, were expressed in the C-SU-iPS-1 clones, indicating that they maintained pluripotency (Fig. 3a). The SCID mouse results of the initiation of teratoma formation revealed all three germ layers (Fig. 3b), and whole-exome capture sequencing and bioinformatics analysis demonstrated that these cells maintained their normal genome well after gene-targeting correction. Furthermore, STR results suggested that the corrected C-SU-iPS-1 and patient iPS (SU-iPS-1) cells shared the same origin (SU-PBMCs) (Fig. 3c).
Pluripotency of gene-corrected α-Thal iPSCs (C-SU-iPS-1). a Immunostaining for the pluripotent markers SSEA-4, TRA-1-81, SOX2, and OCT4. b The SCID mouse results of the initiation of teratoma formation for all three germ layers. c STR analysis showed the same origin of C-SU-iPS-1 and SU-iPS cells with PBMCs
Characterization of the gene-corrected α-Thal iPSCs and the detection of off-target results
The gene-corrected clones maintained a normal karyotype (46, XY) and pluripotent genes, such as SOX2 and NANOG (Fig. 4a). To investigate the effects of gene correction on exome integrity in iPSCs, exome sequencing was performed on the SU-iPS-1 and C-SU-iPS-1 cells. The data showed no obvious mutations in these sites (Fig. 4b, c).
No obvious genome change was detected in SU-iPS-1 and C-SU-iPS-1 cells. a The gene-corrected clones maintained pluripotent genes, such as NANOG, OCT4, and SOX2. b Copy number variations (CNVs), as well as single-nucleotide variations (SNVs) (c) and indels (d), were detected in the SU-iPS-1 and C-SU-iPS-1 cells (e) with exome sequencing, which showed no obvious mutations in these sites
To address the off-target effects of CRISPR/Cas9 by using gene-corrected iPSCs, we used the latest high-fidelity SpCas9-HF1 to target patient-specific iPSCs. Genomic DNA from C-SU-iPS-1 cell lines, the correctly targeted single colony, was examined by whole-exon sequencing. Using the human (hg19) genome as a reference sequence, we identified 24,149 indels (< 50 bp insertions/deletions) in the uncorrected iPSCs and 24,305 in the corrected iPSCs (Table S2) and 7 CNVs in the uncorrected iPSCs and 2 CNVs in the corrected iPSCs (Table S3).
To determine whether these filtered indels were de novo mutations caused by our Cas9/gRNA editing system, we extracted ± 30 bp sequences flanking each filtered indel and used Blastn (e-value cutoff, 1000) to compare them to the potential gRNA off-target host genome sites predicted by sequence similarity at 0–7 mismatches and to Hb-CS on-target sequences. Without any mismatches to the gRNA targets, we found no off-target site around the extracted 60, 600, and 1200 bp sequences of the filtered indels. Within the extracted 50-bp sequences, we found no off-target site, even with 7 mismatches at alignment lengths > 12 nucleotides from PAM NRG (which must be 100% matched).
Together, these data strongly suggest that none of the indels detected in the cells with the excised uncorrected iPSC genome lie within 50 bp of the targets of any potential off-target site, as predicted by search criteria allowing up to 7 mismatches. By expanding the searching sequences up to 600 or 1200 bp, relatively rare off-target sites were identified, including various numbers of mismatches and aligned lengths. With a perfect match to the last 12 bp seed sequence plus PAM NGG, none of the indels fell within the search area of 60–1200 DNA sequences. Our overall interpretation of these data supports the preceding surveyor assay results in these cells as well as in other cell types and establishes, by a very stringent analysis, that no off-target effects on the host genome are elicited by our CRISPR/Cas9 system, although off-target mutagenesis in the noncoding regions of the chromosome may be undetected. The absence of recurring mutations and the fact that none of the mutations reside in any putative off-target sites according to the bioinformatics predictions strongly suggest that these mutations randomly accumulated during regular cell expansion and are not direct results of off-target activities by Cas9.
Differentiation of C-iPSCs into HSCs and erythrocytes
FACS analysis showed that 10.79% of the CRISPR/Cas9-corrected C-SU-iPS-1 cells differentiated into CD34+ haematopoietic progenitor cells (HPCs) compared with 4.39% of the parental cell line (SU-iPS-1) and 12.75% of the wild-type iPSCs (Fig. 5a). To determine whether the differences in differentiation efficiency among the cells were caused by their ability to maintain their pluripotency, we measured the expression levels of pluripotency-related genes in cells that had differentiated for 21 days. However, compared with the HbH-CS iPS cell lines, the gene-repaired HbH-CS cell lines did not show any significant differences except for the number of erythroid (E) cells based on the CFU assay (Fig. 5b, c), while the expression levels of LD2, CD34, CD71, HBB, EPOR, HBA, and HBG showed significant differences based on real-time quantitative PCR (Fig. 5d).
Differentiation of C-iPSCs into HSCs and erythrocytes. a Flow cytometry analysis of CRISPR/Cas9-corrected C-SU-iPS-1 cells, the parental cell line (SU-iPS-1) and wild-type iPSCs using the surface marker CD34 after differentiation for 14 days. b, c Compared with the Hb-CS iPS cell lines, the gene-repaired Hb-CS cell lines did not show any significant difference in haematopoietic differentiation efficiency based on the colony-forming assay but showed differences using real-time quantitative PCR (d)
Discussion
There is a high prevalence of thalassemia in South China and Mediterranean countries. HbH-CS disease has a severe phenotype (e.g., thalassemia intermedia and splenomegaly) compared with deletional Hb H disease (–SEA/-α3.7, –SEA/-α4.2) [14, 15]. Hb-CS has an α-chain with 172 amino acids instead of the normal length of 141 amino acids [1]. The elongated α-chain results from a TAA→CAA single base substitution in the termination codon of the α-globin-2 gene, which results in its translation as an amino acid and allows read-through of the normally untranslated 3′ flanking region of the α-globin mRNA until the next in-phase termination codon is reached [14]. However, a Hb-CS carrier does not have severe clinical symptoms. Therefore, we decided on the strategy of gene repair on the Hb-CS mutation locus of the α2-globin gene. We expected that after correction of the Hb-CS mutation locus, the corrected α2-globin gene combined with the remaining two deletions of the a-globin gene (--/aa) could alleviate the symptoms of thalassaemia compared with HbH-CS disease, which does not require blood transfusions.
Technology utilizing human induced pluripotent stem cells (iPSCs) has considerable potential to provide improved cellular models of human disease [16,17,18]. The CRISPR/Cas9 complex consists of the Cas9 endonuclease and a 100-nucleotide single-guide RNA (sgRNA) [17]. This experiment combined the CRISPR/Cas9 technology of the successful repair of iPSC lines with gRNA highly active filtering techniques and the design of improved oligos to increase the efficiency of editing [19]. Our results indicate that the CRISPR/Cas9 gene editing assay is precise and efficient and does not affect the cells’ genetic stability when used for HbH-CS-iPS cell line correction. They also demonstrated that CRISPR/Cas9 gene editing could restore the capacity for iPS cell pluripotency and differentiation. Thus, this nucleotide editing technique provides promising potential material to treat Hb-CS thalassemia.
Recently, engineered nucleases, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat CRISPR/Cas9, have been widely used to generate double-strand DNA breaks (DSBs) to increase the efficiency of standard homologous recombination [20,21,22]. ZFNs have been used to target the HBB gene in α-thalassemia [23]. However, traditional approaches for correcting mutations have a low efficiency and leave a residual footprint [24, 25], which leads to a number of safety concerns in clinical applications. Therefore, we utilized single-strand oligodeoxynucleotides (ssODNs), which have been reported to show efficacy as templates, to correct the HBA mutation in α2-Thal patient-specific iPSCs and the high-fidelity CRISPR/Cas9 nuclease to achieve a seamless correction of the TAA→CAA mutation in Hb-CS thalassemia patient-specific iPSCs with remarkable efficiency, which is different from previous studies [14, 26]. Additionally, off-target analysis and whole-exome sequencing results revealed that corrected cells exhibited a minimal mutational load and no off-target mutagenesis.
One important consideration is how many of these detected SNVs were the result of off-target mutagenesis by the CRISPR/Cas9 endonuclease. All the sequences of the SNVs and indel sites were compared with the gRNA target sequence, and none were within a potential off-target region, which is consistent with previous analyses studying predicted off-target sites. Our data support the idea that CRISPR/Cas9 technology can successfully make the gene correction at the Hb CS mutation locus of the α2-globin gene. However, the haematopoietic differentiation efficiency of the corrected HbH-CS cell lines showed inconsistent results based on the CFU assay and real-time quantitative PCR, and according to Fig. 5d, the expression of important target genes (HBB, EPOR, and HBA), particularly that of HBA, is dramatically lower in C-iPSC-derived cells than in noncorrected cells, which suggests that haematopoietic differentiation efficiency of the corrected HbH-CS cells was affected by other genetic and environmental factors. Furthermore, there is no clear genotype-phenotype correlation associated with the iPSC phenotype, since iPS cells with identical genotypes do not necessarily show the same severity. The variable genetic and phenotypic characterization of many existing iPS cell lines limits their potential use for research and therapy [19]. Kilpinen et al. described the systematic generation, genotyping, and phenotyping of 711 iPSC lines derived from 301 healthy individuals by the human induced pluripotent stem cells initiative, outlined the major sources of genetic and phenotypic variation in iPS cells, and established their suitability as models of complex human traits and cancer; these researchers found that 5–46% of the variation in different iPS cell phenotypes, including differentiation capacity and cellular morphology, arises from differences between individuals [27]. Furthermore, there is no standard iPSC haematopoietic differentiation and culture system for HbH-CS cells. A recent study showed that the heterogeneity of hPSCs exists in different protocols in different cell stocks of different laboratories [28]. As a result, further study on improving the differentiation efficiency of corrected HbH-CS cells may be needed.
In conclusion, we successfully corrected the HBA2 gene of the Hb-CS mutation in α-Thal iPSCs by the combination of single-strand oligodeoxynucleotides (ssODNs) and CRISPR/Cas9 gRNA. The iPSCs retained both pluripotency and multidifferentiation abilities after correction. This strategy provides promising potential material for the treatment of Hb-CS thalassemia disease. Moreover, our results also indicated that the haematopoietic differentiation efficiency of the corrected Hb-CS cell lines might be affected by other genetic and environmental factors, which suggests that the utility of iPS cells in the clinical translation of gene therapy for thalassemia needs further study.
Animals and ethics statement
The experiments regarding animal research were approved by the Institutional Review Board at the Third Affiliated Hospital of Guangzhou Medical University. The experiments using human cells and mice were approved by the ethics committee of the Third Affiliated Hospital of Guangzhou Medical University.
References
Harteveld CL, Higgs DR (2010) Alpha-thalassaemia. Orphanet J Rare Dis 5:13
Komvilaisak P, Jetsrisuparb A, Fucharoen G, Komwilaisak R, Jirapradittha J, Kiatchoosakun P (2018) Clinical course of homozygous hemoglobin constant spring in pediatric patients. J Pediatr Hematol Oncol 40(5):409–412
Chen FE, Ooi C, Ha SY, Cheung BM, Todd D, Liang R et al (2000) Genetic and clinical features of hemoglobin H disease in Chinese patients. N Engl J Med 343(8):544–550
Thompson AA, Walters MC, Kwiatkowski J, Rasko JEJ, Ribeil JA, Hongeng S, Magrin E, Schiller GJ, Payen E, Semeraro M, Moshous D, Lefrere F, Puy H, Bourget P, Magnani A, Caccavelli L, Diana JS, Suarez F, Monpoux F, Brousse V, Poirot C, Brouzes C, Meritet JF, Pondarré C, Beuzard Y, Chrétien S, Lefebvre T, Teachey DT, Anurathapan U, Ho PJ, von Kalle C, Kletzel M, Vichinsky E, Soni S, Veres G, Negre O, Ross RW, Davidson D, Petrusich A, Sandler L, Asmal M, Hermine O, de Montalembert M, Hacein-Bey-Abina S, Blanche S, Leboulch P, Cavazzana M (2018) Gene therapy in patients with transfusion-dependent beta-thalassemia. N Engl J Med 378(16):1479–1493
Yannaki E, Papayannopoulou T, Jonlin E, Zervou F, Karponi G, Xagorari A, Becker P, Psatha N, Batsis I, Kaloyannidis P, Tahynopoulou V, Constantinou V, Bouinta A, Kotta K, Athanassiadou A, Anagnostopoulos A, Fassas A, Stamatoyannopoulos G (2012) Hematopoietic stem cell mobilization for gene therapy of adult patients with severe beta-thalassemia: results of clinical trials using G-CSF or plerixafor in splenectomized and nonsplenectomized subjects. Mol Ther 20(1):230–238
Sadelain M, Boulad F, Galanello R, Giardina P, Locatelli F, Maggio A, Rivella S, Riviere I, Tisdale J (2007) Therapeutic options for patients with severe beta-thalassemia: the need for globin gene therapy. Hum Gene Ther 18(1):1–9
Liu J, Higgins CA, Whitehouse JC, Harris SJ, Crawford H, Christiano AM et al (2018) Hair follicle dermal cells support expansion of murine and human embryonic and induced pluripotent stem cells and promote haematopoiesis in mouse cultures. Stem Cells Int 2018:8631432
Razaq MA, Taylor S, Roberts DJ, Carpenter L (2017) A molecular roadmap of definitive erythropoiesis from human induced pluripotent stem cells. Br J Haematol 176(6):971–983
Unternaehrer JJ, Daley GQ (2011) Induced pluripotent stem cells for modelling human diseases. Philos Trans R Soc Lond Ser B Biol Sci 366(1575):2274–2285
Liu YL, Yang Y, Kang XJ, Lin B, Yu Q, Song B, Gao G, Chen Y, Sun X, Li X, Bu L, Fan Y (2017) One-step biallelic and scarless correction of a beta-thalassemia mutation in patient-specific iPSCs without drug selection. Mol Ther-Nucl Acids 6:57–67
Lucarelli G, Clift RA, Galimberti M, Angelucci E, Giardini C, Baronciani D, Polchi P, Andreani M, Gaziev D, Erer B, Ciaroni A, D'Adamo F, Albertini F, Muretto P (1999) Bone marrow transplantation in adult thalassemic patients. Blood. 93(4):1164–1167
Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14(2):178–192
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al (2011) Integrative genomics viewer. Nat Biotechnol 29(1):24–26
Niu XH, He WY, Song B, Ou ZH, Fan D, Chen YC, Fan Y, Sun X (2016) Combining single strand oligodeoxynucleotides and CRISPR/Cas9 to correct gene mutations in beta-thalassemia-induced pluripotent stem cells. J Biol Chem 291(32):16576–16585
Kattamis AC, Camaschella C, Sivera P, Surrey S, Fortina P (1996) Human alpha-thalassemia syndromes: detection of molecular defects. Am J Hematol 53(2):81–91
Jang YY, Ye ZH (2016) Gene correction in patient-specific iPSCs for therapy development and disease modeling. Hum Genet 135(9):1041–1058
Smith C, Abalde-Atristain L, He C, Brodsky BR, Braunstein EM, Chaudhari P, Jang YY, Cheng L, Ye Z (2015) Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther 23(3):570–577
Raja JV, Rachchh MA, Gokani RH (2012) Recent advances in gene therapy for thalassemia. J Pharm Bioallied Sci 4(3):194–201
Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, Uchida N, Hendel A, Narla A, Majeti R, Weinberg KI, Porteus MH (2016) CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 539(7629):384–389
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science. 339(6121):819–823
Hockemeyer D, Wang HY, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R (2011) Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29(8):731–734
Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC et al (2009) Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27(9):851–U110
Chang CJ, Bouhassira EE (2012) Zinc-finger nuclease-mediated correction of alpha-thalassemia in iPS cells. Blood. 120(19):3906–3914
Ma N, Liao BJ, Zhang H, Wang LL, Shan YL, Xue YT, Huang K, Chen S, Zhou X, Chen Y, Pei D, Pan G (2013) Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. J Biol Chem 288(48):34671–34679
Ma N, Shan Y, Liao B, Kong G, Wang C, Huang K, Zhang H, Cai X, Chen S, Pei D, Chen N, Pan G (2015) Factor-induced reprogramming and zinc finger nuclease-aided gene targeting cause different genome instability in beta-thalassemia induced pluripotent stem cells (iPSCs). J Biol Chem 290(19):12079–12089
Xie F, Ye L, Chang JC, Beyer AI, Wang JM, Muench MO, Kan YW (2014) Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24(9):1526–1533
Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ (2017) Common genetic variation drives molecular heterogeneity in human iPSCs (vol 546, pg 370, 2017). Nature. 546(7660):686
Tsankov AM, Akopian V, Pop R, Chetty S, Gifford CA, Daheron L, Tsankova NM, Meissner A (2015) A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol 33(11):1182–1192
Acknowledgements
We would like to thank the clinical cytogenetics laboratory for aiding in the data collection. We are also grateful to the individual included in this study as well as his family. We would also like to express our gratitude for the helpful comments received from our reviews. We thank the Laboratory Animal Center of Guangzhou Medical University (Guangzhou, China) for providing us with animal breeding sites.
Funding
This study was supported by the National Natural Science Foundation of China (grant no.: 81571448), the Science and Information Technology of Guangzhou Key Project (grant no.: 201508020258), the Guangdong Province Science and Technology Project (grant no.: 2016B030229008), and the Youth Project by Education of Guangdong Province (2015KQNCX132/B16036095).
Author information
Authors and Affiliations
Contributions
Yi X and Xf S carried out the study design; Yu X, Yu C, Do L, Bi S, Yi Y, Di L, Ya X, and Ze X performed the experiments; Ne L and Di C performed the statistical analysis; Yi X and Xf S wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The experimental procedures and animal conditions were approved by the Animal Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yingjun, X., Yuhuan, X., Yuchang, C. et al. CRISPR/Cas9 gene correction of HbH-CS thalassemia-induced pluripotent stem cells. Ann Hematol 98, 2661–2671 (2019). https://doi.org/10.1007/s00277-019-03763-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-019-03763-2