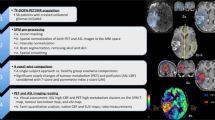
Abstract
Purpose
Oligodendrogliomas are heterogeneous tumors in terms of imaging appearance, and a deeper understanding of the histopathological tumor characteristics in correlation to imaging parameters is needed. We used PET-to-MRI-to-histology co-registration with the aim of studying intra-tumoral 11C-methionine (MET) uptake in relation to tumor perfusion and the protein expression of histological cell markers in corresponding areas.
Methods
Consecutive histological sections of four tumors covering the entire en bloc-removed tumor were immunostained with antibodies against IDH1-mutated protein (tumor cells), Ki67 (proliferating cells), and CD34 (blood vessels). Software was developed for anatomical landmarks-based co-registration of subsequent histological images, which were overlaid on corresponding MET PET scans and MRI perfusion maps. Regions of interest (ROIs) on PET were selected throughout the entire tumor volume, covering hot spot areas, areas adjacent to hot spots, and tumor borders with infiltrating zone. Tumor-to-normal tissue (T/N) ratios of MET uptake and mean relative cerebral blood volume (rCBV) were measured in the ROIs and protein expression of histological cell markers was quantified in corresponding regions. Statistical correlations were calculated between MET uptake, rCBV, and quantified protein expression.
Results
A total of 84 ROIs were selected in four oligodendrogliomas. A significant correlation (p < 0.05) between MET uptake and tumor cell density was demonstrated in all tumors separately. In two tumors, MET correlated with the density of proliferating cells and vessel cell density. There were no significant correlations between MET uptake and rCBV, and between rCBV and histological cell markers.
Conclusions
The MET uptake in hot spots, outside hotspots, and in infiltrating tumor edges unanimously reflects tumor cell density. The correlation between MET uptake and vessel density and density of proliferating cells is less stringent in infiltrating tumor edges and is probably more susceptible to artifacts caused by larger blood vessels surrounding the tumor. Although based on a limited number of samples, this study provides histological proof for MET as an indicator of tumor cell density and for the lack of statistically significant correlations between rCBV and histological cell markers in oligodendrogliomas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oligodendrogliomas are glial tumors believed to originate from oligodendrocytes, and are further classified based on molecular tumor features [1]. Classical oligodendrogliomas are characterized by isocitrate dehydrogenase (IDH) gene mutations and codeletions on chromosome 1p/19q. In the absence of diagnostic molecular testing, oligodendrogliomas are classified as oligodendrogliomas NOS (not-otherwise-specified), while IDH mutated non-codeleted gliomas are classified as astrocytomas [2]. Oligodendrogliomas are most common around the age of 35–44 [3]. Current median survival time for patients with oligodendrogliomas WHO grade III is 12–14 years, and longer for patients with oligodendroglioma WHO grade II [2]. Primary treatment is maximal safe resection, followed by radiotherapy and chemotherapy for high-risk patients [4, 5]. Based on their chemosensitivity, it has recently been suggested that chemotherapy may be the optimal primary treatment for oligodendrogliomas [6].
The histopathological diagnosis of oligodendrogliomas is based on the presence of neoplastic cells with morphological characteristics resembling oligodendrocytes. Oligodendroglial tumors are highly cellular lesions consisting of relatively small but densely packed tumor cells [2, 7, 8]. The tumor periphery consists of less cellular areas but with diffuse infiltrative growth and clustering of tumor cells around the perikarya of preexistent neurons, under the pial surface, and surrounding small cortical vessels [2]. Oligodendrogliomas are further characterized by branching networks of capillaries (chicken-wire pattern) that set them apart from astrocytomas.
Contrast-enhanced magnetic resonance imaging (MRI) is widespread available and routinely used for radiological tumor diagnosis, showing superior soft-tissue resolution. Key features in oligodendrogliomas are the presence of calcifications and cortical thickening with typical involvement of both the cortex and the subcortical white matter [9]. Oligodendroglial tumors show and a more solid, infiltrative, or mixed growth pattern [10]. In general, the differentiation of heterogeneous glioma tissue from surrounding edema on MRI is unreliable [11], and MRI tends to underestimate the actual tumor extent of oligodendrogliomas [10].
Advanced MRI methods such as perfusion-weighted imaging (PWI) and PET with amino acid tracers provide complementary diagnostic information in gliomas [12, 13]. In case histopathological diagnosis is based on partial tumor resection or biopsy with subsequent risk for misclassification and undergrading [14], amino acid PET provides important additional diagnostic accuracy since hot spot areas generally represent the most malignant tumor part [15, 16]. Studies using serial stereotactic biopsies have confirmed the superiority of PET with 11C-methionine (MET) over CT or MRI to delineate the tumor [17, 18]. During recent years, 18F-fluoroethyl-L-tyrosine (FET) has received increased attention and MET has been replaced by FET for the clinical management of gliomas in many centers due to the logistic advantages of FET [12, 19]. FET PET-MRI-guided diagnostic biopsy showed a specificity of 53% for MRI alone versus 94% for combined PET-MRI [20].
The uptake of amino acid tracers is known to reflect the transport mediated by amino acid carriers over the endothelial cell membrane, and to correlate with the microvessel density of the tumor [21]. However, few studies have directly compared the amino acid uptake with histological tumor features. All studies so far have been based on multiple biopsies or partial tumor resections and have focused on single hot spot regions in the tumor [20, 21]. As for today, a deeper understanding of how intra-tumoral variations in tracer uptake and tumor perfusion correlate with specific histological features of the tumor is lacking. It remains unclear whether the tracer uptake measured by PET, or tumor perfusion measured by PWI, outside hot spot areas and in the tumor periphery is a measure for cellular proliferation, tumor cell density, or vessel density. This information is valuable in a number of clinical situations where PET and advanced MRI are part of the diagnostic work-up for patients with gliomas, such as radiotherapy planning, evaluation of response to therapy, and detection of recurrence [12, 21]. We have recently described a new method for co-registration of MR images and histological images in en bloc surgically removed gliomas, allowing region-by-region comparisons of the entire tumor with infiltrating tumor edge [22]. Here, we further developed our technique to perform region-by-region correlations between PET, MRI perfusion and histological images. Our aim was to provide an exact correlation between changes in MET uptake, tumor perfusion, and histological tumor characteristics of oligodendrogliomas, covering the entire tumor volume, including the infiltrating tumor edge.
Materials and methods
Tumor samples
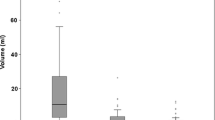
In our recently published co-registration study, we included five patients with suspected diffuse low-grade glioma (DLGG) that were suitable for en bloc tumor resection [22]. Four of these five patients had histopathological diagnosis of oligodendrogliomas (Figs. 1, 2, 3, and 4) and were included in the present study. The institutional review board approved the protocol for the present analysis, and written informed consent was obtained prior to patient participation. A summary of the clinical characteristics of these patients and histopathological diagnoses is shown in Table 1.
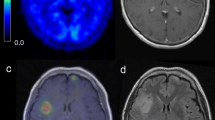
Patient 1. a T2-weighted FLAIR MRI shows a hyperintense slightly heterogeneous tumor in the right frontal lobe. b MET PET shows the hotspot region of the tumor. c DSC perfusion MRI with rCBV greyscale map shows predominantly low or normal perfusion in the region corresponding to the PET hotspot. d Co-registration of MRI, PET and corresponding histological images with defined ROIs (ROI1, red and orange; ROI2 yellow, ROI3 green)
Patient 2. a T2-weighted FLAIR MRI shows a hyperintense tumor with small cystic regions in the left frontal lobe. b MET PET shows the hotspot region of the tumor. c DSC perfusion MRI with rCBV greyscale map shows higher perfusion in the region corresponding to the PET hotspot. d Co-registration of MRI, PET, and corresponding histological images with defined ROIs (ROI1, red and orange; ROI2 yellow, ROI3 green)
Patient 3. a T2-weighted FLAIR MRI shows a hyperintense tumor in the left frontal lobe. b MET PET shows the hotspot regions of the tumor. c DSC perfusion MRI with rCBV greyscale map shows low or normal perfusion in the region corresponding to the PET hotspot. d Co-registration of MRI, PET, and corresponding histological images with defined ROIs (ROI1, red and orange; ROI2 yellow). There was no identifiable ROI3 in this sequence
Patient 4. a T2-weighted FLAIR MRI shows a left frontal hyperintense tumor with minimal contrast enhancement on T1-weighted images (not shown). b MET PET shows a large hotspot region in the tumor. c DSC perfusion MRI with rCBV greyscale map shows areas with low, normal, and high perfusion in the region corresponding to the PET hotspot. d Co-registration of MRI, PET, and corresponding histological images with defined ROIs (ROI1, red and orange; ROI2 yellow). There was no identifiable ROI3 in this sequence. As shown, ROI1 covers almost the entire tumor volume
PET and MRI acquisition
MRI was performed according to our original study protocol [23], using a 3-Tesla scanner (Achieva, Philips Healthcare, Best, The Netherlands), including perfusion sequences. Morphological imaging included axial and coronal T2-weighted fluid attenuated inversion recovery (FLAIR) scans (0.45 × 0.45 × 6 mm3 voxel size). Dynamic susceptibility contrast (DSC) perfusion imaging was performed as previously described [24], with a T2*W single-shot gradient echo-EPI sequence (1.7 × 2.3 × 5.0 mm3). Whole-brain coverage was obtained using these parameters.
PET scanning was performed on a Discovery ST PET/CT (General Electric Medical Systems) in a 3D acquisition mode. This scanner has 24 detector rings with 420 detectors resulting in 47 image planes with a plane separation of 3.27 mm and a total axial field of view of 157 mm. Images were reconstructed using ordered-subsets expectation maximation (OSEM; 2 iterations, 15 subsets, 128 × 128 matrix) with a 2.14 mm FWHM post-filter applying appropriate corrections such as scatter, randoms, and attenuation correction based on a low-dose CT scan. Voxel size of the resulting images was 1.95 × 1.95 × 3.27 mm and the spatial resolution 6 mm. The tracer 11C-Methionine (4.0 MBq/kg) was injected 25 min before start. Scan time was 10 min, 1 frame.
PET and MRI were performed at a mean of 34 days (range, 20-52 days) respectively 28 days (range, 5-53 days) before surgery.
Tissue preparation and immunohistochemistry
Figure 5 illustrates the design of the study with the different steps that have been performed. Preparation of the en bloc tumor tissues was performed as previously described [22]. After resection, the tumor surfaces (anterior, posterior, medial, and lateral) were color-marked for spatial orientation. The en bloc tissue sample was fixed in formalin, subsequently cut into consecutive tissue slices, again fixed in formalin, and embedded in paraffin wax. Serial microsections were rehydrated prior to immunohistochemical staining with antibodies against the tumor cell marker IDH1-mutated protein (IDH1-R132H), the proliferation marker Ki67 and the hematopoietic progenitor cell antigen CD34 (Fig. 5, steps 1–2).
Illustration of the study design. Step 1: En bloc tumor resection of 8–12 tissue slices of 6–8 mm thickness. Steps 2–3: Development of software for anatomical landmark-based co-registration of consecutive microsections (4 μm) for immunohistochemical staining of for tumor cells (IDH1), proliferating cells (Ki67), and blood vessels (CD34). Step 4: Quantification of protein expression in each histological sub-image (500 × 500 pixels). The original image of CD34 protein expression (left) was overlaid with the identified objects (red) in CellProfiler. Steps 5–7: Manual co-registration of MRI, PET, perfusion maps and corresponding histological images. Selection of ROIs on PET covering hot spot areas (ROI1), areas outside hot spots (ROI2) and tumor periphery (ROI3). Analysis of correlations between MET uptake and quantified protein expression, MET uptake and rCBV, and rCBV and quantified protein expression. Note: ROIs in the figures indicate their specific location but not their exact volume
Co-registration of consecutive histological images
Software was developed for anatomical landmark-based co-registration of consecutive histological images of various immunostainings. In summary, a landmark-based multi-step elastic image registration was designed using an in-house Matlab application (The MathWorks, Inc., Natick, MA, USA) and implemented allowing a region-by-region comparison of histological assessment [25]. A set of 5–10 anatomical landmarks was found accurate enough for consecutive co-registration of serial histological images (Fig. 5, step 3). Each histological image was divided into sub-images (500 × 500 pixels) for region-by-region quantification of protein expression of histological cell markers, using an additional in-house Matlab application (see Supplementary file for software description).
Quantification of immunohistochemical stainings
Automated region-by-region analysis of protein expression was performed using an in-house built pipeline in the CellProfiler software [26] (see Supplementary file). We created a pipeline of individual modules that were able to estimate localization, intensity, shape, and size of protein expression for the three histological cell markers (IDH1-mutated protein as a marker for tumor cells, Ki67 as a marker for proliferating cells and CD34 as a marker for blood vessels). Each separate sub-image (500 × 500 pixels) traveled through the pipeline and was processed by each module to quantify the absolute count per square of protein expression. In the final analysis, mean values of protein expression were calculated in each region of interest defined on MET PET (see under PET analysis). This allowed region-by-region comparison between MET uptake, tumor perfusion, and protein expression (Fig. 5, step 4).
Co-registration of histology, PET, and MRI
As a first step, we imported original co-registrations of MRI and histological images (denoted as co-registration 1) created in our previous study [22] into Carimas software (kindly provided by the Turku PET center, Finland). We then imported the T2-FLAIR images that were co-registered with the co-registration 1 image using the Normalized Mutual Information (NMI) tool by Carimas [27, 28]. Following this step, we imported both MET PET scans and MRI perfusion maps (cerebral blood volume, CBV) into the project and co-registered these to the FLAIR images using the same NMI tool (denoted as co-registration 2). This way, all co-registered images had similar orientation (Fig. 5, step 5).
PET analysis
Analysis of MET PET was performed using Carimas 2.9 software (Turku PET center, Finland). MET uptake in regions of interest (ROIs), defined as described below, was calculated as tumor-to-normal tissue (T/N) ratio, i.e., the ratio between the maximum standardized uptake value (SUV), reflecting the value of the pixel with highest radioactivity in the ROIs and the mean uptake in the contralateral normal brain. The normal tissue reference region was drawn as a 1-cm-thick cortical ROI at the level of the basal ganglia, ranging from the occipital to the frontal cortex in the contralateral hemisphere. Additionally, we defined a deep white matter reference region in the contralateral hemisphere above and lateral to the ventricle.
Serial ROIs were systematically selected on all MET PET scans. We defined three different types of ROIs based on MET uptake in the entire tumor volume: ROI1 = areas with highest MET uptake (hotspot), ROI2 = areas with medium MET uptake, and ROI3 = areas with lower MET uptake located in the infiltrating tumor border (Fig. 5, step 6).
MRI perfusion analysis
DSC perfusion was analyzed as previously described [24], with calculation of CBV maps using nordicICE software (NordicNeuroLab AS, Bergen, Norway). Relative CBV (rCBV) was calculated as tumor-to-normal tissue (T/N) ratio by dividing the mean CBV in ROIs defined on PET, with the mean CBV within a reference area located in normal appearing white matter of the contralateral hemisphere above and lateral to the ventricle.
Pre-processing and area definition
To compare quantified protein expression of histological cell markers within the ROIs defined on PET, we applied a grid using Adobe Photoshop CC (Adobe Systems Software). The grid was placed on top of co-registration 2, enabling a detailed and direct comparison between radiological and histological images. All histological sub-images (500 × 500 pixels) within one specific ROI were selected using the grid. This procedure was applied for each histological image of all four tumors. In the final analysis, we used the mean count of protein expression per ROI for each of the three histological cell markers (Fig. 5, step 7).
Statistical analysis
Statistical analysis was performed for the whole resection in each patient separate. The correlation between different variables (MET, rCBV, IDH1 count, Ki67 count, CD34 count) within the ROIs for each tumor was analyzed using non-linear correlation (Spearman rho; two-tailed p values with 95% confidence intervals) in JMP software (version 13).
Results
We made an effort to systematically identify the three types of ROIs (ROI1–3) in all PET scans (Figs. 1, 2, 3, and 4). Thus, 84 representative ROIs were selected in four tumors (47 ROI1, 21 ROI2, 16 ROI3). These 84 ROIs covered a total number of 2134 histological sub-images (size, 500 × 500 pixels) in which protein expression of three histological cell markers (IDH1, Ki67, and CD34) was quantified. Due to technical reasons, a number of immunostainings within the 84 selected ROIs were missing or not suitable for quantification (31 missing for Ki67; 15 missing for CD34). In the final analysis, the following parameters were included: MET (n = 84), rCBV (n = 84), IDH1 counts (n = 84), Ki67 counts (n = 53) and CD34 counts (n = 69). The results for each tumor are described below, followed by a summary of the data.
Patient 1
A 33-year-old man had a non-enhancing heterogeneous tumor in the right frontal pole on MRI. En bloc resection was performed as described previously [22] and histopathological examination showed an oligodendroglioma WHO grade II with IDH1 mutation, 1p/19q codeletion, and Ki67 < 5% (Table 1). A total of 29 ROIs were selected on PET; 16 areas with highest MET uptake (ROI1), seven areas with medium uptake (ROI2), and six areas with lower uptake in the tumor periphery (ROI3) (Fig. 1).
The MET uptake and rCBV values and the quantified protein counts (mean number of counts per ROI) in all 29 ROIs, as well as the statistical correlations between the various parameters are presented in Table 2. As shown, there was a strong correlation between MET uptake and tumor cell density (MET-IDH1: r = 0.91; p < 0.0001), MET uptake and vessel density (MET-CD34: r = 0.73; p < 0.0001), and MET uptake and proliferation (MET-Ki67: r = 0.71; p = 0.0465) in this tumor. No significant correlation was found between MET uptake and tumor perfusion (MET-rCBV: r = 0.06; p = 0.74). In addition, rCBV showed no significant correlations with histological cell markers.
Patient 2
A 50-year-old woman was diagnosed with a tumor in the left frontal pole with heterogeneous signal characteristics on MRI (Fig. 2). The tumor was removed en bloc and histopathological examination showed oligodendroglioma WHO grade II with densely packed IDH1-labeled tumor cells located mainly in the grey matter [22]. Molecular analysis showed IDH1 mutation, Ki67 < 5% and 1p/19q codeletion (Table 1). A total of 17 ROIs were selected of which eight representing regions with highest MET uptake (ROI1), four with medium uptake (ROI2), and five with lower uptake located in the tumor periphery (ROI3). The MET uptake, rCBV value, and protein expression of histological markers in these ROIs are presented in Table 2. There was a statistically significant correlation between MET uptake and IDH1 count (MET-IDH1: r = 0.51; p = 0.0345). As shown in Table 2, there were no significant correlations between MET uptake with tumor perfusion and with expression of Ki67 and CD34. We observed that several of the ROI3 in this tumor were located adjacent to or partially overlapping with the cortex, with inherent higher perfusion (Table 2).
Patient 3
A 39-year-old man was examined showing a non-enhancing, slightly heterogeneous tumor and en bloc tumor resection was performed. Histopathological examination showed a WHO grade II glial tumor with exclusively oligodendrocytic differentiation. Molecular analysis showed IDH1 mutation but no 1p19q codeletion. In spite of the intact 1p19q chromosomes, the tumor was morphologically diagnosed as an oligodendroglioma based on its characteristic oligodendroglial phenotype throughout the entire resection (Table 1). A total of 23 ROIs were selected, of which 14 in hot spot regions (ROI1), five in areas with medium uptake (ROI2), and four in areas with lower uptake in the tumor periphery (ROI3) (Fig. 3) (Table 2). There was a significant correlation between MET uptake and tumor cell count (MET-IDH1: r = 0.44; p = 0.0371), proliferation count (MET-Ki67: r = 0.69; p = 0.0095), and vessel count (MET-CD34: r = 0.67; p = 0.0005). No significant correlations were present between tumor perfusion and histological cell markers.
Patient 4
A 53-year-old man was diagnosed with a left frontal tumor showing minimal contrast enhancement on MRI (Fig. 4). En bloc resection was performed, with some loss of white matter tissue on the medial/inferior side of the tumor. Histopathological examination showed IDH1-mutated codeleted oligodendroglioma WHO grade III, Ki67 proliferation rate was 25% (Table 1) [22]. A total of 15 ROIs were identified on PET, of which nine in the hot spot (ROI1), five with medium uptake (ROI2), and due to loss of white matter tissue during en bloc resection only one representative ROI3 with lower MET uptake located in the tumor periphery. Statistical analysis showed a strong correlation between MET uptake and IDH1 (MET-IDH1: r = 0.85; p < 0.0001) (Table 2). No significant correlation was found between MET uptake and tumor perfusion or the expression of other histological markers. Similar to patient 2, we observed that the single ROI3 in this tumor was located adjacent to the cortex, resulting in inherent increased perfusion values.
Summary of findings
MET uptake was consistently correlated with tumor cell density throughout the entire tumor volume in all four oligodendrogliomas. Similar results were obtained using white matter as reference area for MET uptake (data not shown). Tumor perfusion in the ROIs defined by MET uptake did not correlate with MET uptake or with any of the histological cell markers.
In two tumors (patients 1 and 3), MET uptake also correlated with the density of proliferating cells and blood vessels. These two tumors showed consistently decreasing rCBV values at longer distance from the tumor core. In the other tumors (patients 2 and 4), we noticed an unexpected increase of rCBV values in ROI3, which was due to partial volume effects with adjacent cortex and large vessels in the peritumoral region.
Discussion
In the present study, we performed region-by-region comparisons of MET uptake, tumor perfusion, and protein expression of histological cell markers in a series of en bloc-resected oligodendrogliomas. It has been shown that the uptake of amino acid tracers is an indirect measure of microvessel density [29], but studies correlating uptake with proliferation, tumor cell density, and vessel density have shown incomplete or partly contradicting results [30,31,32,33,34,35,36,37]. With the aim to complement previous reports and to provide the proof-of-concept of how MET uptake and rCBV reflect the cellular heterogeneity in oligodendrogliomas, we evaluated the entire volume in en bloc-resected oligodendrogliomas. Our main finding is that MET unanimously reflects the density of tumor cells. By using the antibody against IDH1-mutated protein as a marker for tumor cells, we confirm previously established correlations between MET and cell density, and extend these findings to a correlation between MET and tumor cell density in oligodendrogliomas [21].
In addition, we found a significant correlation between MET uptake and microvessel density in two of four oligodendrogliomas, one codeleted and one non-codeleted tumor. Several studies have indicated that the generally higher MET uptake in oligodendrogliomas compared to astrocytomas is related to higher cell density, but also to higher microvessel density in these tumors [21, 38]. Indeed, MET uptake has been correlated with microvessel density in oligodendrogliomas independent of malignancy grade [29]. Our results confirm previous findings and show that MET uptake is a measure for microvessel density also in regions outside the hot spot and in peritumoral areas. In this context, the lack of significant correlations between MET-CD34 and MET-Ki67 in two tumors (patient 2 and patient 4) needs further explanation. In one tumor (case 4), we were able to include just one single ROI3 representing tumor periphery, due to minor loss of peritumoral tissue during the en bloc resection. In the other tumor (patient 2), several ROI3 were located adjacent to or partially overlapping with the cortex, with inherent higher perfusion. Selected ROIs were defined on PET and not on perfusion maps, since hot spot identification on rCBV maps in non-enhancing gliomas is challenging with high inter-observer variability [39, 40]. Therefore, we could not control for this artifact. It is likely though that the ROIs3 with increased count for microvessels and proliferating cells are not true histological representations of the tumor periphery, explaining the lack of statistically significant correlations between MET-CD34 and MET-Ki67 in these tumors.
Expression of the Ki67 antigen used as a proliferation marker is present in cycling cells [41], and correlates with malignancy in gliomas [42]. The PET literature regarding MET and FET uptake and proliferation in gliomas is not conclusive [20, 30,31,32,33, 43]. Fractions of cycling tumor cells in the tumor can be found at the tumor center as well as near the inner limits of the MRI-defined abnormalities, which may explain some of the inconsistencies between studies [44]. In the present study, we used absolute values of Ki67 count, instead of the proliferation ratio between cycling cells and tumor cells. The significant MET-Ki67 correlations in two of four oligodendrogliomas are consistent with previous reports and exemplify that the relationship between MET uptake and intra-tumoral cell proliferation in oligodendrogliomas is not unambiguous. One would expect that it is more difficult to establish statistically significant correlations between MET uptake and proliferation in low-grade tumors with low Ki67 index (< 5%). However, this was not the case. Significant MET-Ki67 correlations were found in low-grade oligodendrogliomas but not in the anaplastic oligodendroglioma with Ki67 index of 25%.
The development and maintenance of an adequate blood supply by cell proliferation and angiogenesis are essential for tumor growth and invasion [37, 45, 46]. Previous reports have shown that the rCBV measured by MRI perfusion is a measure for cell density [37], tumor proliferation [47], and vascularization [48] in glioma. In this study, we did not find any significant correlations between rCBV and the markers for tumor cell density, proliferation, or vessel cell density. It should be reminded, however, that the ROIs used for quantifying rCBV values were based on MET PET and not on perfusion maps. Our particular study design might also explain the lack of significant correlation between MET uptake and rCBV. Overall, we clearly demonstrate that MET PET has advantages over PWI, which is in line with previous studies [49, 50], but in contrast to others [51].
In conclusion, we further developed a previously described co-registration of histological and radiological images for region-by-region comparison of MET uptake, tumor perfusion, and histological cell markers in en bloc removed oligodendroglioma. Our key finding is the strong correlation between MET uptake and tumor cell density, in the entire tumor volume as well as the infiltrating border. MET uptake is correlated to the density of microvessels and proliferating cells as well, but this correlation is somewhat less stringent and probably more susceptible to measurement artifacts in regions covering the tumor periphery. The novelty of the present study lies in the use of en bloc-resected tumors, representing the concept of supra-total tumor resections. Previous studies have all been based on limited, less representative tumor tissue [52]. Our model also illustrates a novel way of evaluating imaging parameters with respect to the intra-tumoral heterogeneity of gliomas.
Limitations of the study
One limitation of this study is the small number of patients, which is however inherent to the methodological design of the study. Only a selection of gliomas can be surgically removed by en bloc tumor resection. It should also be noted that we applied four separate analyses of intra-tumoral heterogeneity, which means that comprehensive data were not pooled for inter-tumoral analysis. Instead, we analyzed multiple ROIs in each tumor and aimed to identify common patterns between tumors. Future studies with larger tumor samples and including astrocytic tumor subtypes are needed to confirm our data. In addition, there are some technical limitations to be considered. Both MET uptake and rCBV were measured in vivo before surgery and compared with ex vivo markers. Therefore, anesthesia or surgery may have affected microvessel contractility [48]. We encountered no significant difficulties during fusion of conventional MRI and PET images using the Carimas software. The Normalized Mutual Information (NMI) tool used in this study has previously proven to be highly accurate, with only a 1.2% failure [28]. In our previous work, we provided a detailed description of the limitations occurring during the manual process of histology-to-radiology co-registration [22]. Deformations and shrinkage that occur when tissue is processed for histology (i.e., formalin fixation, sectioning, and staining) make the procedure of co-registration challenging. Potential mismatch between PET/MRI and histological parameters has been minimized by applying a grid to the en bloc tumor resections to select specific areas, and by calculating mean values within relatively large ROIs to account for inaccuracies. However, the selection of ROIs located adjacent to or partially overlapping the cortex has probably influenced our results. Finally, a fixed pipeline was performed in CellProfiler to prevent bias, but not all histological stainings were done simultaneously, which could cause batch-related variations with regard to relative immunohistochemical staining intensities.
References
Louis DN, Perry A, Reifenberger G, Deimling AV, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–20. https://doi.org/10.1007/s00401-016-1545-1.
Wesseling P, van den Bent M, Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:809–27. https://doi.org/10.1007/s00401-015-1424-1.
Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro-Oncology. 2014;16:896–913. https://doi.org/10.1093/neuonc/nou087.
Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18:e315–e29. https://doi.org/10.1016/S1470-2045(17)30194-8.
Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, et al. Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med. 2016;374:1344–55. https://doi.org/10.1056/NEJMoa1500925.
Alattar AA, Brandel MG, Hirshman BR, Dong X, Carroll KT, Ali MA, et al. Oligodendroglioma resection: a surveillance, epidemiology, and end results (SEER) analysis. J Neurosurg. 2017;128(4):1076–83. https://doi.org/10.3171/2016.11.JNS161974.
Miller CR, Dunham CP, Scheithauer BW, Perry A. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24:5419–26. https://doi.org/10.1200/JCO.2006.08.1497.
Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131:397–406. https://doi.org/10.1043/1543-2165(2007)131[397:G]2.0.CO;2.
Smits M. Imaging of oligodendroglioma. Br J Radiol. 2016;89:20150857. https://doi.org/10.1259/bjr.20150857.
Jenkinson MD, du Plessis DG, Smith TS, Joyce KA, Warnke PC, Walker C. Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain. 2006;129:1884–91. https://doi.org/10.1093/brain/awl108.
Jansen EP, Dewit LG, van Herk M, Bartelink H. Target volumes in radiotherapy for high-grade malignant glioma of the brain. Radiother Oncol. 2000;56:151–6.
Langen KJ, Galldiks N, Hattingen E, Shah NJ. Advances in neuro-oncology imaging. Nat Rev Neurol. 2017;13:279–89. https://doi.org/10.1038/nrneurol.2017.44.
Herholz K, Coope D, Jackson A. Metabolic and molecular imaging in neuro-oncology. Lancet Neurol. 2007;6:711–24. https://doi.org/10.1016/S1474-4422(07)70192-8.
Jackson RJ, Fuller GN, Abi-Said D, Lang FF, Gokaslan ZL, Shi WM, et al. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro-Oncology. 2001;3:193–200. https://doi.org/10.1093/neuonc/3.3.193.
Soffietti R, Baumert BG, Bello L, von Deimling A, Duffau H, Frenay M, et al. Guidelines on management of low-grade gliomas: report of an EFNS-EANO task force. Eur J Neurol. 2010;17:1124–33. https://doi.org/10.1111/j.1468-1331.2010.03151.x.
Albert NL, Weller M, Suchorska B, Galldiks N, Soffietti R, Kim MM, et al. Response assessment in neuro-oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro-Oncology. 2016;18:1199–208. https://doi.org/10.1093/neuonc/now058.
Mosskin M, Ericson K, Hindmarsh T, von Holst H, Collins VP, Bergstrom M, et al. Positron emission tomography compared with magnetic resonance imaging and computed tomography in supratentorial gliomas using multiple stereotactic biopsies as reference. Acta Radiol. 1989;30:225–32.
Pirotte B, Goldman S, Dewitte O, Massager N, Wikler D, Lefranc F, et al. Integrated positron emission tomography and magnetic resonance imaging-guided resection of brain tumors: a report of 103 consecutive procedures. J Neurosurg. 2006;104:238–53. https://doi.org/10.3171/jns.2006.104.2.238.
Langen KJ, Stoffels G, Filss C, Heinzel A, Stegmayr C, Lohmann P, et al. Imaging of amino acid transport in brain tumours: positron emission tomography with O-(2-[(18)F]fluoroethyl)-L-tyrosine (FET). Methods. 2017;130:124–34. https://doi.org/10.1016/j.ymeth.2017.05.019.
Lopez WO, Cordeiro JG, Albicker U, Doostkam S, Nikkhah G, Kirch RD, et al. Correlation of (18)F-fluoroethyl tyrosine positron-emission tomography uptake values and histomorphological findings by stereotactic serial biopsy in newly diagnosed brain tumors using a refined software tool. Onco Targets Ther. 2015;8:3803–15. https://doi.org/10.2147/OTT.S87126.
Langen K-J, Rapp M, Sabel M, Galldiks N. Positron-emission-tomography in diffuse low-grade gliomas. Diffuse low-grade gliomas in adults. Cham: Springer; 2017. p. 263–86.
Zetterling M, Roodakker KR, Berntsson SG, Edqvist PH, Latini F, Landtblom AM, et al. Extension of diffuse low-grade gliomas beyond radiological borders as shown by the coregistration of histopathological and magnetic resonance imaging data. J Neurosurg. 2016;125(5):1155–66. https://doi.org/10.3171/2015.10.JNS15583.
Berntsson SG, Falk A, Savitcheva I, Godau A, Zetterling M, Hesselager G, et al. Perfusion and diffusion MRI combined with 11C-methionine PET in the preoperative evaluation of suspected adult low-grade gliomas. J Neuro-Oncol. 2013;114:241–9. https://doi.org/10.1007/s11060-013-1178-3.
Falk A, Fahlstrom M, Rostrup E, Berntsson S, Zetterling M, Morell A, et al. Discrimination between glioma grades II and III in suspected low-grade gliomas using dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging: a histogram analysis approach. Neuroradiology. 2014;56:1031–8. https://doi.org/10.1007/s00234-014-1426-z.
Hoffmann A, Bredno J, Wendland MF, Derugin N, Hom J, Schuster T, et al. Validation of in vivo magnetic resonance imaging blood-brain barrier permeability measurements by comparison with gold standard histology. Stroke. 2011;42:2054–60. https://doi.org/10.1161/STROKEAHA.110.597997.
Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. https://doi.org/10.1186/gb-2006-7-10-r100.
Studholme C, Hawkes D, Hill D. A normalized entropy measure for multimodality image alignment. Med Imaging. 1998;3338:132–43. https://doi.org/10.1117/12.310835.
Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24:25–35. https://doi.org/10.1118/1.598130.
Kracht LW, Friese M, Herholz K, Schroeder R, Bauer B, Jacobs A, et al. Methyl-[11C]- l-methionine uptake as measured by positron emission tomography correlates to microvessel density in patients with glioma. Eur J Nucl Med Mol Imaging. 2003;30:868–73. https://doi.org/10.1007/s00259-003-1148-7.
Dhermain FG, Hau P, Lanfermann H, Jacobs AH, van den Bent MJ. Advanced MRI and PET imaging for assessment of treatment response in patients with gliomas. Lancet Neurol. 2010;9:906–20. https://doi.org/10.1016/S1474-4422(10)70181-2.
Chung JK, Kim YK, Kim SK, Lee YJ, Paek S, Yeo JS, et al. Usefulness of 11C-methionine PET in the evaluation of brain lesions that are hypo- or isometabolic on 18F-FDG PET. Eur J Nucl Med Mol Imaging. 2002;29:176–82.
Langen KJ, Muhlensiepen H, Holschbach M, Hautzel H, Jansen P, Coenen HH. Transport mechanisms of 3-[123I]iodo-alpha-methyl-L-tyrosine in a human glioma cell line: comparison with [3H]methyl-L-methionine. J Nucl Med. 2000;41:1250–5.
Tanaka K, Yamamoto Y, Maeda Y, Yamamoto H, Kudomi N, Kawai N, et al. Correlation of 4′-[methyl-(11)C]-thiothymidine uptake with Ki-67 immunohistochemistry and tumor grade in patients with newly diagnosed gliomas in comparison with (11)C-methionine uptake. Ann Nucl Med. 2016;30:89–96. https://doi.org/10.1007/s12149-015-1035-x.
Kinoshita M, Arita H, Okita Y, Kagawa N, Kishima H, Hashimoto N, et al. Comparison of diffusion tensor imaging and (11)C-methionine positron emission tomography for reliable prediction of tumor cell density in gliomas. J Neurosurg. 2016;125:1136–42. https://doi.org/10.3171/2015.11.JNS151848.
Arita H, Kinoshita M, Kagawa N, Fujimoto Y, Kishima H, Hashimoto N, et al. (1)(1)C-methionine uptake and intraoperative 5-aminolevulinic acid-induced fluorescence as separate index markers of cell density in glioma: a stereotactic image-histological analysis. Cancer. 2012;118:1619–27. https://doi.org/10.1002/cncr.26445.
Okita Y, Kinoshita M, Goto T, Kagawa N, Kishima H, Shimosegawa E, et al. (11)C-methionine uptake correlates with tumor cell density rather than with microvessel density in glioma: a stereotactic image-histology comparison. NeuroImage. 2010;49:2977–82. https://doi.org/10.1016/j.neuroimage.2009.11.024.
Sadeghi N, D'Haene N, Decaestecker C, Levivier M, Metens T, Maris C, et al. Apparent diffusion coefficient and cerebral blood volume in brain gliomas: relation to tumor cell density and tumor microvessel density based on stereotactic biopsies. AJNR Am J Neuroradiol. 2008;29:476–82. https://doi.org/10.3174/ajnr.A0851.
Chan AS, Leung SY, Wong MP, Yuen ST, Cheung N, Fan YW, et al. Expression of vascular endothelial growth factor and its receptors in the anaplastic progression of astrocytoma, oligodendroglioma, and ependymoma. Am J Surg Pathol. 1998;22:816–26.
Friedman SN, Bambrough PJ, Kotsarini C, Khandanpour N, Hoggard N. Semi-automated and automated glioma grading using dynamic susceptibility-weighted contrast-enhanced perfusion MRI relative cerebral blood volume measurements. Br J Radiol. 2012;85:e1204–11. https://doi.org/10.1259/bjr/13908936.
Ulmer S, Liess C, Kesari S, Otto N, Straube T, Jansen O. Use of dynamic susceptibility-contrast MRI (DSC-MRI) to assess perfusion changes in the ipsilateral brain parenchyma from glioblastoma. J Neuro-Oncol. 2009;91:213–20. https://doi.org/10.1007/s11060-008-9701-7.
Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5.
Wharton SB, Hamilton FA, Chan WK, Chan KK, Anderson JR. Proliferation and cell death in oligodendrogliomas. Neuropathol Appl Neurobiol. 1998;24:21–8.
Bette S, Gempt J, Delbridge C, Kirschke JS, Schlegel J, Foerster S, et al. Prognostic value of O-(2-[18F]-fluoroethyl)-L-tyrosine-positron emission tomography imaging for histopathologic characteristics and progression-free survival in patients with low-grade glioma. World Neurosurg. 2016;89:230–9. https://doi.org/10.1016/j.wneu.2016.01.085.
Gerin C, Pallud J, Deroulers C, Varlet P, Oppenheim C, Roux FX, et al. Quantitative characterization of the imaging limits of diffuse low-grade oligodendrogliomas. Neuro-Oncology. 2013;15:1379–88. https://doi.org/10.1093/neuonc/not072.
Shweiki D, Neeman M, Itin A, Keshet E. Induction of vascular endothelial growth factor expression by hypoxia and by glucose deficiency in multicell spheroids: implications for tumor angiogenesis. Proc Natl Acad Sci U S A. 1995;92:768–72.
Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310.
Price SJ, Green HA, Dean AF, Joseph J, Hutchinson PJ, Gillard JH. Correlation of MR relative cerebral blood volume measurements with cellular density and proliferation in high-grade gliomas: an image-guided biopsy study. AJNR Am J Neuroradiol. 2011;32:501–6. https://doi.org/10.3174/ajnr.A2312.
Hu LS, Eschbacher JM, Dueck AC, Heiserman JE, Liu S, Karis JP, et al. Correlations between perfusion MR imaging cerebral blood volume, microvessel quantification, and clinical outcome using stereotactic analysis in recurrent high-grade glioma. AJNR Am J Neuroradiol. 2012;33:69–76. https://doi.org/10.3174/ajnr.A2743.
Cicone F, Filss CP, Minniti G, Rossi-Espagnet C, Papa A, Scaringi C, et al. Volumetric assessment of recurrent or progressive gliomas: comparison between F-DOPA PET and perfusion-weighted MRI. Eur J Nucl Med Mol Imaging. 2015;42:905–15. https://doi.org/10.1007/s00259-015-3018-5.
Filss CP, Cicone F, Shah NJ, Galldiks N, Langen KJ. Amino acid PET and MR perfusion imaging in brain tumours. Clin Transl Imaging. 2017;5:209–23. https://doi.org/10.1007/s40336-017-0225-z.
Dandois V, Rommel D, Renard L, Jamart J, Cosnard G. Substitution of 11C-methionine PET by perfusion MRI during the follow-up of treated high-grade gliomas: preliminary results in clinical practice. J Neuroradiol. 2010;37:89–97. https://doi.org/10.1016/j.neurad.2009.04.005.
Pauleit D, Floeth F, Hamacher K, Riemenschneider MJ, Reifenberger G, Muller HW, et al. O-(2-[18F]fluoroethyl)-L-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain. 2005;128:678–87. https://doi.org/10.1093/brain/awh399.
Acknowledgements
The authors thank Professor Jens Sörensen at the Department of Surgical Sciences, section of Radiology, Uppsala University Hospital, for supervision, Dr. Anders Berglund for statistical advice, the SciLifeLab BioImage Informatics Facility, Uppsala University, for help with setting up the pipelines in CellProfiler and the Turku PET center, Finland, for providing the Carimas software.
Funding
This work was supported by research funds from the Bissen Brain Walk Stiftelsen (A.S.), Erik, Karin och Gösta Selanders Stiftelse (A.S.), Hanna Eklund Stiftelsen (A.S.), and the Uppsala County Council (A.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the principles of the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed written consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Roodakker, K.R., Alhuseinalkhudhur, A., Al-Jaff, M. et al. Region-by-region analysis of PET, MRI, and histology in en bloc-resected oligodendrogliomas reveals intra-tumoral heterogeneity. Eur J Nucl Med Mol Imaging 46, 569–579 (2019). https://doi.org/10.1007/s00259-018-4107-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-018-4107-z