Abstract
Trichoderma reesei displays a high capability to produce extracellular proteins and therefore is used as a platform for the expression of heterologous genes. In a previous study, an expression cassette with the constitutive tef1 promoter and the cbh1 terminator compatible with flow cytometry analysis was developed. Independent transformants obtained by a random integration into the genome of a circular plasmid containing the expression cassette showed a wide range of fluorescence levels. Whole genome sequencing was conducted on eight of the transformed strains using two next-generation sequencing (NGS) platforms: Illumina paired-end sequencing and Oxford Nanopore. In all strains, the expression plasmid was inserted at the same position in the genome, i.e., upstream of the tef1 gene, indicating an integration by homologous recombination. The different levels of fluorescence observed correspond to different copy numbers of the plasmid. Overall, the integration of a circular plasmid with the green fluorescence protein (egfp) transgene under the control of tef1 promoter favors multicopy integration and allows over-production of this heterologous protein on glucose. In conclusion, an expression system based on using the tef1 promotor could be one of the building blocks for improving high-value heterologous protein production by increasing the copy number of the encoding genes into the genome of the platform strain.
Key points
• Varied eGFP levels from tef1 promoter and cbh1 terminator expression.
• Whole genome sequencing on short and long reads platforms reveals various plasmid copy numbers in strains.
• Plasmids integrate at the same genomic site by homologous recombination in all strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trichoderma reesei is a powerful platform to produce a cocktail of native enzymes, mainly used not only in the paper, textile, and feed industries but also in the second-generation bioethanol process. More recently, its high secretion capacity, ease of cultivation, and classification as a Generally Recognized as Safe (GRAS) microorganism make it a suitable choice for the production of heterologous proteins. These heterologous proteins can be either fungal or bacterial enzymes to modify the composition of the original cocktails (Sun et al. 2018; Arai et al. 2023; Wohlschlager et al. 2021) or high-value pharmaceutical proteins (Landowski et al. 2016; Jiang et al. 2019; Arai et al. 2023). Currently, the improvement of heterologous protein expression in T. reesei is based on upgraded strains, the development of promoters, and construction of fusion protein (Nevalainen and Peterson 2014; Singh et al. 2015). However, productivity and titers remain a major challenge for the industrialization of these processes.
One way to increase productivity is to integrate multiple copies of the gene of interest. This approach has been mainly explored in yeasts fungus such as Saccharomyces cerevisiae (Sakai et al. 1991; Qi et al. 2022), Yarrowia lipolytica (Novikova et al. 2021), Pichia pastoris (Deng et al. 2002), Kluyveromyces lactis (Oguro et al. 2015), and Lypomyces starkeyi (Sakai et al. 1990). Most of these multi-integration systems are based on targeting sequences present in multiple copies in the genome (Zheng et al. 2022), on the approach of iterative transformations with marker recycling (Jensen et al. 2014), on the use of a modified selective marker to screen transformants displaying multicopy integration (Semkiv et al. 2016), or on the introduction of genetic construction with multiple copies of the gene (Zheng et al. 2014).
Cases of unintentional multicopy integration have been described in filamentous fungi, but only a few examples of on-purpose approaches have been implemented (Takeda et al. 2018; Plüddemann and van Zyl 2003). Araki et al. (2021) have developed a multicopy integration strategy in Aspergillus sojae that relies on the construction of an attenuated selection marker, claiming 18 to 46 integrated copies depending on the target gene. In T. reesei, increased β-carotene production has been achieved by iterative integration of genes encoding key enzymes of β-carotene synthesis, but the copy number is below three and the impact on productivity remains limited (Li et al. 2023).
In a previous work, a flow cytometry experiment was performed to measure the fluorescence of germinating conidia originating from transformants expressing an enhanced green fluorescent protein eGFP under the control of various promoters ectopically integrated into the genome of the T. reesei strain Cl847 (Mathis et al. 2020). Among the nine screened promoters, transformants with the tef1 promoter (Nakari-Setälä and Penttilä 1995) exhibited the highest egfp expression level with the fastest kinetic of fluorescence production. Interestingly, a wide range of egfp gene expression levels was also observed in these transformants. Therefore, the tef1 promoter appeared as the only one compatible with cytometry, meaning an early and strong expression of fluorescence during the first steps of germination. We therefore decided to probe the transgene integration sites for these transformants to identify a variety of sites enabling the modulation of the expression levels of a given target gene. A similar strategy was previously applied by Qin et al. (2018) with the random integration of an expression cassette comprising genes encoding a lipase and the fluorescent protein DsRed1 in T. reesei, but only two strains with the highest expression of both transgenes were investigated to identify the integration sites.
Strikingly, our analysis revealed a single site and with multicopy insertion of the expression plasmid in all strains analyzed. The genomic insertion site is localized upstream of the tef1 locus with a copy number ranging from 5 to 11. This result suggests that the use of the tef1 promoter may favor multicopy insertion and therefore promote a higher expression for heterologous genes.
Materials and methods
Strains and culture conditions
The T. reesei hyperproducer mutant Cl847 (Durand et al. 1988) used in this study was obtained from several rounds of mutagenesis of the RutC30 strain (ATCC 56765). Cl847 is available at the Collection Nationale de Cultures de Microorganismes (CNCM) at the Institut Pasteur in Paris, France (CNCM MA 6-10W). Transformants were cultivated in liquid PD (potato dextrose) or solid PDA (potato dextrose agar) at 30 °C using frozen spores as inoculum. After transformation (Penttilä et al. 1987), three purification steps were carried out, (i.e., three specific propagation and conidiation steps), followed by cryopreservation (PD glycerol 15%) and another propagation and conidiation. This process, covering four propagations, allows us to check the stability of egfp expression over a long period of time.
For the germination time course, solutions of fresh spores or from cryotubes stored in PD plus glycerol 15% were propagated in PD medium at 25 °C, 125 rpm for 2 to 4 h, then at 15 °C in static overnight, before returning at 25 °C for 6 h (Mathis et al. 2020). For each transformant, three independent sub-cultures of these conidial suspensions were grown in a PD medium for germination to carry out a flow cytometry experiment.
Flow cytometry acquisitions
All analyses were conducted using a Cyflow Space cytometer, equipped with an MLS Blue 480–50 V2 laser emitting at 488 nm with a power of 50 mW (SYSMEX, Kobe, Japan). The optical detector range for FL1 (green fluorescence channel) was 520/20. Acquisition conditions were described by Mathis et al. (2020). All assays in the FCM (flow cytometry) and quantifications were performed as technical triplicates on independent cultures (biological triplicates). FCM parameters were as follows: speed 6, FSC (forward scatter) 125, log3, SSC (side scatter) 200 log3, FL1 450 log4. Once defined, the cytometer settings were maintained throughout the study and for all samples. The use of controls (calibrated beads for size or non-fluorescent controls for gating) ensures that fluorescence and size responses are repeatable and comparable between samples. Fluorescence measurements are provided by the instrument after converting the number of photons captured into an electrical signal that can be interpreted by electronics.
Genome sequencing, sequence alignment, and analyses
Genomic DNA for next-generation sequencing (NGS) was extracted according to the protocol of the Joint Genome Institute (https://1000.fungalgenomes.org/documents/Martin_genomicDNAextraction_AK051010.pdf). Library preparation and Illumina sequencing in 2 × 150 pb paired-end read mode were performed by Eurofins MGW (https://eurofinsgenomics.eu/) for the 8 transformants. Reads were paired and trimmed using Geneious Prime® (Biomatters Ltd, Auckland, New Zealand), and their quality was assessed by FastQC analysis performed on the Galaxy platform (The Galaxy Community 2024). The Cl847 genome is not available in literature, contrary to RutC30 (ATCC 56765), its closest relative. Mapping was achieved using the Geneious mapper on a RutC30 genome which was reconstructed in silico from QM6a (Li et al. 2017) by inserting previously identified chromosomal rearrangements and mutations (Vitikainen et al. 2010; Le Crom et al. 2009; Koike et al. 2013). Whole genome alignments were performed using Geneious Prime (Kearse et al. 2012) with the Geneious mapper (Geneious Prime®, Biomatters Ltd) configured for “medium/low sensitivity” and “deletion and structural variation” settings. Average coverage ranges from 46 and 61, and pairwise identity is superior to 99% for all strains.
Four strains (E1, E3; E4, and E5) were also sequenced using long-read sequencing technologies. DNA libraries for long-read sequencing were prepared with the Ligation kits LSK109 and NBD104 (Oxford Nanopore Technologies, Oxford, UK) according to the manufacturer’s recommendations. The library was sequenced on a flo-min106 (R9.4.1) flowcell on a GridION instrument (Oxford Nanopore Technologies, Oxford, UK). Basecalling, demultiplexing, and trimming have been done with Guppy3.2.6 (Oxford Nanopore Technologies, Oxford, UK). Approximately 200,000 reads giving a total base number of 500 million were obtained per library with an average read length between 10 and 12.5 kb. After the quality filter, around 96% of the reads were kept for the following analysis. Sequencing data from Illumina and GridION platforms are available at NCBI at Bioproject number PRJNA1032401.
Results
Kinetics of fluorescence during germination in independent transformants
In a previous work, transformants of the Cl847 strain obtained by ectopic integration of an egfp under the control of the tef1 promoter (Nakari-Setälä and Penttilä 1995) and the cbh1 terminator showed a high level of fluorescence in conidia increasing with germination time. Cl847 was thus kept as the strain of interest in the present study, also due to its high relevance for industrial purposes, in order to maximize the technological readiness level (TRL) of the results. We decided to take advantage of this feature to assess the variability among 9 independent transformants (E1 to E9) compared to the reference strain Cl847 by following the fluorescence kinetics from the time point 0 (T0h) to 24 h (T24h) in flow cytometry (FCM). The gating parameters allowing a relevant discrimination between fluorescent and auto-fluorescent spores have been already established by Mathis et al. (2020). Gating regions and cytograms at 0, 16, and 24 h are shown in Supplementary data, Fig. S1. The behavior of the CL847 control strain has been previously described in Mathis et al. (2020), i.e., a profile with an increase in the size of the events associated with the swelling of the conidia due to the germination process (FCS size axis) correlated with a slight increase in autofluorescence (FL1 axis).
All transformants displayed a pattern similar to the control strain over time for the FCS axis. In contrast, the conidial cloud is already distributed between the non-fluorescent (S) and fluorescent spores (F) with 43 to 74% of the total population present in the F gate depending on the transformants, with E5 (74%) and E2 (67%) having the highest number of spores in F at T0h. At T16h and T24h, the cytograms highlight a general increase in fluorescence for all transformants, but different profiles of homogeneity, spread, and intensity are observed. Most of the transformants (E1, E2, E4-E6, E8, E9) have a homogeneous population that is almost entirely detected in the F acquisition region. On the contrary, the spore population of E3 and E7 splits in two during the kinetic for some transformants with a plume in the AF region for E3 and the detection of two equivalent populations (50/50) present from T0h for E7. The heterogeneity observed in E7 suggests that it is a heterokaryotic strain. The genetically different nuclei of E7 probably originate from independent integration events leading to different levels of egfp expression. In the following analysis, this transformant was eliminated.
Fluorescence quantification over time highlights differences between transformants
Kinetic fluorescence quantification was performed according to Mathis et al. (2020), measuring the ratio of fluorescence intensity to spore size (F/S) using the events in the SF gating region (Fig. 1).
As expected, Cl847 exhibits a low and constant basal autofluorescence (> 0.1) throughout the kinetic confirming that the fluorescence intensity of the transformants is due to egfp expression. At T0h, the F/S ratio of the transformants and the control strain is similar whereas a significant signal difference from the background is observed for three strains (E2, E4, and E9) at t + 2 h and for all strains at t + 16 h. Interestingly, at t + 24 h, various sets of ratio levels are detected depending on the strain, with FS values ranging from 2.8 (E3) to 6.9 (for E4), i.e., a factor of 2.5. The rank of transformants in respect of size-normalized fluorescence emission at t + 24 h is E3 < E5 and E9 < E1 and E8 < E2 and E6 < E4.
The variability between transformants is probably related to the integration sites of the expression plasmid in the genome of the strains or to the integrated copy number of the egfp. To validate this assumption, we decided to sequence the genome of the 8 transformants using the Illumina short paired-ends reads technology.
Fluorescence quantification differences seem to be linked to the expression cassette copy number
The Cl847 mutant was obtained from the RutC30 strain by six steps of mutagenesis (Durand et al. 1988). As the RutC30 genome has been previously sequenced (Koike et al. 2013; Le Crom et al. 2009; Jourdier et al. 2017), this genome was used as the reference for all alignment procedures. One of the challenges in identifying the insertion site within the genome is the presence of the tef1 gene in both the genomic DNA and the plasmid. Initially, a whole genome alignment was conducted on the RutC30 genome using the Geneious mapper, a tool capable of detecting structural rearrangements. However, this approach did not reveal any insertion sites. To advance this investigation, a strategy was developed to pinpoint the integration sites through the collection of hybrid paired reads, following the methodology previously employed by Takeda et al. (2018). Paired reads could be classified as follows (Fig. 2A): reads mapping both to the genome (PR1) or to the plasmid (PR2), one read mapped to the plasmid and the other one to the genome (hybrid paired reads, PR3), and those where one mapping to either the plasmid or the genome and the second one mapped nowhere (PR4). The PR4 reads include the chimeric reads, i.e., those that straddle the genome and the plasmid. To recover the PR3 group (Fig. 2B), a first alignment was performed on the genome with and the unused paired reads were collected to be mapped on the plasmid. In both steps, only paired reads that were both mapped were aligned. The unused paired reads generated from this analysis were expected to contain mainly hybrid paired reads. Finally, the PR3 group was aligned to the plasmid without restriction in the mapping of both reads of a pair, and a de novo assembly was performed with the unused reads unpaired. Contigs that were not AT-rich or homopolymers were mapped to the genome, but no insertion sites could be identified using this pipeline.
Strategy for identifying the integration sites of the plasmid. A Classification of paired-end reads localization according to their localization in the transformant genomes. PR1, reads mapping to the genome; PR2, reads mapping to the plasmid; PR3 or hybrid reads, one read mapping to the plasmid and the other one to the genome; PR4, reads with one mapping to either the plasmid or the genome whereas the second one mapped nowhere. B Pipeline to recover the hybrid paired reads and localize plasmid integration site
Interestingly, mapping the reads onto the plasmid revealed significantly higher coverage compared to the genome, suggesting the possibility of multicopy integration. To ascertain the number of copies present within the genome, an alignment of all reads was performed against the plasmid, and an estimate of the copy number of hph, ptef1, and egfp elements was obtained by comparing their coverage with the genome (Table 1). The egfp copy number is mostly in agreement with the fluorescence quantification with E2, E4, and E6 (6.1, 9.8, and 7.1) having the highest copy number, whereas E5, E8, and E9 have the lowest (4.8, 3.7, and 3.5). In contrast, no correlation was observed between fluorescence measurements and copy number for strains E1 and E3. This result could be explained by recombination mechanisms during plasmid integration. This hypothesis is supported by the high variability in the coverage of certain elements (263 to 424 for the tef1 promoter in strain E1) and by the variable number of copies of the plasmid elements (5.3 copies of the tef1 promoter for 6.9 copies of egfp in strain E3). Additionally, a linear regression model between the GFP expression levels from Fig. 1 and the egfp gene copy number from Table 1 shows a general trend of correlation; however, the adjusted R2 remains moderate (0.48), and the model is barely significant (p-value = 0.0581). Nonetheless, it is the E3 strain that singularly jeopardizes the correlation. Indeed, if the same model is constructed without E3, the trend becomes highly significant (p-value = 0.0023), and the adjusted R2 is much improved (0.86). According to Fig. 3, E3 is the strain for which the plasmid integration event was the most random, with numerous recombination phases, as the entire plasmid is found only once, unlike E1, for instance. We can therefore hypothesize that despite a good coverage rate, there is little functional GFP in E3. Nevertheless, a positive linear correlation can still be posited between the number of egfp gene copies obtained through sequencing and the protein expression measured by flow cytometry.
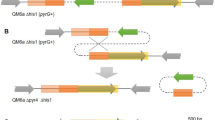
Annotation of different expression plasmid elements inserted into the genome of transformed strains. A Plasmid vector for transformation and native chromosome. B Multicopy integrations as observed for 4 transformants. Abbreviations: P promoter, T terminator, E egfp, H hygromycin; green arrows indicate functional copies of egfp; black arrows indicate dynamics of insertion of the circular plasmid into the genome
Multicopy integration, which may have occurred either at one site or at multiple localizations within the genome, could explain the reason for the failure to identify the integration sites using short-read sequencing platform.
Identification of a unique integration site in all strains with long-read sequencing
In a second attempt to identify the integration site, a long-read sequencing (LRS) was performed using Oxford Nanopore Technology (ONT). We hypothesized that long reads might overlap the inserted plasmid and the flanking regions. There is no sequence homology in the vector as significant as that of the tef1 promoter. All other regions (promoter, terminator, ori, marker, etc.) comprising the vector do not originate from T. reesei, except for the cbh1 terminator located downstream of egfp. Among the transformants studied, none exhibit insertion at the cbh1 locus. The scientific literature concurs that for precise locus insertion, the sequence must be flanked by 1 kb of homology on both sides (Ma et al. 2023). In the absence of CRISPR-like tools or strains deficient in homologous repair systems, locus insertion occurs in 1 out of 10 cases (Schuster et al. 2012). In our study, this insertion frequency is 8 out of 8. The egfp containing plasmid consistently integrated upstream of the tef1 promoter, even without two homologous flanks.
Four (E1, E3, E4, and E5) of the eight transformants were selected as representative of the range of egfp expression for this experiment. To identify the overlapping reads, a double mapping to plasmid and genome was performed, and the chimeric reads plasmid/genome were recovered. As the plasmid contained sequences also present in the genome (cbh1 terminator and tef1 promoter), a filtering pipeline of the chimeric reads is necessary to select the truly overlapping ones which results in about 50 reads per strain with a minimum read size range of 1150 to 4230 and a maximum of 67,240 to 109,977 (Fasta files of this analysis available in Supplementary Data File 1).
As one of the disadvantages of ONT is the high error rate, especially at the ends of the reads (van Dijk et al. 2023), it may be inefficient to analyze the sequences by searching for nucleotide similarity. Therefore, to reconstruct the insertion event, a functional annotation of the selected reads was carried out using the “annotate from” tool of the Geneious software with a similarity parameter of 60% and the plasmid sequence and the genome as annotation sources.
As previously suggested by the Illumina experiment analysis, we detected a multicopy insertion of the plasmid. Surprisingly, the insertion site was identical in all four transformants and was localized upstream of the tef1 gene. Unfortunately, none of the selected reads included both the flanking regions and the multiple plasmid insertions. Nevertheless, a detailed study of the reads allowed us to reconstruct the inserts (Fig. 3). For the four strains, at the 3′ border, the tef1 gene is always associated with a tef1 promoter of the same size as the plasmid, and at 5′ border, the first annotated plasmidic region at the junction with the genome is a tef1 promoter. We cannot assign the plasmidic or genomic source of these two tef1 promoters. Nevertheless, a tef1 promoter sequence of the same size as the plasmid one is probably essential for the viability of the cells since neither of them is found truncated upstream of tef1.
The reconstructed insertion events highlight two types of process: a cyclic multicopy insertion of the plasmid with (E3, E4, and E5) or without (E1) recombination. In addition, some copies of the expression copy are truncated, resulting in partial or no expression of the egfp (E3 and E4). We, therefore, infer the copy number of the expression cassette by considering only the complete copy with promoter and terminator. As expected, the intact copy number correlates with the egfp expression level detected in the FCM experiment with 8 copies for the E4, 7 for the E1, 5 for the E5, and 4 for the E3.
Discussion
In this paper, we describe the finding of the insertion site of an uncut plasmid containing an egfp expression cassette with the tef1 promoter and the cbh1 terminator, in the genome of eight transformed strains. Strikingly, the localization of the circular plasmid was identical in all strains, upstream of the tef1 gene. This integration site bias may be explained by the construction of an expression cassette with the tef1 promoter without the tef1 terminator, which would favor homologous recombination in the promoter region. In T. reesei, only a single copy of the gene is present in the genome (Nakari et al. 1993). Although no tef1 knockout experiment has been reported in T. reesei, we could infer from other fungal species (Cottrelle et al. 1985; Silar et al. 2000) that inactivation of tef1 is probably lethal to the cells. Therefore, we can assume that only strains with an insertion that conserved an intact tef1 gene and promoter were able to survive. It should be noted that the transformants examined in this study were not selected for a high fluorescence phenotype, but were randomly selected, suggesting that multicopy and tef1 upstream insertion should occur in the majority of strains. Other teams have reported the use of a similar expression cassette, namely a tef1 promotor with a cbh1 terminator (Uzbas et al. 2012; Nakari-Setälä and Penttilä 1995; Dashtban and Qin 2012), but none of them mentioned multicopy (> 2 copy) integration. These differences could be explained by the low number of transformants tested (Nakari-Setälä and Penttilä 1995), the choice to select transformants with only one copy (Uzbas et al. 2012), or the transgene sequence itself.
In this study, we have shown that an efficient system for multicopy integration can be achieved by using an expression cassette with the tef1 promoter without the tef1 terminator and by using a circular plasmid. As already mentioned by Nakari-Setälä and Penttilä (1995), producing the heterologous proteins on glucose prevents contaminating proteins, since the hydrolytic enzymes produced by T. reesei are mostly repressed in the presence of glucose. An important step for the production of heterologous proteins on an industrial scale could be the coupling of the tef1 promoter for constitutive expression on glucose and the multicopy integration reported here. Finally, the tef1 region seems to be a recombination hotspot. It shall be recalled that tef1 has proven to be an essential gene (Cottrelle et al. 1985; Silar et al. 2000), which is constitutively expressed (Nakari et al. 1993). Recently, a study has demonstrated that the chromatin state of promoters of essential genes is actively maintained as open to ensure their transcription (Fan et al. 2021). Thus, we may hypothesize that the chromatin state of tef1 promoter favors homologous recombination.
Data availability
Sequencing data from Illumina and GridION platforms are available at NCBI at Bioproject number PRJNA1032401. The reviewer link is https://dataview.ncbi.nlm.nih.gov/object/PRJNA1032401?reviewer=er177l2n4vj3flgpi0r4e8muf.
References
Arai T, Wada M, Nishiguchi H, Takimura Y, Ishii J (2023) Inducer-free recombinant protein production in Trichoderma reesei: secretory production of endogenous enzymes and heterologous nanobodies using glucose as the sole carbon source. Microb Cell Fact 22:103. https://doi.org/10.1186/s12934-023-02109-y
Araki Y, Yuzuki M, Masakari Y, Sato A, Gomi K, Hara S (2021) High-level heterologous protein production using an attenuated selection marker in Aspergillus sojae. J Gen Appl Microbiol 67:77–80. https://doi.org/10.2323/jgam.2020.05.002
Cottrelle P, Thiele D, Price VL, Memet S, Micouin JY, Marck C, Buhler JM, Sentenac A, Fromageot P (1985) Cloning, nucleotide sequence, and expression of one of two genes coding for yeast elongation factor 1 alpha. J Biol Chem 260(5):3090–3096. https://doi.org/10.1016/S0021-9258(18)89477-7
Dashtban M, Qin W (2012) Overexpression of an exotic thermotolerant β-glucosidase in Trichoderma reesei and its significant increase in cellulolytic activity and saccharification of barley straw. Microb Cell Fact 11:63. https://doi.org/10.1186/1475-2859-11-63
Deng N, Su KY, Wang XZ, Long QX, Yang L, Yu ZY (2002) The expression of humanized Fab fragment of the anti-HBsAg antibody in methylotropic yeast Pichia pastoris. Sheng Wu Gong Cheng Xue Bao Sep 18(5):546–50 Chinese
Durand H, Clanet M, Tiraby G (1988) Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb Technol 10:341–346. https://doi.org/10.1016/0141-0229(88)90012-9
Fan K, Moore JE, Zhang X-O, Weng Z (2021) Genetic and epigenetic features of promoters with ubiquitous chromatin accessibility support ubiquitous transcription of cell-essential genes. Nucl Acids Res 49:5705–5725. https://doi.org/10.1093/nar/gkab345
Jensen NB, Strucko T, Kildegaard KR, David F, Maury J, Mortensen UH, Forster J, Nielsen J, Borodina I (2014) EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res 14:238–248. https://doi.org/10.1111/1567-1364.12118
Jiang H, Horwitz AA, Wright C, Tai A, Znameroski EA, Tsegaye Y, Warbington H, Bower BS, Alves C, Co C, Jonnalagadda K, Platt D, Walter JM, Natarajan V, Ubersax JA, Cherry JR, Love JC (2019) Challenging the workhorse: comparative analysis of eukaryotic micro-organisms for expressing monoclonal antibodies. Biotechnol Bioeng 116:1449–1462. https://doi.org/10.1002/bit.26951
Jourdier E, Baudry L, Poggi-Parodi D, Vicq Y, Koszul R, Margeot A, Marbouty M, Bidard F (2017) Proximity ligation scaffolding and comparison of two Trichoderma reesei strains genomes. Biotechnol Biofuels 10:151. https://doi.org/10.1186/s13068-017-0837-6
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Koike H, Aerts A, LaButti K, Grigoriev IV, Baker SE (2013) Comparative genomics analysis of Trichoderma reesei strains. Ind Biotechnol 9:352–367. https://doi.org/10.1089/ind.2013.0015
Landowski CP, Mustalahti E, Wahl R, Croute L, Sivasiddarthan D, Westerholm-Parvinen A, Sommer B, Ostermeier C, Helk B, Saarinen J, Saloheimo M (2016) Enabling low cost biopharmaceuticals: high level interferon alpha-2b production in Trichoderma reesei. Microb Cell Fact 15:104. https://doi.org/10.1186/s12934-016-0508-5
Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, Martin J, Druzhinina IS, Mathis H, Monot F, Seiboth B, Cherry B, Rey M, Berka R, Kubicek CP, Baker SE, Margeot A (2009) Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci U S A 106:16151–16156. https://doi.org/10.1073/pnas.0905848106
Li W-C, Huang C-H, Chen C-L, Chuang Y-C, Tung S-Y, Wang T-F (2017) Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol Biofuels 10:170. https://doi.org/10.1186/s13068-017-0825-x
Li J, Chen Y, Gao A, Wei L, Wei D, Wang W (2023) Simultaneous production of cellulase and β-carotene in the filamentous fungus Trichoderma reesei. J Agric Food Chem 71:6358–6365. https://doi.org/10.1021/acs.jafc.3c00690
Ma C, Liu J, Tang J, Sun Y, Jiang X, Zhang T, Feng Y, Liu Q, Wang L (2023) Current genetic strategies to investigate gene functions in Trichoderma reesei. Microb Cell Fact 22:97. https://doi.org/10.1186/s12934-023-02104-3
Mathis H, Margeot A, Bouix M (2020) Optimization of flow cytometry parameters for high-throughput screening of spores of the filamentous fungus Trichoderma reesei. J Biotechnol 321:78–86. https://doi.org/10.1016/j.jbiotec.2020.05.015
Nakari T, Alatalo E, Penttilä ME (1993) Isolation of Trichoderma reesei genes highly expressed on glucose-containing media: characterization of the tef1 gene encoding translation elongation factor 1 alpha. Gene. 136(1–2):313–8. https://doi.org/10.1016/0378-1119(93)90486-m
Nakari-Setälä T, Penttilä M (1995) Production of Trichoderma reesei cellulases on glucose-containing media. Appl Environ Microbiol 61:3650–3655. https://doi.org/10.1128/aem.61.10.3650-3655.1995
Nevalainen H, Peterson R (2014) Heterologous expression of proteins in Trichoderma. In: Gupta VG, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy M (eds) Biotechnology and biology of Trichoderma. Elsevier, Amsterdam, pp 89–102. https://doi.org/10.1016/B978-0-444-59576-8.00007-2
Novikova LA, Yovkova V, Luzikov VN, Barth G, Mauersberger S (2021) Recombinant Yarrowia lipolytica strains for the heterologous expression of multi-component enzyme systems: expression of mammalian steroidogenic proteins. J Biotechnol 339:42–52. https://doi.org/10.1016/j.jbiotec.2021.07.012
Oguro Y, Yamazaki H, Shida Y, Ogasawara W, Takagi M, Takaku H (2015) Multicopy integration and expression of heterologous genes in the oleaginous yeast, Lipomyces starkeyi. Biosci Biotechnol Biochem 79:512–515. https://doi.org/10.1080/09168451.2014.982504
Penttilä M, Nevalainen H, Rättö M, Salminen E, Knowles J (1987) A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164. https://doi.org/10.1016/0378-1119(87)90110-7
Plüddemann A, van Zyl WH (2003) Evaluation of Aspergillus niger as host for virus-like particle production, using the hepatitis B surface antigen as a model. Curr Genet 43:439–446. https://doi.org/10.1007/s00294-003-0409-0
Qi H, Yu L, Li Y, Cai M, He J, Liu J, Hao L, Xu H, Qiao M (2022) Developing multi-copy chromosomal integration strategies for heterologous biosynthesis of caffeic acid in Saccharomyces cerevisiae. Front Microbiol 13:851706. https://doi.org/10.3389/fmicb.2022.851706
Qin L, Jiang X, Dong Z, Huang J, Chen X (2018) Identification of two integration sites in favor of transgene expression in Trichoderma reesei. Biotechnol Biofuels 11:142. https://doi.org/10.1186/s13068-018-1139-3
Sakai A, Shimizu Y, Hishinuma F (1990) Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl Microbiol Biotechnol 33:302–306. https://doi.org/10.1007/BF00164526
Sakai A, Ozawa F, Higashizaki T, Shimizu Y, Hishinuma F (1991) Enhanced secretion of human nerve growth factor from Saccharomyces cerevisiae using an advanced delta-integration system. Biotechnology (n y) 9:1382–1385. https://doi.org/10.1038/nbt1291-1382
Schuster A, Bruno KS, Collett JR, Baker SE, Seiboth B, Kubicek CP, Schmoll M (2012) A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol Biofuels 5:1. https://doi.org/10.1186/1754-6834-5-1
Semkiv MV, Dmytruk KV, Sibirny AA (2016) Development of a system for multicopy gene integration in Saccharomyces cerevisiae. J Microbiol Methods 120:44–49. https://doi.org/10.1016/j.mimet.2015.10.023
Silar P, Rossignol M, Haedens V, Derhy Z, Mazabraud A (2000) Deletion and dosage modulation of the eEF1A gene in Podospora anserina: effect on the life cycle. Biogerontology 1:47–54. https://doi.org/10.1023/A:1010029723081
Singh A, Taylor LE, Vander Wall TA, Linger J, Himmel ME, Podkaminer K, Adney WS, Decker SR (2015) Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol Adv 33:142–154. https://doi.org/10.1016/j.biotechadv.2014.11.009
Sun N, Qian Y, Wang W, Zhong Y, Dai M (2018) Heterologous expression of Talaromyces emersonii cellobiohydrolase Cel7A in Trichoderma reesei increases the efficiency of corncob residues saccharification. Biotechnol Lett 40:1119–1126. https://doi.org/10.1007/s10529-018-2564-x
Takeda AK, Udagawa H, Uemura Y, Takagi S (2018) Identifying the inserted locus of randomly integrated expression plasmids by whole-genome sequencing of Aspergillus strains. Biosci Biotechnol, Biochem 82:1880–1888. https://doi.org/10.1080/09168451.2018.1506312
The Galaxy Community (2024) The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucl Acids Res 52:W83–W94. https://doi.org/10.1093/nar/gkae410
Uzbas F, Sezerman U, Hartl L, Kubicek CP, Seiboth B (2012) A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl Microbiol Biotechnol 93:1601–1608. https://doi.org/10.1007/s00253-011-3674-8
van Dijk EL, Naquin D, Gorrichon K, Jaszczyszyn Y, Ouazahrou R, Thermes C, Hernandez C (2023) Genomics in the long-read sequencing era. Trends Genet 39:649–671. https://doi.org/10.1016/j.tig.2023.04.006
Vitikainen M, Arvas M, Pakula T, Oja M, Penttilä M, Saloheimo M (2010) Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics 11:441. https://doi.org/10.1186/1471-2164-11-441
Wohlschlager L, Csarman F, Zrilić M, Seiboth B, Ludwig R (2021) Comparative characterization of glyoxal oxidase from Phanerochaete chrysosporium expressed at high levels in Pichia pastoris and Trichoderma reesei. Enzyme Microb Technol 145:109748. https://doi.org/10.1016/j.enzmictec.2021.109748
Zheng J, Guo N, Zhou H-B (2014) A simple strategy for the generation of multi-copy Pichia pastoris with the efficient expression of mannanase. J Basic Microbiol 54:1410–1416. https://doi.org/10.1002/jobm.201400208
Zheng H, Wang K, Xu X, Pan J, Sun X, Hou J, Liu W, Shen Y (2022) Highly efficient rDNA-mediated multicopy integration based on the dynamic balance of rDNA in Saccharomyces cerevisiae. Microb Biotechnol 15:1511–1524. https://doi.org/10.1111/1751-7915.14010
Author information
Authors and Affiliations
Contributions
HM, AM, and FB designed the study. HM and FB conducted the experiments and analyzed the data. DN performed the bioinformatics analysis of the ONT data. HM and FB wrote the manuscript. All authors have read and approved the final manuscript for submission.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mathis, H., Naquin, D., Margeot, A. et al. Enhanced heterologous gene expression in Trichoderma reesei by promoting multicopy integration. Appl Microbiol Biotechnol 108, 470 (2024). https://doi.org/10.1007/s00253-024-13308-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00253-024-13308-x