Abstract
The methylotrophic bacterium Methylorubrum extorquens AM1 has the potential to become a platform organism for methanol-driven biotechnology. Its ethylmalonyl-CoA pathway (EMCP) is essential during growth on C1 compounds and harbors several CoA-activated dicarboxylic acids. Those acids could serve as precursor molecules for various polymers. In the past, two dicarboxylic acid products, namely mesaconic acid and 2-methylsuccinic acid, were successfully produced with heterologous thioesterase YciA from Escherichia coli, but the yield was reduced by product reuptake. In our study, we conducted extensive research on the uptake mechanism of those dicarboxylic acid products. By using 2,2-difluorosuccinic acid as a selection agent, we isolated a dicarboxylic acid import mutant. Analysis of the genome of this strain revealed a deletion in gene dctA2, which probably encodes an acid transporter. By testing additional single, double, and triple deletions, we were able to rule out the involvement of the two other DctA transporter homologs and the ketoglutarate transporter KgtP. Uptake of 2-methylsuccinic acid was significantly reduced in dctA2 mutants, while the uptake of mesaconic acid was completely prevented. Moreover, we demonstrated M. extorquens-based synthesis of citramalic acid and a further 1.4-fold increase in product yield using a transport-deficient strain. This work represents an important step towards the development of robust M. extorquens AM1 production strains for dicarboxylic acids.
Key points
• 2,2-Difluorosuccinic acid is used to select for dicarboxylic acid uptake mutations.
• Deletion of dctA2 leads to reduction of dicarboxylic acid uptake.
• Transporter-deficient strains show improved production of citramalic acid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Given the limited availability and global concern about the sustainability of fossil resources, the development of sustainable production processes for prevalent chemicals is indispensable. Currently, a new field of biotechnological processes is emerging that focuses on the use of alternative carbon sources. To be sustainable, these carbon sources must be derivable from renewable sources and must not compete with food production. Methanol, whose production share from renewable sources and waste streams starts to increase (Roode-Gutzmer et al. 2019), offers itself as an alternative raw material for biotechnological processes of the future. Moreover, its use as feedstock minimizes the risk of contamination during biotechnological applications and reduces the cost of downstream processing by using a defined minimal medium.
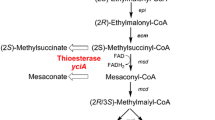
A group of widely used chemicals that are currently mainly produced from fossil raw materials are polyamides and polyesters. Biotechnologically produced dicarboxylic acids can serve as sustainable platform chemicals for the production of those polymers (Jang et al. 2012). Although pathways for microbiological production of dicarboxylic acids from classical feedstocks have been known for many years (Lee et al. 2011; Alonso et al. 2014), methanol-based routes for carboxylic acid production have been described only recently for the methylotrophic bacterium Methylorubrum extorquens AM1 (DSM 1338), e.g., for itaconic acid, 3-hydroxypropionic acid, mesaconic acid, and 2-methylsuccinic acid (Sonntag et al. 2014; Yang et al. 2017; Lim et al. 2019). The latter two are directly derived from the ethylmalonyl-CoA pathway (EMCP) by enzymatic hydrolysis (Fig. 1). The EMCP as an anaplerotic pathway is essential for methylotrophic growth of M. extorquens. It regenerates glyoxylate, which is needed for replenishment of the serine cycle and includes two CO2-fixing steps (Erb et al. 2007; Peyraud et al. 2009). The pathway is also essential for the assimilation of C2 compounds during growth on e.g. acetate in microorganisms having no glyoxylate pathway as M. extorquens (Erb et al. 2010; Okubo et al. 2010). The EMCP consists of 11 enzymes and includes several CoA-bound dicarboxylic acid intermediates with high biotechnological potential (Alber 2011). Mixtures of mesaconic acid and 2-methylsuccinic acid were produced from respective CoA-precursors by introducing the thioesterase YciA originating from Escherichia coli (Sonntag et al. 2014). The high carbon flux via the EMCP enabled remarkable product titers to be achieved without further modification of metabolic pathways. In both studies of Sonntag and colleagues (2014; 2015), the concentration of dicarboxylic acid products in supernatant was reduced remarkably (i.e., 45% or 60% over a period of 20 h, respectively) as soon as the cells were reaching the stationary growth phase. This behavior is probably attributable to product reuptake. An adjustment of the cobalt concentration in the growth medium from 12.6 µM to 0.2 µM did not only lead to a sixfold increase in product yield due to biomass formation-limiting inhibition of certain EMCP enzymes, but also decreased the reuptake of products in stationary growth phase (Sonntag et al. 2015). Despite the lack of obvious product reuptake under these low-cobalt conditions in the late cultivation phase, it remains unclear whether product uptake continuously proceeds during the acid production phase. This would resemble a partial futile cycle, leading to loss of resources and therefore decreased product yield. The phenomenon of product reuptake was also described for 3-hydroxypropionic acid, another EMCP-derived product from M. extorquens AM1, even when using a low cobalt concentration of 1.26 µM in the medium (Yang et al. 2017). These observations make it worth to investigate the product uptake mechanism and to prevent it on a molecular level.
Overview of ethylmalonyl-CoA pathway (EMCP) in M. extorquens AM1. The EMCP is interlaced with the serine cycle, the PHB cycle, and the tricarboxylic acid cycle. Cleavage of (2R/3S)-methylmalyl-CoA by malyl-CoA/beta-methylmalyl-CoA lyase (mcl) releases glyoxylate, which replenishes the serine cycle (indicated by boxes). Further genes of EMCP are: β-ketothiolase (phaA); acetoacetyl-CoA reductase (phaB); crotonase (croR); crotonyl-CoA carboxylase/reductase (ccr); ethylmalonyl-CoA epimerase (epi); ethylmalonyl-CoA mutase (ecm); methylsuccinyl-CoA dehydrogenase (msd); mesaconyl-CoA dehydratase (mcd); propionyl-CoA carboxylase (pcc); methylmalonyl-CoA mutase (mcm). Expression of heterologous thioesterase-encoding gene yciA leads to hydrolysis of (2S)-methylsuccinyl-CoA and mesaconyl-CoA (bold blue arrows). The corresponding products, namely 2-methylsuccinic acid and mesaconic acid, are released into the supernatant
The main targets for prevention of product uptake are the dicarboxylic acid import proteins. It has been observed that a 30-fold reduction in sodium concentration in the growth medium resulted in ceasing of product uptake (Sonntag et al. 2015), suggesting that carriers of the DctA family may be involved, which are known to be sodium-dependent (Janausch et al. 2002). A transposon mutant study by Van Dien and coworkers identified a mutant with a disrupted dctA homolog that was no longer able to use succinate as sole carbon source (Van Dien et al. 2003). The affected protein was referred to as GenBank entry A33597. Since this mutant strain was promising for dicarboxylic acid production, the gene encoding the protein with the highest similarity to A33597 was deleted by Sonntag et al. (2015). Surprisingly, the strain did not show the phenotype previously described. Sonntag et al. (2015) speculated that the deleted gene might not be identical to the gene mutated in the Van Dien study, as the GenBank entry A33597 in question represents a Sinorhizobium meliloti protein sequence and therefore cannot be unambiguously assigned.
Another possibility of preventing the reuptake of products is to convert them to non-metabolizable molecules. For example, the hydration product of mesaconic acid, namely (S)-citramalic acid, would not only fulfill this criterion, but would also extend the product spectrum of M. extorquens AM1 by a chiral, enantiopure product. The mesaconase/fumarate hydratase from Paraburkholderia xenovorans (Kronen et al. 2015) seems promising for heterologous expression for this purpose.
In our study, we aimed at reducing reuptake of dicarboxylic acids at the molecular level. Using a simple and straightforward selection approach, we identified a mutant (partial deletion of dctA2) with reduced uptake of mesaconic acid and 2-methylsuccinic acid. We confirmed our results by construction of deletion mutants and were able to rule out the involvement of other DctA transporters. In addition, we have successfully implemented the production of a new dicarboxylic acid product from M. extorquens AM1, whose production also benefits from the transport deficiency. These new insights into dicarboxylic acid transport contribute to the development of M. extorquens AM1 towards a comprehensive production platform for methanol-based biotechnology.
Material and methods
Bacterial strains and growth conditions
Escherichia coli DH5α (Hanahan 1985; Grant et al. 1990) cultures were grown in LB medium (Bertani 1951) at 37 °C. For cultivation of M. extorquens AM1 (DSM 1338, Peel and Quayle 1961), minimal medium was prepared as described before (Peyraud et al. 2009), containing 123 mM methanol, 31 mM sodium succinate or 5 mM sodium acetate as carbon source, respectively. CoCl2 concentration in the medium was set to 12.6 µM (Kiefer et al. 2009; Sonntag et al. 2014). For solid medium, agar–agar at 15 g/L was added. For cultivation of M. extorquens AM1 in liquid medium, 5 mL precultures in methanol minimal medium were grown in test tubes for 48 h at 30 °C and 180 rpm on a rotary shaker. Main cultures of 20 mL were subsequently inoculated to an OD600 of 0.1 in 100 mL shake flasks and incubated under the same conditions and in the same medium as the precultures. If required, tetracycline-hydrochloride at 10 μg/mL, kanamycin sulfate at 30 μg/mL (for E. coli), or kanamycin sulfate at 50 μg/mL (for M. extorquens AM1) was added to the medium. For high-resolution measurements of growth curves, 1 ml of main cultures were incubated in a BioLector® microbioreactor system in 48-well Flowerplates® (m2p-labs GmbH, Baesweiler, Germany) at 30 °C and 1000 rpm. For complementation experiments, glyoxylate was added to a final concentration of 370 mg/L (5 mM). Depending on the specific production experiment, heterologously expressed genes encoding thioesterase YciA from either E. coli (YciAEc) or H. influenza (YciAHI) and a gene encoding a fumarate hydratase (mesaconase) from Paraburkholderia xenovorans (MesaPx) was used. The two thioesterases produce similar amounts of products with slightly different ratios. For YciAHI, a crystal structure is available (3BJK, Willis et al. 2008).
Chemicals
All chemicals were purchased from Carl Roth (Karlsruhe, Germany), VWR International (Darmstadt, Germany), or Merck (Darmstadt, Germany). Solvents for chromatography were purchased in LC–MS grade quality.
Genome sequencing
DNA of M. extorquens AM1 wild type and mutant strains was sequenced by Illumina sequencing (MiSeq; 2 × 250 bp; 1–2 million PE-Reads; GenXPro, Frankfurt, Germany). Paired and cleaned Illumina reads were trimmed with BBDuk Trimmer (JGI, Berkeley, USA) to a quality cut-off of 20. Mapping to the reference genome (NCBI NC_012807–NC_012811, annotation date 04/11/2021) and calling for SNPs was done with Geneious Prime (Biomatters, Auckland, New Zealand). Assembling qualities and base coverages are listed in Online Resource Table S1.
DNA cloning and plasmid construction
All standard plasmid cloning procedures were performed in E. coli DH5α. Plasmid DNA was purified with GeneJET Plasmid Miniprep Kit from Thermo Scientific (Waltham, USA). Polymerase chain reactions (PCR) were performed with Q5 Polymerase from New England Biolabs (Frankfurt, Germany) according to the manufacturer’s protocol. Subsequently, PCR products were purified using the DNA Clean & Concentrator Kit from Zymo Research Europe (Freiburg, Germany). Oligonucleotides were purchased from Merck (Darmstadt, Germany), restriction enzymes and T4 ligase from NEB. All genetic constructs were confirmed by Sanger sequencing at Eurofins Scientific (Luxembourg, Luxembourg). Transformation of M. extorquens AM1 was done as described before (Toyama et al. 1998). Genomic DNA of M. extorquens AM1 was purified with GenElute™ Bacterial Genomic DNA Kit from Merck (Darmstadt, Germany).
Generation of dicarboxylic acid transporter deletion mutants
Knockout of the genes encoding potential carboxylic acid importers (dctA1: MEXAM1_RS15430, dctA2: MEXAM1_RS10985 and dctA3: MEXAM1_RS20450) and the ketoglutarate permease KgtP (kgtP: MEXAM1_RS24315) were carried out with allelic exchange vector pCM184 carrying a kanR antibiotic resistance cassette (Marx and Lidstrom 2002). Vector pCM184_∆dctA1 was constructed as described in the publication by Sonntag et al. (2015), in which dctA1 is referred to as dctA. The deletion vectors pCM184_∆dctA2, pCM184_∆dctA3 and pCM184_∆kgtP were designed accordingly in two constitutive cloning steps. First, 500 bp of genomic upstream flanking region of the respective gene of interest was amplified while introducing restriction sites at both ends of the PCR product. The resulting fragment was digested with according restriction enzymes and ligated to equally digested pCM184 (Table 1). The resulting vectors carrying the upstream fragment were subsequently treated in the same manner to introduce the previously digested downstream fragment to result in the final allelic exchange vectors. After restreaking M. extorquens AM1 transformants on solid methanol medium containing kanamycin, the correct integration of resistance marker was verified with respective primer pairs, binding inside and outside of the cassette. Screening for single recombination mutants and removal of the resistance marker with cre-expression vector pCM157 was achieved on solid methanol medium containing tetracycline as described by Marx and Lidstrom (2002). Elimination of pCM157 was performed by cultivating strains overnight in medium without antibiotics and screening single colonies for tetracycline sensitivity. Deletion of the complete ORF was double checked by PCR of genomic DNA. All used oligonucleotides are listed in Table 1. For multiple gene deletions, the described procedure was performed several times in succession. Single, double and triple deletion mutants were simultaneously created in M. extorquens AM1 wild type as well as in the ∆cel strain (Delaney et al. 2013).
Dicarboxylic acid analysis
For quantification of dicarboxylic acid products, the supernatant of M. extorquens AM1 cultures was centrifuged for 5 min at 16.000 g and passed through a 0.22 μm PDVF-syringe filter (Carl Roth, Karlsruhe, Germany). For the quantification of mesaconic acid and 2-methylsuccinic acid, 10 µL of the cell free supernatant was chromatographed on a 150 × 4.6 mm Rezex™ ROA-Organic Acid H + (8%) column (Phenomenex, Aschaffenburg, Germany) at 30 °C oven temperature in a SLC10-A HPLC system (Shimadzu, Duisburg, Germany). 5 mM H2SO4 in MilliQ water was used as mobile phase. Samples were analyzed with a SPD20A UV–Vis detector at 205 nm as described in Sonntag et al. (2014). Retention times of the analytes are listed in Table 2. For the quantification of samples containing additional dicarboxylic acid products (e.g., citramalic acid), which all have similar retention times, an alternative LC–MS/MS setup was used. Chromatography of 1 µL of the respective samples was done on a 150 × 4.6 mm Luna Omega 3 µm PS C18 100 Å column (Phenomenex, Aschaffenburg, Germany) in a Nexera X2 UHPLC system (Shimadzu, Duisburg, Germany). Separation was performed isocratically at 12% [v/v] acetonitrile and 88% [v/v] ddH2O, both containing 0.2% [v/v] formic acid at 40 °C oven temperature. The column was washed after every run by raising the acetonitrile content in the mobile phase to 95%. Mesaconic acid, 2-methylsuccinic acid, citramalic acid as well as other potential EMCP-derived carboxylic acid products (Table 2) were analyzed on a LCMS-8045 system (Shimadzu). The analytes were negatively ionized with APCI or ESI ion source, fragmented and finally quantified by comparing the results to calibration curves of corresponding standards. Quantification was performed with the LabSolutions software (Shimadzu). The retention times and the manufacturers of the standard substances are listed in Table 2. Fragmentation of (2S,3R)-2-hydroxy-3-methylsuccinic acid and (S)-citramalic acid for unambiguous identification is shown in Online Resource Fig. S1.
Transcriptome analysis
A whole transcriptome analysis was done for M. extorquens AM1 harboring pCM160_RBS_yciAHI or pCM160, respectively. Samples were taken after 22.75, 31.75, 47, 51.25 and 70.5 h of cultivation. For stabilization of RNA, one volume of growing bacterial culture (> 106 cells) was mixed with two volumes of RNAprotect Bacteria Reagent from QIAGEN (Hilden, Germany) and incubated for 5 min. Cells were centrifuged for 10 min at 5000 g to remove supernatant and stored pelleted at − 80 °C. RNA was isolated and analyzed by GenXPro (Frankfurt, Germany) via Illumina sequencing (2–5 million reads; 2 × 75 bp). Raw read counts were normalized by CPM (counts per million) method to compensate for varying sample sequencing depth. For graphical representation, transcript levels were scaled to the count of the respective gene in the control strain at the first sampling point. The transcriptome dataset was deposited at NCBI (accession number GSE199961).
Results
Search for dicarboxylic acid transporter candidates by transcriptome analysis
M. extorquens AM1 naturally produces small amounts of dicarboxylic acids derived from the EMCP pathway. Sonntag and colleagues succeeded in increasing the amounts of released 2-methylsuccinic acid and mesaconic acid to a combined titer of 0.13 g/L by expressing the thioesterase encoding gene yciA from E. coli heterologously in a methanol minimal medium containing 12.6 µM CoCl2 (Sonntag et al. 2014, 2015). However, at the end of the exponential growth phase, the acid concentrations started to decrease again, which is probably caused by product-reuptake and metabolization. It is unclear whether product uptake also occurs during the exponential growth phase, which could reduce the overall yield. To address this question, we investigated the dicarboxylic acid importers of M. extorquens AM1. A number of candidates with homology to the multispecies dicarboxylic acid transporter DctA (Janausch et al. 2002) were identified by BLAST analysis by Sonntag et al. (2014). In the referred study (Sonntag et al. 2014), unexpectedly and in contrast to other publications (Van Dien et al. 2003), a dctA knockout mutant (MEXAM1_RS15430) was still able to grow on succinate. Therefore, it was questioned whether dctA (hereinafter referred to as dctA1) from both studies was in fact the identical ORF. Sonntag et al. also identified two other dctA homologs (MEXAM1_RS10985 and MEXAM1_RS20450, referred to as dctA2 and dctA3 in our study) and an additional dicarboxylic acid transporter encoding gene (kgtP MEXAM1_RS24315, Seol and Shatkin 1991) in the genome sequence, but these were not further investigated (Sonntag et al. 2014). To monitor expression levels of the homolog candidates and to potentially identify additional dicarboxylic acid importers in our study, M. extorquens AM1 cells expressing an alternative thioesterase gene from Haemophilus influenzae (yciAHI) were analyzed on a whole transcriptome level together with an empty vector control strain. Additionally, product titers were determined. Small amounts of the EMCP derived carboxylic acids (2S,3R)-2-hydroxy-3-methylsuccinic acid, crotonic acid, ethylmalonic acid, methylmalonic acid and succinic acid were detected, but their titers were irrelevant and insufficient for quantification (< 5 mg/L, also present in the empty vector control culture). M. extorquens AM1 cells harboring plasmid pCM160_RBS_yciAHI released up to 104 ± 8 mg/L 2-methylsuccinic acid and up to 85 ± 9 mg/L mesaconic acid in the production phase, followed by clear decreases of 2-methylsuccinic acid and mesaconic acid titers (Fig. 2a).
Transcriptome analysis of dicarboxylic acid production strain to identify candidates for product uptake factors. a–b Growth kinetics and time-dependent concentration of mesaconic acid and 2-methylsuccinic acid in supernatant of M. extorquens AM1 + pCM160_RBS_yciAHI (a) or M. extorquens AM1 + pCM160 (b) growing in methanol minimal medium. Other EMCP-derived carboxylic acid products with titers insufficient for quantification (< 5 mg/L) are not displayed. c Normalized relative gene expression of dctA1, dctA2, dctA3, and kgtP for M. extorquens AM1 + pCM160_RBS_yciAHI. d Normalized relative gene expression of dctA1, dctA2, dctA3, and kgtP for M. extorquens AM1 + pCM160. The transcript levels are scaled to the transcript count of the respective gene in the control strain M. extorquens AM1 + pCM160 at 23 h of cultivation. Error bars represent standard deviations from three independent replicates. An additional visualization of the data in form of products per OD600 can be found in Online Resource Fig. S2
The relative expression levels of dicarboxylic acid transporter candidates identified by Sonntag and colleagues (2014) were analyzed in the production strain as well as in cells harboring an empty control vector. Hydrolysis of EMCP-esters by a heterologous thioesterase is a strong intervention in the primary carbon metabolism of the cell. This is reflected by the different growth kinetics of production and control strain (Fig. 2a,b). Therefore, we decided not to perform an analysis of differential gene expression. Instead, relative gene expression was determined for each strain separately, before comparing the kinetic patterns of both strains. The gene dctA1, which has been already deleted in the work of Sonntag et al. (2015), showed a higher relative gene expression rate compared to control strains (Fig. 2c,d). This effect is particularly noticeable in the early sampling points, whereas expression was comparably strong during the main acid production phase (e.g., equivalent timepoints 47 h in production strains vs. 23 h or 32 h in control strains). Furthermore, moderately increased expression levels at later time points in the production strain were observed for dctA2, dctA3, and kgtP but at least for dctA2 and kgtP a similar pattern was found in the non-producing reference strain.
We could not identify any additional genes with acid transporter annotation within the transcriptome data that were significantly higher expressed in cells sampled at the late stages of cultivation (51 h or 71 h) compared to the earlier sampling time points in the data set. Therefore, the transcriptome analysis of M. extorquens AM1 harboring pCM160_RBS_yciAHI yielded no candidates for potential dicarboxylic acid uptake factors.
Investigation of 2,2-difluorosuccinic acid as potential selection agent for dicarboxylic acid transport mutants
Since the transcriptome analysis did not provide clear hints for genes with clear upregulation before the phase of product reuptake, a more direct approach was considered to identify cells incapable of dicarboxylic acid uptake. For selection for respective M. extorquens cells with an uptake defect, 2,2-difluorosuccinic acid (DFS), a presumably cytotoxic dicarboxylic acid with structural similarity to the target products, was used (Fig. 3).
To investigate the effect of DFS, the growth of M. extorquens AM1 in minimal medium containing different carbon sources and different concentrations of DFS was monitored in a microbioreactor system. Although the differences between replicates growing on succinate medium were quite high, the results showed, that the addition of 2, 5, or 10 mg/L DFS had no significant effect on growth (Fig. 4a). In contrast, the addition of DFS to acetate or methanol containing cultures resulted in suppression of growth and prolonged lag phases (Fig. 4b,c). The addition of 2 mg/L DFS to the methanol medium resulted only in a slight delay in growth. In turn, the addition of 5 mg/L resulted in a greater delay and a reduction of the maximum cell density, and the addition of 10 mg/L DFS completely inhibited growth (Fig. 4c). Although maximum cell densities were lower, similar effects were observed when cells were cultured in acetate medium (Fig. 4b). Since an active EMCP in M. extorquens AM1 is required for the use of methanol or acetate as sole carbon source (Peyraud et al. 2009), we assumed that DFS might target the EMCP. This assumption was reinforced by the fact, that the DFS-mediated growth defect in methanol medium could be partially compensated by the addition of glyoxylate (Fig. 4d). The provision of glyoxylate is the essential function of the EMCP during growth on C1 or C2 compounds (Fig. 1, Peyraud et al. 2009) and the rescue of EMCP mutants by glyoxylate addition was already demonstrated many years ago (Salem and Quayle 1971; Chistoserdova and Lidstrom 1996).
Investigation of toxic effects of DFS on M. extorquens AM1. a–c Growth of M. extorquens AM1 in succinate, acetate, or methanol minimal medium with different additives. Cultures were grown in a microbioreactor. Growth was monitored by scattered light signal. 2,2-Difluorosuccinic acid (DFS) was added at the start of cultivation in concentrations of 2 mg/L, 5 mg/L, or 10 mg/L. M. extorquens AM1 culture without DFS was used as a negative control. For better readability, figure part b is scaled differently. d Complementation experiment, in which 370 mg/L of glyoxylate was added to cultures containing 5 mg/L or 10 mg/L of DFS, respectively. Colored areas represent standard deviations from three independent replicates
When DFS was added to an M. extorquens AM1 culture expressing the E. coli yciA gene, we observed a growth delay but at the same time the production of 2-methylsuccinic acid and mesaconic acid was raised to a maximum measured combined product titer of 153 mg/L ± 2 mg/L (Fig. 5). Even though the measured time points can only give an indication of the complete production kinetics, the product titers were clearly increased compared to the control experiment without DFS addition. This experiment provided further indication of an EMCP inhibitory effect, as EMCP flux limitation leads to increased dicarboxylic acid production in thioesterase expression strains (Sonntag et al. 2015).
Growth kinetics and time-dependent combined concentration of dicarboxylic acid products (mesaconic acid and 2-methylsuccinic acid) in supernatant of M. extorquens AM1 harboring pCM160_RBS_yciAEc in methanol minimal medium (filled symbols) and in methanol minimal medium with addition of 5 mg/L 2,2-difluorosuccinic acid (DFS) after 5 h of cultivation (empty symbols). Other EMCP-derived carboxylic acid products with titers insufficient for quantification (< 5 mg/L) are not displayed. Error bars represent standard deviations from two independent replicates. An additional visualization of the data in form of products per OD600 can be found in Online Resource Fig. S3
Altogether, our investigations showed DFS to be a suitable selection agent for dicarboxylic acid uptake mutants. However, the mutant selection has to be performed under conditions, which require a functional EMCP, as this pathway seems to contain the molecular target for the compound.
Identification of potential dicarboxylic acid uptake transporters by using DFS
To isolate DFS-resistant strains, an M. extorquens AM1 culture was transferred to solid minimal medium containing acetate or methanol as carbon source and additional 5 mg/L DFS. After 18 days of incubation, a small number of colonies could be observed (one colony on methanol medium, 20 colonies on acetate medium). One strain from each medium was isolated. Restreaking the cells on solid succinate, methanol, or acetate minimal medium resulted in wild-type-like growth behavior. Isolated M. extorquens DFS mutant 1 (isolated from acetate-DFS minimal medium) and DFS mutant 2 (isolated on methanol-DFS minimal medium) were transformed with plasmid pCM160_RBS_yciAEc and tested for dicarboxylic acid production along with an M. extorquens AM1 wild type strain also containing plasmid pCM160_RBS_yciAEc. None of the strains showed a growth defect in liquid methanol minimal medium (Fig. 6a-c). Although a slight decrease in mesaconic acid concentration was still observed in the culture of DFS mutant 1 after 40 h of cultivation, it showed significantly reduced product reuptake compared to the wild type control strain. DFS mutant 2 showed production/uptake kinetics similar to the wild type control. The genomes of the wild type control strain, DFS mutant 1 and DFS mutant 2 were sequenced to identify any new mutations. A total of 11 identical mutations were found in all three strains, which differ from the NCBI reference genome sequence (Online Resource Table S2). In DFS mutant 1, an additional 12 bp deletion within the dicarboxylic acid transporter gene dctA2 (MEXAM1_RS10985) could be identified (Fig. 6d). Although the dctA2 gene product is probably involved in product reuptake, a residual uptake was still observed in DFS mutant 1. In DFS mutant 2, a single point mutation was identified causing an amino acid exchange in a SLC13 family permease (MEXAM1_RS18205, Fig. 6e). This mutation was probably responsible for the DFS resistance phenotype. Nevertheless, the strain did not show a decreased reuptake of mesaconic acid and 2-methylsuccinic acid behavior (Fig. 6c) observed for DFS mutant 1.
Phenotypes and genotypes of DFS-resistant mutants. a–c Growth kinetics and time-dependent concentration of mesaconic acid and 2-methylsuccinic acid in supernatant of M. extorquens AM1 wild type + pCM160_RBS_yciAEc (a), DFS mutant 1 + pCM160_RBS_yciAEc (b), and DFS mutant 2 + pCM160_RBS_yciAEc (c), growing in methanol minimal medium. Other EMCP-derived carboxylic acid products with titers insufficient for quantification (< 5 mg/L) are not displayed. Error bars represent standard deviations from three independent replicates. d Gene locus region of MEXAM1_RS10985-MEXAM1_RS1097, in which DFS mutant 1 has a 12 bp deletion within the dicarboxylic acid transporter MEXAM1_RS10985 (≙ dctA2). e Gene locus region MEXAM1_RS18205-MEXAM1_RS18230, in which a mutation in a SLC13 family permease encoding gene could be identified in the genome of DFS mutant 2. An additional visualization of the data in form of products per OD600 can be found in Online Resource Fig. S4
Mesaconic and 2-methylsuccinic acid production behavior of strains lacking one or several transporter-encoding genes
The 12 bp deletion in the dctA2 gene in DFS mutant 1 conferred a reduced reuptake of mesaconic acid and 2-methylsuccinic acid, yet the reuptake was only partially prevented. This could indicate that the transporter protein variant lacking four amino acids is still partially active or that more than one import protein is involved in the reuptake of the products. Aiming at complete suppression of reuptake, we constructed single, double and triple deletion strains lacking one, two or three putative dicarboxylic acid transporter genes (start to stop codon). Besides dctA2, this deletion approach involved also dctA1 and dctA3. Additionally, a ∆kgtP knockout strain was constructed and analyzed. To also obtain strains with high process suitability, we introduced the deletions not only in M. extorquens AM1, but also in M. extorquens AM1 ∆cel. Since the latter lacks the cellulose synthesis operon, it has been shown to be more suitable for downstream applications as biofilm formation and cell clumping are reduced (Delaney et al. 2013). Although we attempted to achieve a triple dctA deletion by testing different sequences of consecutive gene deletions, not all combinations could be achieved in both, the wild type and the ∆cel strain. Nevertheless, all possible combinations could be achieved in one of the two strains. Since production behavior did not differ significantly between both strains, the phenotypes of all combinations of transporter gene deletions could be analyzed. Since we observed that YciAHI resulted in higher product titers compared to YciAEc (e.g., Fig. 2 versus Fig. 5) and a crystal structure for YciAHI is available that may facilitate further developments (3BJK, Willis et al. 2008), the YciAHI thioesterase was chosen over the E. coli enzyme for the experiment. It was introduced into all constructed transporter deletion strains to investigate the effects on product reuptake during shake flask cultivations with methanol minimal medium. Single deletions of dctA1, dctA3, or kgtP had no effect on the production or reuptake behavior of mesaconic acid or 2-methylsuccinic acid compared to the reference strains (Fig. 7a,b). Only the effect caused by complete deletion of the dctA2 gene is clearly evident and it is comparable to the effect caused by the 12 bp deletion in the dctA2 gene in DFS mutant 1. The same applies to strains with double or triple deletions: Only in strains containing at least the dctA2 deletion, a reduced dicarboxylic acid uptake could be observed after cell growth stopped, whereas additional deletions did not lead to additional phenotypes. In all ∆dctA2 strains, product levels increased during the exponential growth phase of the cells as in the control strains. The product concentrations per biomass during the growth phase and therefore also the maximum product levels determined at the end of the exponential growth phase were, however, not higher than those observed in the production strain without transporter gene deletions (Fig S5a). Once the cells entered the stationary phase, the consumption of 2-methylsuccinic acid was clearly reduced while the mesaconic acid decrease was almost completely prevented. Therefore, in addition to a higher maintenance of the product levels, the ratio between the two products changed in the course of the stationary phase.
Growth kinetics and time-dependent concentration of mesaconic acid and 2-methylsuccinic acid in supernatant of M. extorquens AM1 cells without and with single, double or triple transporter deletions. Strains heterologously express thioesterase encoding gene yciAHI in methanol minimal medium. Other EMCP-derived carboxylic acid products with titers insufficient for quantification (< 5 mg/L) are not displayed. Error bars represent standard deviations from three independent replicates. Strains were constructed based on either a M. extorquens AM1 wild type or b M. extorquens AM1 ∆cel strain. An additional visualization of the data in form of products per OD600 can be found in Online Resource Fig. S5
Investigation of transporter deletion effects on citramalic acid production
An alternative way to reduce reuptake of dicarboxylic acid products is by converting them to non-metabolizable products. By introduction of a mesaconase/fumarate hydratase from P. xenovorans (Kronen et al. 2015) in addition to YciAEc, we were able to convert mesaconic acid to citramalic acid by enzymatic hydration (Fig. 8a). Analytical evidence for the conversion is given in Online Resource Fig. S1. In contrast to mesaconic acid and 2-methylsuccinic acid, citramalic acid is most probably not taken up and metabolized by the cell, as its concentration stayed stable during the stationary phase (Fig. 8b,c). However, cell density values and product measurements of M. extorquens AM1 and M. extorquens AM1 ∆cel strains carrying pCM160_yciAEc_RBS_mesaPx showed high standard deviations for the biological replicates. The maximal citramalic acid concentration measured was 164 ± 71 mg/L or 170 ± 49 mg/L, respectively (Fig. 8b,c, left graphs). Use of corresponding triple dctA deletion strains resulted in clearly higher citramalic acid concentrations of approximately 249 ± 5 mg/L or 241 ± 4 mg/L, respectively (Fig. 8b,c, right graphs). Moreover, in both strain backgrounds, the transporter gene deletions obviously reduced the variations observed within three independent transformants.
Production of citramalic acid with strains lacking DctA transporters. a Reaction scheme for hydration of mesaconic acid to (S)-citramalic acid catalyzed by mesaconase/fumarate hydratase (from Kronen et al. 2015). Absolute stereochemistry and double bond geometry are labeled in gray. b Growth kinetics and time-dependent concentration of mesaconic acid, 2-methylsuccinic acid and citramalic acid in supernatant of M. extorquens AM1 wild type and triple dctA transporter deletion strain expressing thioesterase encoding gene yciAHI and mesaconase in methanol minimal medium. c Growth kinetics and time-dependent concentration of mesaconic acid, 2-methylsuccinic acid and citramalic acid in supernatant of ∆cel and ∆cel triple dctA transporter deletion strain expressing thioesterase encoding gene yciAHI and mesaconase from P. xenovorans in methanol minimal medium cultures. Other EMCP-derived carboxylic acid products with titers insufficient for quantification (< 5 mg/L) are not displayed. Error bars represent standard deviations from three independent transformants. An additional visualization of the data in form of products per OD600 can be found in Online Resource Fig. S6
Discussion
In this study, we focused on the reduction of product reuptake in M. extorquens AM1 strains producing dicarboxylic acids as 2-methylsuccinic acid and mesaconic acid. This will serve as a basis for the development of stable and more productive strains for the future production of dicarboxylic acids from methanol.
In the past, product reuptake in the later cultivation phases reduced the product yield (Sonntag et al. 2014, 2015). It was postulated that there are two possible modes for product uptake: 1) it may be triggered upon transition from exponential to stationary growth phase or 2) the product uptake takes place over the entire course of cultivation and is masked by the high production rates during the early cultivation stages (Sonntag et al. 2014). In the second case, the productivity in the exponential growth phase would be reduced by the presence of a futile cycle. By repressing the reimport of products, one could eliminate this cycle and thus achieve higher productivity.
To identify potential import factors, we performed transcriptome analysis of a strain harboring plasmid pCM160_RBS_yciAHI, encoding the thioesterase YciA from Haemophilus influenzae. However, we could not identify genes that were clearly upregulated in stationary growth phase compared to early sampling points in the exponential growth phase and which were annotated to encode for transporters. When specifically analyzing the time-dependent expression levels of the dctA homologs and kgtP (targets suggested by Sonntag and colleagues (2014)) and comparing them with a control strain, we found slightly increased levels only for dctA1. After concluding that the transcriptome analysis was no appropriate approach to identify relevant transport factors, we chose a straightforward mutant selection approach using a cytotoxic dicarboxylic acid.
In preliminary tests, DFS toxicity surprisingly applied only to methanol- and acetate-grown and not to succinate-grown cells. As its toxicity could furthermore be compensated by supplementing the cells with glyoxylate, we concluded that DFS toxicity is not only mediated by the EMCP but directly targets it. Since DFS has a high structural similarity to the dicarboxylic acids that constitute the EMCP intermediates in their CoA-activated form, DFS might act as competitive inhibitor. Glyoxylate supplementation has previously been shown to fully restore methylotrophic growth of Δccr, Δepi, and Δecm strains and to partially restore growth of Δmsd strains (Schada von Borzyskowski et al. 2018), so we can speculate that DFS acts on one or several of the corresponding EMCP enzymes.
Using a simple selection approach, we were able to isolate DFS-resistant M. extorquens AM1 mutants from DFS-containing methanol minimal medium or acetate minimal medium. In one of the two mutants selected for further investigations, DFS resistance was accompanied by reduced uptake of mesaconic acid and 2-methylsuccinic acid. DFS mutant 2, in which we identified a mutation in a putative dicarboxylic acid transporter encoding gene (SLC13 family), was resistant to DFS. Nevertheless, the strain showed 2-methylsuccinic acid and mesaconic acid uptake behavior comparable to a non-mutated control strain. In contrast, mutation or deletion of dctA2, led to strong reduction of product reuptake. The corresponding mutant strain, DFS mutant 1, not only exhibited the DFS resistance phenotype we selected for, but in addition showed clearly reduced reuptake of mesaconic acid and 2-methylsuccinic acid. Since the strain was still able to grow on succinate, we assume that dctA2 is not identical to the open reading frame designated as A33597 in Van Dien’s study (Van Dien et al. 2003).
Although we did not perform complementation experiments with dctA2, strong indications for a dicarboxylic acid uptake function of DctA2 exist. The fact that also a decreased sodium concentration resulted in a ceasing of dicarboxylic acid reuptake (Sonntag et al. 2015) is in line with this assumption, as DctA transporters are sodium-dependent (Janausch et al. 2002). The finding of identical acid uptake phenotypes with a dctA2 deletion strain is an additional argument for DctA2 to be the causal factor for the observed reuptake defect. It furthermore demonstrated that the removal of four amino acids from the protein in the DFS mutant 1 probably results in a complete loss of function with respect to mesaconic acid and 2-methylsuccinic acid uptake. Possible explanations for the residual uptake activity observed for dctA2 mutants were contributions of DctA2 homologs DctA1 and DctA3. After we found out that single deletions of dctA1 or dctA3 did not cause changes to the product uptake kinetic, we initially hypothesized that these factors might nevertheless be responsible for the residual uptake activity seen for the dctA2 mutants. A conceivable scenario would be that transcription of their genes is activated when DctA2 is absent. However, we ruled out this possibility by testing mutant strains in which all three dctA genes were deleted. Only combinations of deletions that included the dctA2 deletion resulted in clearly reduced product reuptake. The fact that product yields were maintained in stationary phase but that no product increase per biomass could be observed during growth for the dctA2 mutants compared to the wild type (Fig. S5a), may suggest that uptake is likely to only occur if a threshold of product concentration is reached or carbon becomes limiting. Another aspect revealed from the analysis of the production kinetics of the dctA2 mutant strains is based on the observation that the reduced reuptake does not apply to the two products in the same way. Expression of yciA from H. influenzae in M. extorquens AM1 wild type strain in our experiments usually resulted in release of similar amounts of the two dicarboxylic acid products, with a slightly higher level of 2-methylsuccinic acid. The dctA2 deletion seems especially to prevent mesaconic acid uptake, while 2-methylsuccinic acid is still imported to a certain extent. This observation once more suggests the presence of other, yet unidentified transporters involved at least in 2-methylsuccinic acid uptake. The possibility of passive diffusion of completely protonated acids through the membrane is furthermore ruled out by the different kinetics of the two acids. As the two products have comparable pKa values and show high structural similarity, strong differences in membrane passing rates are not expected.
Besides prevention of product reuptake in the stationary phase, we also aimed at elimination of product uptake potentially taking place in the production phase during exponential growth. In the experiments conducted, the mesaconic acid and 2-methylsuccinic acid production kinetics in the exponential growth phase was the same, both with and without dctA2 deletion. Although the overall product titer was not increased, the process is much more robust when performed in a dctA2 deletion strain as the harvest time for achieving maximum product yield is no longer as time critical.
For the production of citramalic acid, however, the use of the dctA triple deletion strain resulted not only in a significant gain in robustness, but also in production yield. Citramalic acid as a non-natural product of M. extorquens AM1 is not reimported by the cells, as shown by the respective experiment with the control strain. Although a clear explanation for the improvements of citramalic acid production in the DctA mutant strain compared to the control strain cannot be drawn from the experiments, reduced mesaconic acid uptake is probably causal for the observed phenotypes. This reduced uptake might slow down its further metabolization and thereby increase the carbon flux towards citramalic acid. The use of a dctA triple deletion strain for citramalic production led to a 1.4-fold increase in mean product yield and a substantial reduction of the standard deviations. The study that first described mesaconase/fumarate hydratase from P. xenovorans demonstrated its enantioselectivity towards (S)-citramalic acid (Kronen et al. 2015). We therefore assumed that the constructed M. extorquens AM1 strains also produce enantiopure (S)-citramalic acid.
Although we were not able to achieve higher overall yields for mesaconic acid or 2-methylsuccinic acid, the use of a dctA2 deletion strain almost completely prevented the reuptake of mesaconic acid and part of 2-methylsuccinic acid within the stationary growth phase. Furthermore, the product yield of citramalic acid was significantly increased in the dctA2 deletion strain. Therefore, the dicarboxylic acid transporter deletion mutants provided here are excellent starting points for the creation of different dicarboxylic acid production strains.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file.
References
Alber BE (2011) Biotechnological potential of the ethylmalonyl-CoA pathway. Appl Microbiol Biotechnol 89:17–25. https://doi.org/10.1007/s00253-010-2873-z
Alonso S, Rendueles M, Díaz M (2014) Microbial production of specialty organic acids from renewable and waste materials. Crit Rev Biotechnol 35:497–513. https://doi.org/10.3109/07388551.2014.904269
Bertani G (1951) Studies on lysogenesis I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. https://doi.org/10.1128/jb.62.3.293-300.1951
Chistoserdova LV, Lidstrom ME (1996) Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiol 142:1459–1468.https://doi.org/10.1099/13500872-142-6-1459
Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee M-C, Marx CJ (2013) Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One 8:e62957. https://doi.org/10.1371/journal.pone.0062957
Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE (2007) Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA 104:10631–10636. https://doi.org/10.1073/pnas.0702791104
Erb TJ, Frerichs-Revermann L, Fuchs G, Alber BE (2010) The apparent malate synthase activity of Rhodobacter sphaeroides is due to two paralogous enzymes, (3S)-malyl-coenzyme A (CoA)/β-methylmalyl-CoA lyase and (3S)-malyl-CoA thioesterase. J Bacteriol 192:1249–1258. https://doi.org/10.1128/JB.01267-09
Grant SGN, Jessee J, Bloom FR, Hanahan D (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci 87:4645–4649. https://doi.org/10.1073/PNAS.87.12.4645
Hanahan D (1985) Techniques for transformation of E. coli. In: Glover DM (ed) DNA cloning: a practical approach. IRL Press, Oxford, pp 109–135
Janausch IG, Zientz E, Tran QH, Kröger A, Unden G (2002) C4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta - Bioenerg 1553:39–56. https://doi.org/10.1016/S0005-2728(01)00233-X
Jang Y-S, Kim B, Shin JH, Choi YJ, Choi S, Song CW, Lee J, Park HG, Lee SY (2012) Bio-based production of C2–C6 platform chemicals. Biotechnol Bioeng 109:2437–2459. https://doi.org/10.1002/bit.24599
Kiefer P, Buchhaupt M, Christen P, Kaup B, Schrader J, Vorholt JA (2009) Metabolite profiling uncovers plasmid-induced cobalt limitation under methylotrophic growth conditions. PLoS One 4:e7831. https://doi.org/10.1371/journal.pone.0007831
Kronen M, Sasikaran J, Berg IA (2015) Mesaconase activity of class I fumarase contributes to mesaconate utilization by Burkholderia xenovorans. Appl Environ Microbiol 81:5632–5638. https://doi.org/10.1128/AEM.00822-15
Lee JW, Kim HU, Choi S, Yi J, Lee SY (2011) Microbial production of building block chemicals and polymers. Curr Opin Biotechnol 22:758–767. https://doi.org/10.1016/j.copbio.2011.02.011
Lim CK, Villada JC, Chalifour A, Duran MF, Lu H, Lee PKH (2019) Designing and engineering Methylorubrum extorquens AM1 for itaconic acid production. Front Microbiol 10:1–14. https://doi.org/10.3389/fmicb.2019.01027
Marx CJ, Lidstrom ME (2001) Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiol 147:2065–2075. https://doi.org/10.1099/00221287-147-8-2065
Marx CJ, Lidstrom ME (2002) Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques 33:1062–1067. https://doi.org/10.2144/02335rr01
Okubo Y, Yang S, Chistoserdova L, Lidstrom ME (2010) Alternative route for glyoxylate consumption during growth on two-carbon compounds by Methylobacterium extorquens AM1. J Bacteriol 192:1813–1823. https://doi.org/10.1128/JB.01166-09
Peel D, Quayle JR (1961) Microbial growth on C1 compounds. 1. Isolation and characterization of Pseudomonas AM 1. Biochem J 81:465–469. https://doi.org/10.1042/bj0810465
Peyraud R, Kiefer P, Christen P, Massou S, Portais J-C, Vorholt JA (2009) Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc Natl Acad Sci USA 106:4846–4851. https://doi.org/10.1073/pnas.0810932106
Roode-Gutzmer QI, Kaiser D, Bertau M (2019) Renew Methanol Synth Chembioeng Rev 6:209–236. https://doi.org/10.1002/cben.201900012
Salem AR, Quayle JR (1971) Mutants of Pseudomonas AM1 that require glycollate or glyoxylate for growth on methanol or ethanol. Biochem J 124:74P. https://doi.org/10.1042/bj1240074P
Salis HM (2011) The ribosome binding site calculator. Methods Enzymol 498:19–42. https://doi.org/10.1016/B978-0-12-385120-8.00002-4
Schada von Borzyskowski L, Sonntag F, Pöschel L, Vorholt JA, Schrader J, Erb TJ, Buchhaupt M (2018) Replacing the ethylmalonyl-CoA pathway with the glyoxylate shunt provides metabolic flexibility in the central carbon metabolism of Methylobacterium extorquens AM1. ACS Synth Biol 7:86–97. https://doi.org/10.1021/acssynbio.7b00229
Seol W, Shatkin AJ (1991) Escherichia coli kgtP encodes an alpha-ketoglutarate transporter. Proc Natl Acad Sci USA 88:3802–3806
Sonntag F, Buchhaupt M, Schrader J (2014) Thioesterases for ethylmalonyl–CoA pathway derived dicarboxylic acid production in Methylobacterium extorquens AM1. Appl Microbiol Biotechnol 98:4533–4544. https://doi.org/10.1007/s00253-013-5456-y
Sonntag F, Müller JEN, Kiefer P, Vorholt JA, Schrader J, Buchhaupt M (2015) High-level production of ethylmalonyl-CoA pathway-derived dicarboxylic acids by Methylobacterium extorquens under cobalt-deficient conditions and by polyhydroxybutyrate negative strains. Appl Microbiol Biotechnol 99:3407–3419. https://doi.org/10.1007/s00253-015-6418-3
Toyama H, Anthony C, Lidstrom ME (1998) Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol Lett 166:1–7. https://doi.org/10.1111/j.1574-6968.1998.tb13175.x
Van Dien SJ, Okubo Y, Hough MT, Korotkova N, Taitano T, Lidstrom ME (2003) Reconstruction of C3 and C4 metabolism in Methylobacterium extorquens AM1 using transposon mutagenesis. Microbiology 149:601–609. https://doi.org/10.1099/mic.0.25955-0
Willis MA, Zhuang Z, Song F, Howard A, Dunaway-Mariano D, Herzberg O (2008) Structure of YciA from Haemophilus influenzae (HI0827), a hexameric broad specificity acyl-coenzyme A thioesterase. Biochem 47:2797–2805. https://doi.org/10.1021/bi702336d
Yang Y-M, Chen W-J, Yang J, Zhou Y-M, Hu B, Zhang M, Zhu L-P, Wang G-Y, Yang S (2017) Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route. Microb Cell Fact 16:179. https://doi.org/10.1186/s12934-017-0798-2
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was supported by funds of the Federal Ministry of Education and Research (BMBF, FKZ 031B0340A, project Chiramet).
Author information
Authors and Affiliations
Contributions
LP and MB conceived and designed research. LP conducted experiments. EG constructed plasmid pCM160_RBS_yciAHI. LP and MB analyzed data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pöschel, L., Gehr, E. & Buchhaupt, M. Improvement of dicarboxylic acid production with Methylorubrum extorquens by reduction of product reuptake. Appl Microbiol Biotechnol 106, 6713–6731 (2022). https://doi.org/10.1007/s00253-022-12161-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12161-0