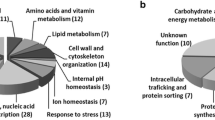
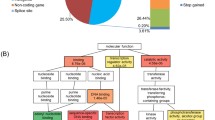
Abstract
The industrial yeast Saccharomyces cerevisiae has a plastic genome with a great flexibility in adaptation to varied conditions of nutrition, temperature, chemistry, osmolarity, and pH in diversified applications. A tolerant strain against 2-furaldehyde (furfural) and 5-hydroxymethyl-2-furaldehyde (HMF) was successfully obtained previously by adaptation through environmental engineering toward development of the next-generation biocatalyst. Using a time-course comparative transcriptome analysis in response to a synergistic challenge of furfural-HMF, here we report tolerance phenotypes of pathway-based transcriptional profiles as components of the adapted defensive system for the tolerant strain NRRL Y-50049. The newly identified tolerance phenotypes were involved in biosynthesis superpathway of sulfur amino acids, defensive reduction-oxidation reaction process, cell wall response, and endogenous and exogenous cellular detoxification. Key transcription factors closely related to these pathway-based components, such as Yap1, Met4, Met31/32, Msn2/4, and Pdr1/3, were also presented. Many important genes in Y-50049 acquired an enhanced transcription background and showed continued increased expressions during the entire lag phase against furfural-HMF. Such signature expressions distinguished tolerance phenotypes of Y-50049 from the innate stress response of its progenitor NRRL Y-12632, an industrial type strain. The acquired yeast tolerance is believed to be evolved in various mechanisms at the genomic level. Identification of legitimate tolerance phenotypes provides a basis for continued investigations on pathway interactions and dissection of mechanisms of yeast tolerance and adaptation at the genomic level.






Similar content being viewed by others
References
Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenço AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, Oliveira AL, Sá-Correia I, Freitas AT (2010) YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res 39:D136–D140
Alenquer M, Tenreiro S, Sá-Correia I (2006) Adaptive response to the antimalarial drug artesunate in yeast involves Pdr1p/Pdr3p-mediated transcriptional activation of the resistance determinants TPO1and PDR5. FEMS Yeast Res 6:1130–1139
Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2
Alriksson B, Horváth IS, Jönsson LJ (2010) Overexpression of Saccharomyces cerevisiae transcription factor and multidrug resistance genes conveys enhanced resistance to lignocellulose derived fermentation inhibitors. Process Biochem 45:264–271
Aranda A, del Olmo ML (2004) Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl Environ Microbiol 70:1913–1922
Argueso JL, Carazzolle MF, Mieczkowski PA, Duarte FM, Netto OVC, Missawa SK, Galzerani F, Costa GGL, Vidal RO, Noronha MF, Dominska M, Andrietta MGS, Andrietta SR, Cunha AF, Gomes LH, Tavares FCA, Alcarde AR, Dietrich FS, McCusker JH, Petes TD, Pereira GAG (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19:2258–2270
Ask M, Bettiga M, Mapelli V, Olsson L (2013) The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22
Blaiseau PL, Thomas D (1998) Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J 17:6327–6336
Blevins WR, Tavella T, Moro SG, Blasco-Moreno B, Close-Mosquera A, Diez J, Carey LB, Albà MM (2019) Extensive post-transcriptional buffering of gene expression in the response to severe oxidative stress in baker’s yeast. Sci Rep 9:11005
Bowman MJ, Jordan DB, Vermillion KE, Braker JD, Moon J, Liu ZL (2010) Stereochemistry of furfural reduction by an aldehyde reductase from Saccharomyces cerevisiae that contributes to in situ furfural detoxification. Appl Environ Microbiol 76:4926–4932
Bradbury JE, Richards KD, Niederer HA, Lee SA, Dunbar PR, Gardner RC (2006) A homozygous diploid subset of commercial wine yeast strains. Antonie Van Leeuwenhoek 89:27–37
Brosnan JT, Brosnan ME (2006) The sulfur-containing amino acids: an overview. J Nutr 136:1636S–1640S
Čáp M, Váchová L, Palková Z (2009) Yeast colony survival depends on metabolic adaptation and cell differentiation rather than on stress defense. J Biol Chem 284:32572–32581
de Witt RN, Kroukamp H, Volschenk H (2019) Proteo respons of two natural strains of Saccharomyces cerevisiae with divergent lignocellulosic inhibitor stress tolerance. FEMS Yeast Res 19:1–16
Del Sorbo G, Schoonbeek H, De Waard MA (2000) Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet Biol 30:1–15
Ding MZ, Wang X, Liu W, Cheng JS, Yang Y, Yuan YJ (2012) Proteomic research reveals the stress response and detoxification of yeast to combined inhibitors. PLoS One 7:e43474
ERCC (2005a) The external RNA control consortium: a progress report. Nat Methods 2:731–734
ERCC (2005b) Proposed methods for testing and selecting ERCC external RNA controls. BMC Genomics 6:150
Fernandes L, Rodrigues-Pousada C, Struhl K (1997) Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17:6982–6993
Gibney PA, Lu C, Caudy AA, Hess DC, Botstein D (2013) Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. PNAS 110:E4393–E4402
Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD (2006) Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 71:339–349
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
Heer D, Heine D, Sauer U (2009) Resistance of Saccharomyces cerevisiae to high concentrations of furfural is based on NADPH-dependent reduction by at least two oxireductases. Appl Environ Microbiol 75:7631–7638
Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Earle-Hughes J, Snesrud E, Lee N, Quackenbush J (2000) A concise guide to cDNA microarray analysis. BioTechniques 29:548–562
Jamieson DJ (1998) Oxidative stress response of the yeast Saccharomyces cerevisiae. Yeast 14:1511–1527
Jordan D, Braker JD, Bowman MJ, Vermillion KE, Moon J, Liu ZL (2011) Kinetic mechanism of an aldehyde reductase of Saccharomyces cerevisiae that relieves toxicity of furfural and 5-hydroxymethylfurfural. Biochim Biophys Acta 1814:1686–1694
Jung YH, Kim S, Yang J, Seo J-H, Kim KH (2017) Intracellular metabolite profiling of Saccahromyces cerevisiae evolved under furfural. Microb Biotechnol 10:395–404
Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzae H (2013) Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154:416–429
Lazard M, Dauplais M, Blanquet S, Plateau P (2015) Trans-sulfuration pathway seleno-amino acids are mediators of selenomethioine toxicity in Saccharomyces cerevisiae. J Biol Chem 290:10741–10750
Legras J-C, Galeote V, Bigey F, Camarasa C, Marsit S, Nidelet T, Sanchez I, Couloux A, Guy J, Franco-Duarte R, Marcet-Houben M, Gabaldon T, Schuller D, Sampaio JP, Dequin S (2018) Adaptation of Saccharomyces cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol Biol Evol 35:1712–1727
Lin FM, Tan Y, Yuan YJ (2009) Temporal quantitative proteomics of Saccharomyces cerevisiae in response to a nonlethal concentration of furfural. Proteomics 9:5471–5483
Liu ZL (2018) Understanding the tolerance of the industrial yeast Saccharomyces cerevisiae against a major class of toxic aldehyde compounds. Appl Microbiol Biotechnol 102:5369–5390
Liu ZL, Cotta MA (2015) Technical assessment of cellulosic ethanol production using β-glucosidase producing yeast Clavispora NRRL Y-50464. Bioenerg Res 8:1203–1211
Liu ZL, Moon J (2009) A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10
Liu ZL, Slininger PJ (2007) Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J Microbiol Methods 68:486–496
Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW (2004) Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J Ind Microbiol Biotechnol 31:345–352
Liu ZL, Slininger PJ, Gorsich SW (2005) Enhanced biotransformation of furfural and 5-hydroxy methylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol 121–124:451–460
Liu ZL, Moon J, Andersh AJ, Slininger PJ, Weber S (2008) Multiple gene mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and HMF by ethanologenic yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 81:743–753
Liu ZL, Ma M, Song M (2009) Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Gen Genomics 282:233–244
Liu ZL, Wang X, Weber SA (2018) Tolerant industrial yeast Saccharomyces cerevisiae possess a more robust cell wall integrity signaling pathway against 2-furaldehyde and 5-(hydroxymethyl)-2-firaldehyde. J Biotechnol 276-277:15–24
Liu ZL, Huang X, Zhou Q, Xu J (2019) Protein expression analysis revealed a fine-tuned mechanism of in situ detoxification pathway for the tolerant industrial yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 103:5781–5796
Lushchak VI, Gospodaryov DV (2005) Catalases protect cellular proteins from oxidative modification from Saccharomyces cerevisiae. Cell Biol Int 29:187–192
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660
Mendoza-Cózatl D, Loza-Tavera H, Hernández-Navarro A, Moreno-Sánchez R (2005) Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol Rev 29:653–671
Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO (2009) Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 75:6132–6141
Moon J, Liu ZL (2012) Engineered NADH-dependent GRE2 from Saccharomyces cerevisiae by directed enzyme evolution enhances HMF reduction using additional cofactor NADPH. Enzym Microb Technol 50:115–120
Niino YS, Chakraborty S, Brown BJ, Massey V (1995) A new old yellow enzyme of Saccharomyces cerevisiae. J Biol Chem 270:1983–1991
Okazaki S, Tachibana T, Naganuma A, Mano N, Kuge S (2007) Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell 27:675–688
Orlean P (2012) Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192:775–818
Park SE, Koo HM, Park YK, Park SM, Park JC, Lee OK, Park YC, Seo JH (2011) Expression of aldehyde dehydrogenase 6 reduces inhibitory effect of furan derivatives on cell growth and ethanol production in Saccharomyces cerevisiae. Bioresour Technol 102:6033–6038
Peter J, Chiara MD, Friedrich A, Yue J-X, Pflieger D, Bergström A, Sigwalt A, Barre B, Freel K, Llored K, Cruaud C, Labadie K, Aury J-M, Istace B, Lebrigand K, Barbry P, Engelen S, Lemainque A, Wincker P, Liti G, Schacherer J (2018) Genoe evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556:339–447
Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF, Lidén G (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464
Prasad R, Goffeau A (2012) Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu Rev Microbiol 66:39–63
Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H, Takagi H (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng 113:451–455
Schmitt AP, McEntee K (1996) Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. PNAS 93:5777–5782
Steenwyk J, Rokas A (2017) Extensive copy number variation in fermentation-related genes among Saccharomyces cerevisiae wine strains. G3 (Bethesda) 7:1475–1485
Sundström L, Larsson S, Jönsson LJ (2010) Identification of Saccharomyces cerevisiae genes involved in the resistance to phenolic fermentation inhibitors. Appl Biochem Biotechnol 161:106–115
Święcilo A (2016) Cross-stress resistance in Saccharomyces cerevisiae yeast—new insight in to an old phenomenon. Cell Stress Chaperones 21:187–200
Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sá-Correia I (2006) The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucl Acids Res 34:D446–D451
Thompson OA, Hawkins GM, Gorsich SW, Doran-Peterson J (2016) Phenotypic characterization and comparative transcriptomics of evolved Saccharomyces cerevisiae strains with improved tolerance to lignocellulosic derived inhibitors. Biotechnol Biofiuels 9:200
Thorsen M, Lagniel G, Kristiansson E, Junot C, Nerman O, Labarrem J, Tamás MJ (2007) Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol Genomics 30:35–43
Unrean P, Gätgens J, Klein B, Noack S, Champreda V (2018) Elucidating cellular mechanisms of Saccahromyces cerevisiae tolerant to combined lignocellulosic-derived inhibitor using high-throughput phenotyping and multiomics analyses. FEMS Yeast Res 18:1–10
Wallance-Salina V, Brink DP, Ahrén D, Gorwa-Grauslund MF (2015) Cell periphery-related proteins as major genomic targets behind the adaptative evolution of an industrial yeast Saccharomyces cerevisiae strain to combined heat and hydrolysate stress. BMC Genomics 16:514
Wang Y, Zhang S, Liu H, Zhang L, Yi C, Li H (2015) Changes and roles of membrane compositions in the adaptation of Saccharomyces cerevisiae to ethanol. J Basic Microbiol 55:1–10
Wang X, Liu ZL, Zhang X, Ma M (2017) A new source of resistance to 2-furaldehyde from Scheffersomyces (Pichia) stipitis for sustainable lignocellulose-to-biofuel conversion. Appl Micorbiol Biotechnol 101:4981–4993
Wheeler GL, Trotter EW, Dawes IW, Grant CM (2003) Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J Biol Chem 278:49920–49928
Yang J, Ding MZ, Li BZ, Liu ZL, Wang X, Yuan YJ (2012) Integrated phospholipidomics and transcriptomics analysis of Saccharomyces cerevisiae with enhanced tolerance to a mixture of acetic acid, furfural, and phenol. Omics 16:374–386
Yasokawa D, Murata S, Iwahashi Y, Kitagawa E, Nakagawa R, Hashido T, Iwahashi H (2010) Toxicity of methanol and formaldehyde towards Saccharomyces cerevisiae as assessed by DNA microarray analysis. Appl Biochem Biotechnol 160:1685–1698
Zhang Y, Liu ZL, Song M (2015) ChiNet uncovers rewired transcription subnetworks in tolerant yeast for advanced biofuels conversion. Nucleic Acids Res 43:4393–4407
Zhou Q, Liu ZL, Ning K, Wang A, Zeng X, Xu J (2014) Genomic and transcriptome analysis reveal that MAPK- and phosphatidylinositol-signaling pathways mediate tolerance to 5-hydroxymehyl-2-furaldehyde for industrial yeast Saccharomyces cerevisiae. Sci Rep 4:6556
Acknowledgments
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 59 kb)
Rights and permissions
About this article
Cite this article
Liu, Z.L., Ma, M. Pathway-based signature transcriptional profiles as tolerance phenotypes for the adapted industrial yeast Saccharomyces cerevisiae resistant to furfural and HMF. Appl Microbiol Biotechnol 104, 3473–3492 (2020). https://doi.org/10.1007/s00253-020-10434-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10434-0