Abstract
Colobanthus quitensis (Kunth) Bartl. and Deschampsia antarctica Desv. are the only Magnoliophyta to naturally colonize the Antarctic region. The reason for their sole presence in Antarctica is still debated as there is no definitive consensus on how only two unrelated flowering plants managed to establish breeding populations in this part of the world. In this study, we have explored and compared the rhizosphere and root-endosphere dwelling microbial community of C. quitensis and D. antarctica specimens sampled in maritime Antarctica from sites displaying contrasting edaphic characteristics. Bacterial phylogenetic diversity (high-throughput 16S rRNA gene fragment targeted sequencing) and microbial metabolic activity (Biolog EcoPlates) with a geochemical soil background were assessed. Gathered data showed that the microbiome of C. quitensis root system was mostly site-dependent, displaying different characteristics in each of the examined locations. This plant tolerated an active bacterial community only in severe conditions (salt stress and nutrient deprivation), while in other more favorable circumstances, it restricted microbial activity, with a possibility of microbivory-based nutrient acquisition. The microbial communities of D. antarctica showed a high degree of similarity between samples within a particular rhizocompartment. The grass’ endosphere was significantly enriched in plant beneficial taxa of the family Rhizobiaceae, which displayed obligatory endophyte characteristics, suggesting that at least part of this community is transmitted vertically. Ultimately, the ecological success of C. quitensis and D. antarctica in Antarctica might be largely attributed to their associations and management of root-associated microbiota.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antarctica is a place of extremes [1]. Only two species of Magnoliophyta (flowering plants) managed to establish their presence in this remote and life-challenging region: a member of the family Poaceae—Deschampsia antarctica Desv. (Antarctic hairgrass) and Colobanthus quitensis (Kunth) Bartl. (Antarctic pearlwort), belonging to the family Caryophyllaceae [2]. Their vast distribution in maritime and coastal Antarctica has baffled scientists for decades and was a subject of many debates [2,3,4]. Originally from South America, those two Magnoliophyta were suspected of being migratory relics from the Oligocene-Pliocene colonization event [2], while other data hints towards their more recent arrival during the late Pliocene [5]. Nonetheless, their ecological success in harsh Antarctic conditions is undisputed and attributable mostly to the extensive adaptations to many stress factors like cold, freeze–thaw cycles, UV radiation, drought, flooding, high concentration of sodium, and varying levels of nutrient concentrations from severe deficit to extreme over manuring [4]. To cope with environmental stresses, Antarctic plants have evolved adaptations in terms of molecular, cellular, and physiological functions to boost their cold tolerance by increasing their metabolic rates upon cold acclimation [6], such as stress protein production, upregulation of antioxidant synthesis, and accumulation of compatible solutes like amino acid proline and non-structural carbohydrates [7, 8] and secretion of antifreeze proteins into their apoplasts [9]. They also developed low temperature-efficient photosynthetic and respiratory systems whose maximal activity occurs at 13 °C, and 30% of its maximal photosynthetic capacity is retained at 0 °C [10, 11]. Moreover, such ontogenesis-related features of Antarctic flowering plants like long life cycles, an extended primordium development of leaves and flowers [12], or diverse propagation strategies based on seed production derived from self-pollination and less often cross-pollination (C. quitensis and D. antarctica) or vegetative reproduction (only D. antarctica) enabled continuous habitat range expansion of those plants since 1960, presumably aided by the ongoing global climate warming [13, 14]. However, the geographical and physical isolation of Antarctica still enforces low intra-species genetic diversity by restricting gene flow between Antarctica and South America into those populations [15, 16].
The relations between plants and microbes are now fairly well understood for the flora of temperate and tropical regions [17] but are somewhat under researched for polar regions, especially for Antarctica [18]. It is the consensus that microbial communities associated with the host plants’ roots exert the greatest influence on the plant health and development, most notably those that reside within two distinct rhizocompartments: the rhizosphere and the root endosphere [19]. Rhizospheric microbes inhabit the root-adjacent soil and feed on the root-derived organic exudates, whereas the endosphere community consists largely of plant-specific endosymbionts selectively recruited from the rhizosphere [20]. Some studies indicate that rhizospheric and endospheric microbial communities are not only spatially separated but also display different niche characteristics [21]. Rhizospheric bacteria are mostly engaged in mineral substrate dissolution by secreting organic acids (often gluconic acid), siderophores, or even cyanides consequently providing the plant with bioavailable phosphorus and biogenic metals. Their main role also includes biocontrol of soil-borne pathogens by competing with them for resources with the use of antimicrobial agents like bacteriocin-like peptides, different classes of antibiotics, and lytic enzymes: chitinase or β-1,3-glucanase. Rhizobacteria also have a direct input into root-growth stimulation by secretion of phytohormones such as indole-3-acetic acid. Endospheric bacteria on the other hand are engaged in plant stress responses mainly by reducing the inner-plant stress hormone ethylene levels through ACC deaminase-mediated hydrolysis. Endophytes also have a major contribution in plant-growth enhancement by providing fixed nitrogen, which is the main limiting element in soil environments [22].
As mentioned, studies on the Antarctic plant-associated microbiome (especially the bacterial part) are largely underrepresented in literature [23]. Available but scarce data allow us to draw only limited conclusions on microbiological phenomena connected to Antarctic Magnoliophyta [24,25,26]. This is largely due to the low resolution of the techniques used but also due to low sample diversity (mostly from only one particular site) or even exclusion of a vital part of the community, namely the endosphere, from analysis. Therefore, in this study, we employed cultivation-independent methods to compare root-associated microbial communities of C. quitensis and D. antarctica sampled from sites displaying contrasting edaphic, ecologic, and microclimatic characteristics. Our hypothesis states that the phylogenetic and metabolic diversity of microbial communities associated with native Antarctic plants is greater in sites of high stress-inducing factor intensity than in those providing milder growth conditions. To gain the necessary knowledge on the differences and similarities within the rhizospheric and endosphere microbial communities of Antarctic flowering plants, we made use of the high-throughput 16S rRNA gene fragment targeted sequencing (assessing the bacterial communities phylogenetic diversity) and community-level physiological profiling by Biolog EcoPlates.
Materials and Methods
Sites and Sampling
Samples were collected during the austral summer season of 2017–2018 from three sites on King George Island, South Shetland Islands, maritime Antarctica (Table 1). Several specimens (4–6 per site) were collected with the root adjacent soil with the use of sterile tools into sterile plastic containers and transported frozen (− 20 °C) to the laboratory in the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (IBB, PAS). Additionally, bulk soil samples from those sites were gathered in triplicates (approximately 1.5 kg per site) for component analysis and transported in the same conditions.
Measurement of Soil Components
Soil pH (in 1 M KCl) and salinity (in double-distilled water) were measured with a CPC-411 Elmetron™ multiparameter probe [27]. Phosphates and nitrates were determined spectrophotometrically in a Shimadzu UV 1601 spectrophotometer and in an Epoll-Eco 20 spectrophotometer, respectively. Other elements were determined by atomic absorption spectroscopy [28].
Bacterial Extraction
Bacterial cells were extracted from rhizospheric soil and roots samples from nine C. quitensis individuals (three per site) and respectively the same number of D. antarctica individuals. The following method was devised based on the findings of Lunau et al. [29] regarding the separation of prokaryotic cells from mineral and organic debris and the guidelines provided by Szymańska et al. [30] regarding root-associated microbe isolation. To analyze the microbiome of the root-adjacent soil, a sample of the soil was carefully removed from between the roots with a sterile spatula onto a pre-sterilized aluminums foil piece. Approximately 1 g of the soil was weighed and placed in a 50-mL conical tube containing 20 mL of sterile and cool (4 °C) dilution liquid composed of 0.9% (w/v) NaCl and 10 mM tetrasodium pyrophosphate (Na4P2O7). The suspension was then shaken for 30 min in a Tornado™ Vortexer at 2000 rpm at 4 °C. The tubes were then placed in a VWR Ultrasonic Cleaner USCTH filled with chilled water and sonicated for 60 s. The tubes were vortexed afterwards for 30 s to suspend detached cells. After brief centrifugation (1 min; 1000 rpm; 4 °C), the suspension was submitted to metabolic fingerprinting by the Biolog EcoPlate technique. To detach the rest of the adhering soil, the root system was washed in 60 mL of sterile NaCl/Na4P2O7 solution by shaking for 30 min in the aforementioned shaker (1000 rpm; 4 °C) and then rinsed 3 times in 5 mL of the same sterile and cooled solution by vortexing. Washed roots were sterilized by incubation in a cooled 10% hydrogen peroxide (H2O2) solution for 5 min and then rinsed 3 times with sterile NaCl/Na4P2O7 solution. The so surface-sterilized roots were placed in a pre-cooled sterile mortar. Two and a half milliliter of sterile NaCl/Na4P2O7 was added with 0.6 g of sterile, sharp garnet sand (lysing matrix A, MP Biomedicals) and gently ground with a pestle, allowing the sharp angular garnet pieces to comminute the roots to an amorphous pulp. The pulp was transferred to a 50-mL conical tube containing 20 mL of sterile and cool (4 °C) NaCl/Na4P2O7 solution and submitted to the above-mentioned procedure (shaking, ultrasonication, and vortexing). The resulting supernatant suspension was submitted to metabolic fingerprinting by the Biolog EcoPlate technique and DNA extraction. All reagents used in the bacterial extraction steps were molecular biology grade and were purchased from Sigma Aldrich.
DNA Extraction
Rhizosphere soil DNA was extracted using the PowerSoil® DNA isolation kit (QIAGEN GmbH, Hilden, Germany) according to manufacturer protocol. An approximately 0.2 g of soil was used. DNA solutions were kept at 4 °C for further analysis. The dilution liquid containing endosphere bacteria was passed through a sterile 47-mm Whatman polycarbonate filter (0.22 µm pore size). The DNA from the filter-trapped bacteria was extracted using the PowerWater® DNA isolation kit (QIAGEN, GmbH, Hilden, Germany) according to manufacturer protocol and kept at 4 °C.
16S rRNA Gene Amplification
The phylogenetic study was performed by targeted sequencing and analysis of the prokaryotic 16S ribosomal RNA gene. A fragment of the 16S rRNA gene containing the V3 and V4 variable regions was amplified using gene-specific primers: 16S_V3-F and 16S_V4-R positions 341-357F and 785-805R, respectively, according to Escherichia coli 16S rRNA gene reference sequence [31]. Illumina Nextera XT overhang adapter nucleotide sequences were included in addition to the 16S rRNA gene-specific sequences, which allowed sample indexing and pooling. Each PCR was conducted in triplicates using KAPA HiFi PCR kit (Roche, Basel, Switzerland) in a final volume of 20 µL per reaction according to the manufacturer’s instructions.
Amplicon Sequencing
Obtained PCR products were pooled into 12 samples (2 rhizocompartments × 3 sampling sites × 2 plant species) in equimolar ratio and indexed using Nextera XT barcodes (Illumina, San Diego, CA, USA). Amplicon libraries were sequenced on Illumina MiSeq instrument (Illumina, San Diego, CA, USA) in the DNA Sequencing and Oligonucleotide Synthesis Laboratory (IBB, PAS). Sequencing was conducted in paired-end mode (2 × 300 bp) with the use of a v.3 (600 cycles) chemistry cartridge, which allowed the generation of long paired reads fully covering 16S V3–V4 amplicons.
Phenotype Fingerprinting with Biolog EcoPlate™
The Biolog EcoPlate assay determines the ability of a mixed microbial community to use any of 31 carbon compounds as the sole carbon source (plus a single control well with no-carbon). Microbial communities were characterized for their ability to catabolize 10 different carbohydrates, 9 carboxylic and acetic acids, 4 polymers, 6 amino acids, and 2 amines [32]. Root-associated bacterial suspensions were adjusted with sterile 0.9% NaCl to optical transmittance of 0.9. One hundred microliter aliquots of each suspension were added to each well of EcoPlate microplates (Biolog Inc., Hayward, CA, USA). The plates were incubated in darkness at 10 °C. The temperatures were chosen to accommodate the activity range of the resident microbial communities: the psychrophiles and the psychrotrophs [33]. The color development was read at 590 nm (A590) in a Varioskan plate reader (Thermo Fisher Scientific, Waltham, MA, USA), and cellular respiration was measured kinetically by determining the colorimetric reduction of tetrazolium dye. Data were collected approximately twice a week over a 65-day period. The prolonged incubation of EcoPlates was based on our previous observations [34,35,36]. Data from the 36th day of incubation were used as there was no further color development after this date. Final absorbance data were first blanked against the time zero reading and then blanked against the respective control well containing no carbon source. Readings that had the A590 value of 0.25 or higher were scored as a positive EcoPlate response (PER).
Data Analysis
Raw sequencing data were cleaned, aligned, and classified automatically by the EzBioCloud platform using the PKSSU4.0 database [37]. Chimeric, low quality, and non-target (chloroplast, mitochondrial, and archaeal) amplicons were automatically discarded. The operational taxonomic unit was defined as a group of sequences that exhibit greater than 97% similarity to each other. Illumina reads were deposited in the NCBI Sequence Read Archive (SRA) as BioProject PRJNA726953. All results were compiled using Excel 2016 (MS Office) for Windows. A two-sample t-test was applied to compare different data sets. Variance within the sets was assessed using the f-test beforehand. Correlations between biological and geochemical parameters were calculated using Pearson’s correlation coefficient. Principal component analysis (PCA) was performed using the singular value decomposition method. Data visualization and statistical analysis have been performed using the R software (R v.4.0.2) and the following packages: ggplot2, fmsb, Hmisc, ggpubr, corrplot, and autoplot [38].
Results
Site 1 soil was characterized by high magnesium, very high calcium, and relatively high sodium contents and salinity, and also pH was high (8.3). Soil from site 2 had the highest potassium content among the examined soil samples with high copper and iron concentrations and a low pH (4.3). Site 3 soils had the highest concentration of nitrates, phosphates, manganese, and zinc with the lowest reported pH (4.0) (Fig. 1C).
Sampling site details. A Satellite map displaying the geographical situation of the sampling sites: red circle, King George Island, Maritime Antarctica; 1, sampling site 1, Lions Rump, King George Bay shore; 2, Puchalski Hill; 3, Point Thomas Penguin Rookery, Admiralty Bay shore. B Sampling sites 1–3 landscape and ground photographs. C Sampling sites 1–3 geochemical composition of the soil; N, mg NO3/100 g soil; P, mg P2O5/100 g soil; K, mg K2O/100 g soil; Mg, mg Mg/100 g soil; Ca, mg Ca/100 g soil; Na, mg Na/100 g soil; salinity, g NaCl/L; Mn, mg Mn/kg soil; Zn, mg Zn/kg soil; Cu, mg Cu/kg soil; Fe, mg Fe/kg soil
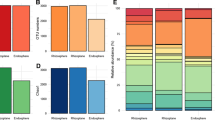
Bacterial operational taxonomic unit (OTU) numbers were always higher in the rhizosphere (av. 3064, sd. 778) compared to the root tissue (av. 2151.3, sd. 797.5). C. quitensis–associated communities had on average higher OTU numbers than D. antarctica–associated communities (soil, 3464.3 vs 2663.7; root, 2766.7 vs 1536). Rhizospheric communities from site 1 displayed the highest phylogenetic diversity for D. antarctica and C. quitensis alike (3637 and 3736 OTUs respectively), while the lowest values were noted for site 3 samples (D. antarctica, 1790; C. quitensis, 2987). C. quitensis endosphere community was least diverse in samples from site 1 (2210 OTUs) whereas for D. antarctica in samples from site 3 (1300 OTUs). The most diverse endospheric community was noted in site 2 samples for both plant species (D. antarctica, 1681 OTUs; C. quitensis, 3468 OTUs). Community response numbers on Biolog EcoPlates were on average lower in the endosphere compared to the rhizosphere for C. quitensis samples (av. 18.4, sd. 6.8 and av. 24.8, sd. 4.4, respectively), while for D. antarctica, those samples were comparable (rhizosphere, av. 26.78, sd. 1.3; endosphere, av. 26.89, sd. 0.7) (Fig. 2).
The rhizosphere community of C. quitensis and D. antarctica was most abundant in the following bacterial phyla: Proteobacteria (av. 24.7%, sd. 3.4 and av. 26.3%, sd. 4.6, respectively), Actinobacteria (av. 16.2%, sd. 12.2 and av. 16.8%, sd. 10.2), Bacteroidetes (av. 11.7%, sd. 2.6 and av. 19.0%, sd. 6.0), Saccharibacteria (av. 6.4%, sd. 1.6 and av. 5.9%, sd. 1.4), Verrucomicrobia (av. 8.1%, sd. 3.1 and av. 6.7%, sd. 2.0), Acidobacteria (av. 8.4%, sd. 2.2 and av. 4.9%, sd. 0.7), and Parcubacteria (av. 4.4%, sd. 3.0 and av. 1.5%, sd. 0.6). The root endosphere of both plants was occupied largely by bacteria of the phylum Proteobacteria (C. quitensis, av. 36.9%, sd. 9.8; D. antarctica, av. 45.6%, sd. 4.9) but also by Bacteroidetes (C. quitensis, av. 15.1%, sd. 2.3; D. antarctica, av. 28.5%, sd. 11.7) and Actinobacteria (C. quitensis, av. 15.1%, sd. 3.1; D. antarctica, av. 12.7%, sd. 3.2). Considerable differences between plant species were noticeable for Acidobacteria (C. quitensis, av. 5.8%, sd. 5.5; D. antarctica, av. 0.9%, sd. 0.6) and Planctomycetes (C. quitensis, av. 3.0%, sd. 1.3; D. antarctica, av. 0.7%, sd. 0.4). The only significant (at p < 0.03) difference in relative abundance was noted for the Chloroflexi bacteria (C. quitensis, av. 7.5%, sd. 2.2; D. antarctica, av. 1.9%, sd. 2.0) (Fig. 3).
Several bacterial families were present in considerable amounts in the rhizosphere, although the occurrence of some of them was highly site specific (Fig. 4A). Relative abundance of the family Micrococcaceae showed severe differences for C. quitensis (av. 6.3%, sd. 10) as well as for D. antarctica (4.6%, sd. 6.4), displaying highest values in site 1 for C. quitensis (17.9%) and in site 2 for D. antarctica (12.0%). High average abundance with concomitant high variations between samples was also observed for the family Xanthomonadaceae (C. quitensis, av. 4.7%, sd. 4.9; D. antarctica, av. 5.0, sd. 3.0). Relative abundance for this family peaked in site 3 for C. quitensis (10.4%) and in site 1 for D. antarctica (7.3%). Most stable levels of relative abundance were noted for the family Chitinophagaceae (C. quitensis, av. 4.5%, sd. 1.1; D. antarctica, av. 7.8%, sd. 1.6) and the candidate family (PAC000016_f) of the phylum Saccharibacteria (C. quitensis, av. 3.6%, sd. 0.1; D. antarctica, av. 2.7%, sd. 1.1).
The endosphere of both plant species was dominated by the family Sphingobacteriaceae (C. quitensis, av. 6.6%, sd. 0.7; D. antarctica, av. 14.9%, sd. 3.7). D. antarctica root endosphere also exhibited a high relative abundance of the family Pseudomonadaceae (av. 9.6%, sd. 3.3), while in C. quitensis, this family for noticeably represented only in site 1 (14.5%), accompanied by the family Oxalobacteraceae (12.7%). Similarly, the family Flavobacteriaceae occurred in considerable abundance in D. antarctica root endosphere only in site 1 (15.5%). Relative abundances of the family Microbacteriaceae was high in site 3 for both C. quitensis (8.6%) and D. antarctica (10.8%). A similar situation was observed for the family Xanthomonadaceae (C. quitensis, 7.7%; D. antarctica, 9.9%). Chitinophagaceae were present in all root samples at comparable levels (C. quitensis, av. 4.6%, sd. 1.1; D. antarctica, av. 4.2%, sd. 0.4). Noteworthy here are the relative abundances of alphaproteobacterial families. The Sphingomonadaceae were present in the roots of C. quitensis (av. 3.8%, sd. 2.3) and D. antarctica (av. 5.4%, sd. 1.6), while the Rhizobiaceae occupied only D. antarctica roots (av. 3.2%; C. quitensis, 0.4%).
Metabolic features of the rhizospheric community revolved mainly around carbohydrate catabolism (Fig. 4B). Highest absorbance value at 590 nm (A590) obtained for the rhizosphere was A590 = 4.93. The most actively catabolized compound was D-mannitol, both in C. quitensis (av. A590 = 3.6, sd. 1.4) and D. antarctica rhizosphere (av. A590 = 2.5, sd. 0.1). The glucose-containing polymer α-cyclodextrin was also readily metabolized (C. quitensis, av. A590 = 2.5, sd. 2.0; D. antarctica, av. A590 = 1.8, sd. 0.3), albeit for C. quitensis, its catabolism was most pronounced in site 1 (A590 = 4.7), similarly for α-D-lactose (C. quitensis, av. A590 = 3.0, sd. 1.8, site 1 A590 = 4.3; D. antarctica, av. A590 = 1.6, sd. 0.3). Other actively catabolized in the rhizosphere compounds included D-cellobiose (C. quitensis, av. A590 = 2.2, sd. 0.7; D. antarctica, av. A590 = 1.4, sd. 0.1), i-erythritol(C. quitensis, av. A590 = 2.2, sd. 0.9; D. antarctica, av. A590 = 1.7, sd. 0.3), L-asparagine (C. quitensis, av. A590 = 1.9, sd. 0.7; D. antarctica, av. A590 = 1.2, sd. 0.1), L-arginine (C. quitensis, av. A590 = 1.9, sd. 0.9; D. antarctica, av. A590 = 1.3, sd. 0.2). The endospheric community displayed similar features as the rhizospheric community. The highest absorbance value at 590 nm (A590) obtained for the endosphere was A590 = 5.86. The main difference between plant species was the more uniform catabolism intensity of carbon sources for D. antarctica, most notably for D-mannitol (C. quitensis, av. A590 = 3.4, sd. 2.4; D. antarctica, av. A590 = 2.8, sd. 0.04). Most high-absorbance values for C. quitensis were obtained for the samples from site 1: α-D-lactose (A590 = 4.53), α-cyclodextrin (A590 = 4.59), i-erythritol (A590 = 4.35), D-xylose (A590 = 3.54) and D-cellobiose (A590 = 3.17). The catabolism intensity of those carbohydrates was more uniform across the endospheric samples of D. antarctica: α-D-lactose (av. A590 = 1.6, sd. 0.4), α-cyclodextrin (av. A590 = 1.8, sd. 0.5), i-erythritol (av. A590 = 2.0, sd. 0.9), D-xylose (av. A590 = 1.4, sd. 0.1), and D-cellobiose (av. A590 = 1.6, sd. 0.4).
Several correlations in the root endosphere of both plants were apparent between biological and geochemical components (Fig. 5). The relative abundance of the family Micrococcaceae displayed significant correlations with the catabolism of several compounds, most notably D-xylose (p = 0.003) and α-D-lactose (p = 0.008). The Pseudomonadaceae and Oxalobacteraceae displayed significant correlations with the same compounds. Phenylethylamine catabolism was positively correlated with the occurrence of Sphingobacteriaceae (p = 0.008) and the Rhizobiaceae (p = 0.02). Negative correlations revolved mainly around the relative abundance of the family Chitinophagaceae. It displayed negative correlations with the catabolism intensity of several compounds, including D-cellobiose (p = 0.03) and D-mannitol (p = 0.04) but also with the geochemical parameters like salinity (p = 0.03) and calcium content (p = 0.03) and the relative abundance of other families like Pseudomonadaceae (p = 0.02) and Oxalobacteraceae (p = 0.03). It showed however positive correlations with heavy metal concentrations (Cu/Fe p = 0.04). Microbacteriaceae and Xanthomonadaceae displayed negative correlations with sodium contents (p = 0.007 and p = 0.002, respectively), while the Oxalobacteraceae showed negative relations with manganese concentrations (p = 0.047).
PCA showcased several phenomena within both plants’ root-associated microbial communities (Fig. 6). The PCA clustering based on the relative abundance of family-rank groups indicated that the rhizosphere community differs in structure from the endosphere community, both for C. quitensis and D. antarctica. The bacterial communities of C. quitensis showed great differences between sampling sites, while those of D. antarctica clustered according to the rhizocompartment of origin. The situation was similar for the EcoPlate-based clustering, mainly for C. quitensis, where no apparent clustering was observed. Rhizospheric and endospheric D. antarctica communities formed a loose cluster in this analysis. PCA based on a combination of phylogenetic and physiological data revealed a clear distinction between C. quitensis and D. antarctica root-associated communities. Two tight clusters emerged, separately harboring the rhizospheric community and the endospheric community of D. antarctica, while for C. quitensis, there were no apparent similarities between the samples.
Principal component analysis (PCA) of biological data. A PCA based on percentage contribution of bacterial sequences identified on a family-rank level. B PCA based on responses obtained for bacterial communities by the Biolog EcoPlate method. C PCA based on a combination of family-rank bacterial sequence percentile contribution and normalized community responses on Biolog EcoPlates. Green dots, Deschampsia antarctica rhizosphere data; blue dots, Deschampsia antarctica endosphere data; red dots, Colobanthus quitensis rhizosphere data; orange dots, Colobanthus quitensis endosphere data
Significant differences in phylogenetic and physiological levels emerged between microbial communities of C. quitensis and D. antarctica (Fig. 7 A and B). In the rhizosphere of D. antarctica, relative abundance of the family Chitinophagaceae was significantly higher (p = 0.049) than in the C. quitensis rhizosphere. The endosphere communities of D. antarctica were significantly richer in sequences of the family Rhizobiaceae (p = 0.00091) and Sphingobacteriaceae (p = 0.0014). Phenylethylamine catabolism was significantly more pronounced in the D. antarctica root endosphere (p = 0.0073).
A Statistically significant differences (p < 0.05) within Deschampsia antarctica and Colobanthus quitensis rhizosphere/endosphere communities based on sequence contribution identified on a phylum taxonomic level; B statistically significant differences (p < 0.05) within Deschampsia antarctica and Colobanthus quitensis rhizosphere/endosphere communities based on community responses on Biolog EcoPlates; C core microbiome of Deschampsia antarctica and Colobanthus quitensis endosphere communities based on sequence contribution (> 1%, red line) identified on a family-rank taxonomic level. Red boxplots, bacterial families present in the roots of both plant species at > 1%; green boxplots, bacterial families present only in the roots of Deschampsia antarctica at > 1%; blue boxplot, bacterial family present only in the roots of Colobanthus quitensis at > 1%
The endospheric core community was based on the relative abundances of family-rank groups (Fig. 7C). Those that displayed an abundance of 1% or higher were scored as core community members. Only five families were at sufficient abundance in the endosphere of both examined plant species: Sphingobacteriaceae (5.9–19.1%), Sphingomonadaceae (1.1–6.7%), Microbacteriaceae (2.3–10.8%), Chitinophagaceae (3.4–5.6%), and Comamonadaceae (1.3–5.4%). Five families were above the 1% threshold only in D. antarctica endosphere samples: Pseudomonadaceae (C. quitensis, 0.1–14.5%; D. antarctica, 5.8–11.6%), Oxalobacteraceae (C. quitensis, 0.1–12.7%; D. antarctica, 2.2–8.9%), Flavobacteriaceae (C. quitensis, 0.03–4.1%; D. antarctica, 1.4–15.5%), Sacharimonas family (C. quitensis, 0.5–1.8%; D. antarctica, 1.2–4.0%), and Rhizobiaceae (C. quitensis, 0.1–0.6%; D. antarctica, 2.1–4.5%). Only one family, the Xanthomonadaceae, made the cut for C. quitensis (C. quitensis, 1.2–7.7%; D. antarctica, 0.4–10.0%).
Discussion
A large body of literature dedicated to root-associated microbiomes indicates that bacterial and fungal communities dwelling in the plant rhizosphere and endosphere are host-specific species [39,40,41]. Nonetheless, the physiological status of this host plant influences the phylogenetic structure and metabolic capabilities of the associated microbiome [42]. This physiological status however is dependent on the edaphic and climatic conditions experienced by the plant [43].
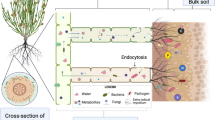
Our results show that Antarctic flowering plants shape their root-associated microbiome differently, resulting in divergent microbial communities. The microbiome of C. quitensis root system bears different characteristics in each of the examined locations. In site 1 characterized by low essential nutrient concentrations (N-P-K) and high salinity and pH, the rhizosphere contained a highly diverse bacterial community, both phylogenetically and metabolically. The family Micrococcaceae (Actinobacteria) seemed to be a vital component of the rhizosphere in such conditions [44]. Members of this family along with other Actinobacteria have been observed in large quantities in salt marsh plants’ rhizosphere, indicating their stress alleviating effect in low water activity substratum [45, 46]. The corresponding C. quitensis endosphere was occupied by a fraction of metabolically active opportunitroph bacteria as indicated by the relatively low OTU numbers accompanied by high numbers of positive EcoPlate responses. Those were mainly the Pseudomonadaceae and Oxalobacteraceae family members. Inoculation with different Pseudomonadaceae strains has improved the salt-tolerance of Zea mays, which was connected to the water-binding exopolysaccharides produced by those bacteria [47], whereas the Oxalobacteraceae were enriched by the same plant species in nitrogen-poor soils, stimulating lateral root growth, consequently increasing nitrogen compound and other resources acquisition [48]. Furthermore, as these two families harbor mostly copiotrophic bacteria that display a multitude of metabolic features [49], their relative abundances in the endosphere were significantly correlated with the catabolism intensity of some of the carbon sources, most notably plant cell wall components: D-cellobiose and D-xylose. Cellulases and xylanases are essential in allowing bacterial entry into plant roots [50, 51]. At site 2, the structure of the root-associated communities diverges considerably from those at site 1. High phylogenetic diversity was accompanied by low metabolic activity and numbers of utilized carbon sources. This indicates that the community consists of either low activity bacteria or that respiratory activity was restricted at this location in C. quitensis rhizosphere. In tundra soils such as this one, nitrate or mineral nitrogen compounds may be deficient as most are bound in organic matter, which decompose extremely slowly under polar conditions [52, 53]. Some hypothesize that in this scenario, plants might exhibit microbivory by releasing proteases into the rhizosphere to liberate the microbe-bound nitrogen but also by destroying the cells of the endosphere microbes through oxidizing agent production on root cell plasma membranes [54]. Furthermore, the examined endosphere contained bacteria that are not usually found in this rhizocompartment, namely those belonging to the phylum Acidobacteria and Gemmatimonadetes [20]. In this regard, active endocytosis has been detected in Arabidopsis thaliana, which internalized and digested non-endophyte microbes [55]. At site 3, the root-associated microbiome of C. quitensis displays yet a different structure. This site is particularly rich in nitrogen, not only in the examined nitrates, but also in ammonia and organic forms [28]. Here the phylogenetic diversity in the rhizosphere community is relatively moderate, but the metabolic diversity is high, while the endosphere community displays moderate phylogenetic diversity and very low microbial activity. The predominant bacterial family in the rhizosphere were the Xanthomonadaceae, which displayed a positive correlation with soil nitrate contents. On several occasions, this bacterial group was observed to increase in numbers when organic and mineral nitrogen fertilization was applied [56, 57]. Together with the Microbacteriaceae and some other low abundance families, they constituted the bulk of the C. quitensis endosphere. The mentioned low activity of these bacteria might be due to host-defense mechanisms, enhanced by the heightened nitrate levels, as they were proved to promote defense signal molecules production like spermine and spermidine [58].
Based on the results of the PCA clustering, the microbial communities of D. antarctica displayed a high degree of similarity between samples within a particular rhizocompartment as compared to the vastly divergent C. quitensis communities. However, the microbiome of D. antarctica was also prone to restructuring enforced by the prevailing abiotic conditions. D. antarctica in the rhizosphere and the endosphere harbored in the majority of cases a phylogenetically low diversity community but highly active in terms of variety of catabolized carbon compounds. An exception was the salt-stressed site 1 community, where no specific bacterial group was enriched in the rhizosphere. However, in the endosphere of this site, Bacteroidetes families were strongly featured: Flavobacteriaceae and Sphingobacteriaceae. The latter was a consistent inhabitant of the D. antarctica endosphere, and their mean relative abundance was significantly higher compared to C. quitensis endosphere. Members of this family were noted to proliferate in salt-stressed plants’ rhizosphere and root tissues [59, 60] and were proven to confer tolerance to osmotically challenging conditions [61]. Those families were also observed in the invasive grass Poa annua L. communities presumably aiding its establishment in Antarctica, especially the Flavobacteriaceae [62]. An interesting case is the relative abundance of the family Rhizobiaceae, known to hold key species of plant beneficial rhizobacteria [63]. The occurrence of Rhizobiaceae and Sphingobacteriaceae was positively correlated with phenylethylamine catabolism intensity. For the Rhizobiaceae, this connection was previously described by [64] and was thought to indicate the formation of nitrogen-fixing bacteroids within the plant host cells. Despite some site-specific anomalies, the root-associated communities of D. antarctica displayed far greater stability across the sampling locations than C. quitensis communities. This could indicate that at least part of the D. antarctica root-associated microbial community is transmitted vertically, either by seeds or vegetatively due to the scattering of turf pieces. Monocotyledons of the Poaceae family were proven to be superior in carrying a beneficial bacteria load with their seeds as compared to other plant species [65]. This load of selected plant beneficial bacteria can be responsible for the ecological success of D. antarctica in the Antarctic region and the wider ecological niche than C. quitensis [66, 67] but also for its relatively low genetic diversity [15]. The variability that is introduced by sexual reproduction might diminish the grass’ compatibility with its associated microbial community which could have evolved since the Pliocene colonization event. Such compatibility loss was frequently observed in cultivars of genetically altered agricultural crops [68, 69]. While the majority of the D. antarctica root microbiome might contain facultative endophytes, dispersing through the soil and colonizing C. quitensis as indicated by the common core microbiome consisting of five bacterial families, this might not apply to the Rhizobiaceae. They seem to be exclusive D. antarctica obligatory root endophytes, as they were not observed in considerable abundance in the rhizospheric soils nor the endospheres of here examined C. quitensis specimens nor the invasive in Antarctica grass P. annua (yet still present in European P. annua samples) [62].
In conclusion, the Antarctic-native flowering plants display different strategies in assembling their root-associated microbiomes. C. quitensis seems to adjust its resident microbial community to the prevailing conditions even making use of microbivory, presumably due to the lack of associated efficient nitrogen fixers. D. antarctica on the other hand is inclined to rely on a fixed subset of bacteria that are presumably vertically passed to the daughter plant. This grass species holds to some obligatory nitrogen-fixing endophytes as well as other taxa that do not colonize C. quitensis roots, yet a shared core microbiome is likely to exist. Consequently, the “enigma” behind the presence of only two flowering plants in Antarctica might be strongly connected to their unique relationships with rhizospheric and root-dwelling bacteria.
Data Availability
Illumina reads were deposited in the NCBI Sequence Read Archive (SRA) as BioProject PRJNA726953.
References
Pearce DA (2012) Extremophiles in Antarctica: life at low temperatures. In: Stan-Lotter H, Fendrihan S (eds) Adaption of microbial life to environmental extremes (pp. 87–118). Springer, Vienna. pp. 87–118
Parnikoza IY, Maidanuk DN, Kozeretska IA (2007) Are Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. migratory relicts? Cytol Genet 41:226–229
Biersma EM, Torres-Díaz C, Molina-Montenegro MA, Newsham KK, Vidal MA, Collado GA, Acuña-Rodríguez IS, Ballesteros GI, Figueroa ChC, Goodall-Copestake WP, Leppe MA, Cuba-Díaz M, Valladares MA, Pertierra LR, Convey P (2020) Multiple late-Pleistocene colonisation events of the Antarctic pearlwort Colobanthus quitensis (Caryophyllaceae) reveal the recent arrival of native Antarctic vascular flora. J Bioge 47(8):1663–1673. https://doi.org/10.1111/jbi.13843
Alberdi M, Bravo LA, Gutiérrez A, Gidekel M, Corcuera LJ (2002) Ecophysiology of Antarctic vascular plants. Phys Plantarum 115(4):479–486. https://doi.org/10.1034/j.1399-3054.2002.1150401.x
Mosyakin SL, Bezusko LG, Mosyakin AS (2007) Origins of native vascular plants of Antarctica: comments from a historical phytogeography viewpoint. Cytol Genet 41(5):308–316
Theocharis A, Clement C, Barka EA (2012) Physiological and molecular changes in plants grown at low temperatures. Planta 235:1091–1105
Zuñiga GE, Alberdi M, Corcuera LJ (1996) Non-structural carbohydrates in Deschampsia antarctica Desv. from South Shetland Islands, maritime Antarctic. Environ Exp Bot 36:393–399
Bravo LA, Ulloa N, Zuñiga GE, Casanova A, Corcuera LJ, Alberdi M (2001) Cold resistance in Antarctic angiosperms. Physiol Plant 111:55–65
Bravo LA, Griffith M (2005) Characterization of antifreeze activity in Antarctic plants. J Exp Bot 56:1189–1196
Chew O, Lelean S, John UP, Spangenberg GC (2012) Cold acclimation induces rapid and dynamic changes in freeze tolerance mechanisms in the cryophile Deschampsia antarctica E. Desv Plant Cell Environ 35:829–837
Xiong FS, Mueller EC, Day TA (2000) Photosynthetic and respiratory acclimation and growth response of Antarctic vascular plants to contrasting temperature regimes. Am J Bot 87:700–710
Park AK, Kim I-S, Do H, Kim H, Choi W, Jo S-W, Shin SC, Lee JH, Yoon H-S, Kim H-W (2019) Characterization and structural determination of cold-adapted monodehydroascorbate reductase, MDHAR, from the Antarctic hairgrass Deschampsia antarctica. Curr Comput-Aided Drug Des 9:537
Convey P (1996) Reproduction of Antarctic flowering plants. Antarctic Sci 8(2):127–134
Cavieres LA, Sáez P, Sanhueza C, Sierra-Almeida A, Rabert C, Corcuera LJ, Alberdi M, Bravo LA (2016) Ecophysiological traits of Antarctic vascular plants: their importance in the responses to climate change. Plant Ecol 217(3):343–358. https://doi.org/10.1007/s11258-016-0585-x
Chwedorzewska KJ, Bednarek PT (2008) Genetic variability in the Antarctic hairgrass Deschampsia antarctica Desv. from maritime Antarctic and sub-Antarctic sites. Polish J Ecol 56:209–216
Giełwanowska I, Androsiuk P, Chwedorzewska K, Szandar K (2015) Genetic variability of Colobanthus quitensis from King George Island (Antarctica). Pol Polar Res 281-295.https://doi.org/10.1515/popore−2015−001
Dixit VK, Misra S, Mishra SK, Joshi N, Chauhan PS (2021) Rhizobacteria‐mediated bioremediation: insights and future perspectives. In Parray JA, Mahmoud AHAE, Sayyed R (eds) Soil bioremediation: an approach towards sustainable technology, pp. 193–211. https://doi.org/10.1002/9781119547976.ch9
Flocco CG, Mac Cormack WP, Smalla K (2019) Antarctic soil microbial communities in a changing environment: their contributions to the sustainability of Antarctic ecosystems and the bioremediation of anthropogenic pollution. In: ed. Castro-Sowinski S, The ecological role of micro-organisms in the Antarctic environment. Springer, Cham pp. 133–161.
Yu P, Hochholdinger F (2018) The role of host genetic signatures on root–microbe interactions in the rhizosphere and endosphere. Front Plant Sci 9:1896. https://doi.org/10.3389/fpls.2018.01896
Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CM, Schenk PM (2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552. https://doi.org/10.3389/fmicb.2017.02552
Beckers B, De Beeck MO, Weyens N, Boerjan W, Vangronsveld J (2017) Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:1–17
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60(4):579–598. https://doi.org/10.1007/s13213-010-0117-1
Arenz BE, Blanchette RA, Farrell RL (2014) Fungal diversity in Antarctic soils. Antarctic terrestrial microbiology. Springer, Berlin, Heidelberg, pp 35–53
Teixeira LC, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, Rosado AS (2010) Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J 4(8):989–1001. https://doi.org/10.1038/ismej.2010.35
Zhang Q, Acuña JJ, Inostroza NG, Duran P, Mora ML, Sadowsky MJ, Jorquera MA (2020) Niche differentiation in the composition, predicted function, and co-occurrence networks in bacterial communities associated with Antarctic vascular plants. Front Microbiol 11:1036. https://doi.org/10.3389/fmicb.2020.01036
Molina-Montenegro MA, Ballesteros GI, Castro-Nallar E, Meneses C, Gallardo-Cerda J, Torres-Díaz C (2019) A first insight into the structure and function of rhizosphere microbiota in Antarctic plants using shotgun metagenomic. Polar Biol 42(10):1825–1835. https://doi.org/10.1007/s00300-019-02556-7
Jones JB. (2001) Laboratory guide for conducting soil tests and plant analysis (No. S593 J65). CRC press.
Tatur A, Myrcha A, Niegodzisz J (1997) Formation of abandoned penguin rookery ecosystems in the maritime Antarctic. Polar Biol 17(5):405–417
Lunau M, Lemke A, Walther K, Martens-Habbena W, Simon M (2005) An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environl Microb 7(7):961–968. https://doi.org/10.1111/j.1462-2920.2005.00767.x
Szymańska S, Płociniczak T, Piotrowska-Seget Z, Złoch M, Ruppel S, Hrynkiewicz K (2016) Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol Res 182:68–79. https://doi.org/10.1016/j.micres.2015.09.007
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41(1):e1–e1
Weber KP, Legge RL (2009) One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J Microbiol Meth 79(1):55–61. https://doi.org/10.1016/j.mimet.2009.07.020
Moyer CL, Morita RY (2007). Psychrophiles and psychrotrophs eLS. https://doi.org/10.1002/9780470015902.a0000402.pub2
Grzesiak J, Zdanowski MK, Górniak D, Świątecki A, Aleksandrzak-Piekarczyk T, Szatraj K, Nieckarz S-K, M, (2015) Microbial community changes along the Ecology Glacier ablation zone (King George Island, Antarctica). Polar Biol 38(12):2069–2083. https://doi.org/10.1007/s00300-015-1767-z
Grzesiak J, Kaczyńska A, Gawor J, Żuchniewicz K, Aleksandrzak-Piekarczyk T, Gromadka R, Zdanowski MK (2020) A smelly business: microbiology of Adélie penguin guano (Point Thomas rookery, Antarctica). Sci Total Environ 714:136714. https://doi.org/10.1016/j.scitotenv.2020.136714
Grzesiak J, Woltyńska A, Zdanowski MK, Górniak D, Świątecki A, Olech MA, Aleksandrzak-Piekarczyk, T. (2021). Metabolic fingerprinting of the Antarctic cyanolichen Leptogium puberulum–associated bacterial community (Western Shore of Admiralty Bay, King George Island, Maritime Antarctica). Microb Ecol, 1-12.https://doi.org/10.1007/s00248-021-01701-2
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Micr 67(5):1613
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. Accessed 27 Mar 2021
Long HH, Schmidt DD, Baldwin IT (2008) Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE 3(7):e2702. https://doi.org/10.1371/journal.pone.0002702
Dawson W, Hör J, Egert M, van Kleunen M, Pester M (2017) A small number of low-abundance bacteria dominate plant species-specific responses during rhizosphere colonization. Front Microbiol 8:975. https://doi.org/10.3389/fmicb.2017.00975
Ladygina N, Hedlund K (2010) Plant species influence microbial diversity and carbon allocation in the rhizosphere. Soil Biol Biochem 42(2):162–168. https://doi.org/10.1016/j.soilbio.2009.10.009
Khare E, Mishra J, Arora NK (2018) Multifaceted interactions between endophytes and plant: developments and prospects. Front Microbiol 9:2732. https://doi.org/10.3389/fmicb.2018.02732
McDowell N, Pockman WT, Allen CD, Breshears DD, Cobb N, KolbT PJ, Sperry J, West A, Williams DG, Yepez EA (2008) Mechanisms of plant survival and mortality during drought: why do some plants survive while others succumb to drought? New Phytol 178(4):719–739. https://doi.org/10.1111/j.1469-8137.2008.02436.x
Gallardo-Cerda J, Levihuan J, Lavín P, Oses R, Atala C, Torres-Díaz C, Barrera A, Molina-Montenegro MA (2018) Antarctic rhizobacteria improve salt tolerance and physiological performance of the Antarctic vascular plants. Polar Biol 41(10):1973–1982
Chen P, Zhang C, Ju X, Xiong Y, Xing K, Qin S (2019) Community composition and metabolic potential of endophytic Actinobacteria from coastal salt marsh plants in Jiangsu. China Front Microbiol 10:1063. https://doi.org/10.3389/fmicb.2019.01063
Gong Y, Bai JL, Yang HT, Zhang WD, Xiong YW, Ding P, Qin S (2018) Phylogenetic diversity and investigation of plant growth-promoting traits of Actinobacteria in coastal salt marsh plant rhizospheres from Jiangsu. China Sys Appl Microbiol 41(5):516–527. https://doi.org/10.1016/j.syapm.2018.06.003
Awad NM, Turky AS, Abdelhamid MT, Attia M (2012) Ameliorate of environmental salt stress on the growth of Zea mays L. plants by exopolysaccharides producing bacteria. J Appl Sci Res , 8(4): 2033–2044.
Yu P, He X, Baer M, Beirinckx, S, Tian, T, Moya, YA, Zhang X, Deichmann M, Frey FP, Bresgen V, Li Ch, Razavi BS, Schaaf G, von Wirén N, Su Z, Bucher M, Tsuda K, Goormachtig S, Chen X Hochholdinger, F. (2021). Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nature Plants, 1-19.https://doi.org/10.1038/s41477-021-00897-
Pershina E, Valkonen J, Kurki P, Ivanova E, Chirak E, Korvigo I, Provorov N, Andronov E (2015) Comparative analysis of prokaryotic communities associated with organic and conventional farming systems. PLoS ONE 10(12):e0145072. https://doi.org/10.1371/journal.pone.0145072
Santos CR, Hoffmam ZB, de Matos Martins VP, Zanphorlin LM, de Paula Assis LH, Honorato RV, Lopes de Oliveira PS, Ruller R, Murakami MT (2014) Molecular mechanisms associated with xylan degradation by Xanthomonas plant pathogens. J Biol Chem 289(46):32186–32200. https://doi.org/10.1074/jbc.M114.605105
Robledo M, Jiménez-Zurdo JI, Velázquez E, Trujillo ME, Zurdo-Piñeiro JL, Ramírez-Bahena MH, Ramos B, Díaz-Mínguez JM, Dazzo F, Martínez-Molina E, Mateos PF (2008) Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. PNAS 105(19):7064–7069. https://doi.org/10.1073/pnas.0802547105
Kielland K (1994) Amino acid absorption by arctic plants: implications for plant nutrition and nitrogen cycling. Ecology 75(8):2373–2383. https://doi.org/10.2307/1940891
Näsholm T, Kielland K, Ganeteg U (2009) Uptake of organic nitrogen by plants. New Phytol 182(1):31–48. https://doi.org/10.1111/j.1469-8137.2008.02751.x
White JF, Kingsley KL, Verma SK, Kowalski KP (2018) Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 6(3):95. https://doi.org/10.3390/microorganisms6030095
Paungfoo-Lonhienne C, Rentsch D, Robatzek S, Webb RI, Sagulenko E, Näsholm T, Schmidt S, Lonhienne TG (2010) Turning the table: plants consume microbes as a source of nutrients. PLoS ONE 5(7):e11915. https://doi.org/10.1371/journal.pone.0011915
Robles-Aguilar AA, Grunert O, Hernandez-Sanabria E, Mysara M, Meers E, Boon N, Jablonowski ND (2020) Effect of applying struvite and organic n as recovered fertilizers on the rhizosphere dynamics and cultivation of lupine (Lupinus angustifolius). Fornt Plant Sci 11:1752. https://doi.org/10.3389/fpls.2020.572741
Höflich G, Tauschke M, Kühn G, Rogasik J (2000) Influence of agricultural crops and fertilization on microbial activity and microorganisms in the rhizosphere. J Agron Crop Sci Crop Sci 184(1):49–54. https://doi.org/10.1046/j.1439-037x.2000.00369.x
Mur LA, Simpson C, Kumari A, Gupta AK, Gupta KJ (2017) Moving nitrogen to the centre of plant defence against pathogens. Ann Bot 119(5):703–709. https://doi.org/10.1093/aob/mcw179
Benidire L, Khalloufi F, Oufdou K, Barakat M, Tulumello J, Ortet P, Heulin T, Achouak W (2020) Phytobeneficial bacteria improve saline stress tolerance in Vicia faba and modulate microbial interaction network. Sci Total Environ 729:139020. https://doi.org/10.1016/j.scitotenv.2020.139020
Wang M, Chen S, Chen L, Wang D (2019) Responses of soil microbial communities and their network interactions to saline-alkaline stress in Cd-contaminated soils. Environ Pollut 252:1609–1621. https://doi.org/10.1016/j.envpol.2019.06.082
Fan D, Subramanian S, Smith DL (2020) Plant endophytes promote growth and alleviate salt stress in Arabidopsis thaliana. Sci Rep 10(1):1–18. https://doi.org/10.1038/s41598-020-69713-
Znój A, Grzesiak J, Gawor J, Gromadka R, Chwedorzewska KJ (2021) Bacterial communities associated with Poa annua roots in central European (Poland) and Antarctic Settings (King George Island). Microorganisms 9(4):811. https://doi.org/10.3390/microorganisms9040811
Spaink HP, Kondorosi A, Hooykaas PJ (eds) (2012) The Rhizobiaceae. Molecular Biology of Model Plant-Associated Bacteria Springer, Dordrecht
Fujihara S, Terakado J, Nishibori N (2006) Accumulation of an aromatic amine, β-phenethylamine, in root nodules of adzuki bean Vigna angularis. Plant Soil 280(1):229–237. https://doi.org/10.1007/s11104-005-3096-4
Truyens S, Weyens N, Cuypers A, Vangronsveld J (2015) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7(1):40–50. https://doi.org/10.1111/1758-2229.12181
Parnikoza I, Kozeretska I, Kunakh V (2011) Vascular plants of the Maritime Antarctic: origin and adaptation. Am J Plant Sci 2(03):381–395. https://doi.org/10.4236/ajps.2011.23044
Olech M (2002) Plant communities on King George Island. In: Beyer L, Bölter M (eds) Geoecology of Antarctic ice-free coastal landscapes. Ecological Studies (Analysis and Synthesis), vol 154. Springer, Berlin, Heidelberg. pp.215–231 https://doi.org/10.1007/978-3-642-56318-8_12
Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LF, de Hollander M, Garcia AA, Ramírez CA, Raaijmakers MR, JM, (2017) Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J 11(10):2244–2257. https://doi.org/10.1038/ismej.2017.85
Zachow C, Müller H, Tilcher R, Berg G (2014) Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima—ancestor of all beet crops—and modern sugar beets. Front Microbiol 5:415. https://doi.org/10.3389/fmicb.2014.00415
Acknowledgements
Samples and data were obtained due to the scientific facility of the H. Arctowski Polish Antarctic Station.
Funding
This work was supported by the National Science Center, Poland (Grant 2016/21/N/NZ9/01536).
Author information
Authors and Affiliations
Contributions
A.Z.: conceptualization, validation, investigation, resources, writing—review and editing, project administration, and funding acquisition. J.G. (Jakub Grzesiak): conceptualization, software, formal analysis, data curation, writing—original draft, and visualization. J.G. (Jan Gawor): validation, investigation, data curation, and writing—review and editing. R.G.: writing—review and editing. K.J.C.: conceptualization, resources, writing—review and editing, and supervision. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Znój, A., Gawor, J., Gromadka, R. et al. Root-Associated Bacteria Community Characteristics of Antarctic Plants: Deschampsia antarctica and Colobanthus quitensis—a Comparison. Microb Ecol 84, 808–820 (2022). https://doi.org/10.1007/s00248-021-01891-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01891-9