Abstract
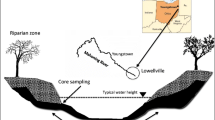
Sediment contaminated with polycyclic aromatic hydrocarbons (PAHs) is widely distributed in aquatic ecosystems. The microbial community structure of riverbank PAH-contaminated sediments was investigated using phospholipid-derived fatty acid (PLFA) analysis. Surface and subsurface riverbank sediment was collected from a highly contaminated site and from an uncontaminated site along the Mahoning River, OH. PAH concentrations, physical sediment characteristics, and other microbial community parameters (biomass as phospholipid phosphate (PLP) and activity) were also measured. PAHs were detected in all samples but were only quantifiable in the contaminated (250 μg/g g−1) subsurface sediment. Subsurface samples from both locations showed very similar PLP values and distribution of PLFAs, with 27–37 % of the microbial community structure being composed of sulfate reducing and other anaerobic bacteria. Principal components analysis indicated no correlation between PAH contamination and PLFA diversity. Although PLP and phospholipid fatty acid measurements of bacterial communities did not reflect the environmental differences among sites, the highly PAH-contaminated sediment showed the highest measured microbial activity (reduction of 1,200 nmol INT g−1 h−1), likely from a population adapted to environmental pollutants, rates that are much higher than measured in many uncontaminated soil and sediment systems. These data warrant further investigation into community structure at the genetic level and indicate potential for bioremediation by indigenous microbes.








Similar content being viewed by others
References
Agency for Toxic Substances and Disease Registry (ATSDR) Division of Toxicology (2010) ToxFAQs for polycyclic aromatic hydrocarbons. ATSDR, Atlanta. http://www.atsdr.cdc.gov/tfacts69.html. Accessed Nov 2010
Baniulyte D, Favila E, Kelly J (2009) Shifts in microbial community composition following surface application of dredged river sediments. Microb Ecol 57:160–169
Bausenwein U, Gattinger A, Langer U, Embacher A, Hartmann H-P, Sommer M, Munch JC, Schloter M (2008) Exploring soil microbial communities and soil organic matter: variability and interactions in arable soils under minimum tillage practice. Appl Soil Ecol 40:67–77
Bhupathirajua VK, Hernandezb M, Landfeara D, Alvarez-Cohen L (1999) Application of a tetrazolium dye as an indicator of viability in anaerobic bacteria. J Microbiol Methods 37:231–243
Björk R, Björkmand M, Andersson M, Klemedtsson L (2008) Temporal variation in soil microbial communities in Alpine tundra. Soil Biology & Biochemistry 40:266–268
Blume E, Bischoff M, Reichert JM, Moormanc T, Konopka A, Turco RF (2002) Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl Soil Ecol 20:171–181
Boschker HTS, Middelburg JJ (2002) Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol Ecol 40:85–95
DeBruyn J, Mead T, Wilhelm S, Sayler G (2009) PAH biodegradative genotypes in Lake Erie sediments: evidence for broad geographical distribution of pyrene-degrading Mycobacteria. Environ Sci Technol 43:3467–3473
Dijkman NA, Boschker HT, Stal LJ, Kromkamp JC (2010) Composition and heterogeneity of the microbial community in a coastal microbial mat a as revealed by the analysis of pigments and phospholipid-derived fatty acids. J Sea Research 63:62–70
Drenovsky RE, Elliot GN, Graham KJ, Scow KM (2004) Comparison of phospholipid fatty acid (PLFA) and total soil fatty acid methyl esters (TSFAME) for characterizing soil microbial communities. Soil Biology & Biochemistry 36:1793–1800
Drijber RA, Doran JW, Parkhurst AM, Lyon DJ (2000) Changes in soil microbial community structure with tillage under long-term wheat-fallow management. Soil Biology & Biochemistry 32:1419–1430
Dunn RM, Mikola J, Bol R, Bardgett RD (2006) Influence of microbial activity on plant-microbial competition for organic and inorganic nitrogen. Plant Soil 289:321–334
Duran M, Haznedarog BZ, Zitomer DH (2006) Microbial source tracking using host specific FAME profiles of fecal coliforms. Water Res 40:67–74
Fang J, Findlay RH (1996) The use of a classic lipid extraction method for simultaneous recovery of organic pollutants and microbial lipids from sediments. J Microbiol Meth 27:63–71
Fang J, Barcelona MJ (1998) Biogeochemical evidence for microbial community change in a jet fuel hydrocarbons-contaminated aquifer. Org Geochem 29:4899–4907
Findlay RH (2004) Determination of microbial community structure using phospholipid fatty acid profiles. In: Kowalchuk GA, Bruijn FJ, Head IM, Akkermans AD, van Elsas JD (eds) Molecular microbial ecology manual, 2nd edn. Kluwer Academic, Norwell, pp 983–1005
Findlay RH, Trexler MB, Guckert JB, White DC (1989) Laboratory study of disturbance in marine sediments: response of a microbial community. Mar Ecol Prog Ser 62:121–133
Foster AL, Munk L, Koski RA, Shanks WC III, Stillings LL (2008) Relationships between microbial communities and environmental parameters at sites impacted by mining of volcanogenic massive sulfide deposits, Prince William Sound, Alaska. Appl Geochem 23:279–307
Frostegård Å, Tunlid A, Bååth E (2011) Use and misuse of PLFA measurements in soils. Soil Biology and Biochemistry 43(8):1621–1625
Glucksman AM, Skipper HD, Brigmon RL, Doming JW (2000) Use of MIDI-FAME technique to characterize groundwater communities. J Appl Microbiol 88:711–719
Haack SK, Garchow H, Odelson DA, Forney LJ, Klug MJ (1994) Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacteria communities. Appl Environ Microbiol 60:2483–2493
Hayes L, Nevin K, Lovley D (1999) Role of prior exposure on anaerobic degradation of naphthalene and phenanthrene in marine harbor sediments. Org Geochem 30:937–945
Helmisaari HS, Derome J, Fritze H, Nieminen T, Palmgren K, Salemaa M, Vanha-Majamaa I (1995) Copper in Scots pine forests around a heavy metal smelter in south-western Finland. Water Air Soil Pollut 85:1727–1732
Hoffman M, Keys DE, Song K-Y, Brown EW, Fry FS, Whittaker P (2008) Evaluation of multiple strains of Enterobacter sakazakii using fatty acid profiles. Food Chem 107:1623–1628
Jeffery S, Harris JA, Rickson RJ, Ritz K (2007) Microbial community phenotypic profiles change markedly with depth within the first centimeter of the arable soil surface. Soil Biol Biochem 39:1226–1229
Johnston GP, Kalik Z, Johnston CG (2009) Effect on carbonyl cyanide m-chlorophenylhydrazone (CCCP) on microbial activity in historically contaminated river sediment. American Society for Microbiology General Meeting, Boston
Joynt J, Bischoff M, Turco R, Konopka A (2006) Microbial community analysis of soils contaminated with lead, chromium and petroleum hydrocarbons. Microb Ecol 51:209–219
Kelly J, Häggblom M, Tate R (2003) Effects of heavy metal contamination and remediation on soil microbial communities in the vicinity of a zinc smelter as indicated by analysis of microbial community phospholipid fatty acid profiles. Biol Fertil Soils 38:65–71
Kleikemper J, Pelz O, Schroth MH, Zeyer J (2002) Sulfate-reducing bacterial community response to carbon source amendments in contaminated aquifer microcosms. Microb Ecol 42:109–118
Langworthy DE, Stapleton RD, Sayler GS, Findlay RH (1998) Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl Environ Microbiol 64:3422–3428
Langworthy DE, Stapleton RD, Sayler GS, Findlay RH (2002) Lipid analysis of the response of a sedimentary microbial community to polycyclic aromatic hydrocarbons. Microb Ecol 43:189–198
Mathew M, Obbard JP (2001) Optimisation of the dehydrogenase assay for measurement of indigenous microbial activity in beach sediments contaminated with petroleum. Biotechnol Lett 23:227–230
Mosher JJ, Levison BS, Johnston CG (2003) A simplified dehydrogenase enzyme assay in contaminated sediment using 2-(p-iodophenyl)-3(p-nitrophenyl)-5-phenyl tetrazolium chloride. J Microb Meth 53:411–415
Mosher JJ, Findlay RH, Johnston CG (2006) Physical and chemical factors affecting microbial biomass and activity in contaminated subsurface sediment. Can J Microbiol 52:397–403
Ohio Environmental Protection Agency (1996) Biological and water quality study of the Mahoning River Basin. OEPA technical report MAS/1995-12-14, pp 1–249
Obbard JP, Ng KL, Xu R (2004) Bioremediation of petroleum contaminated beach sediments: use of crude palm oil and fatty acids to enhance indigenous biodegradation. Water Air Soil Poll 157:149–161
Pennanen T, Frosetgård A, Fritze H, Bååth E (1996) Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal polluted gradients in coniferous forests. Appl Environ Microbiol 62:420–428
Ringelberg DB, Talley JW, Perkins EJ, Tucker SG, Luthy RG, Bouwer EJ, Fredickson HL (2001) Succession of phenotypic, genotypic, and metabolic community characteristics during in vitro bioslurry treatment of polycyclic aromatic hydrocarbon-contaminated sediments. Appl Env Microbiol 67:1542–1550
Ruamps LS, Nunan N, Chenu C (2011) Microbial biogeography at the soil pore scale. Soil Biol Biochem 43:280–286
Ruess L, Chamberlain PM (2010) The fat that matters: soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biology & Biochemistry 42:1898–1910
Slater G, Cowie B, Harper N, Droppo I (2008) Variation in PAH input and microbial community in surface sediments of Hamilton Harbour: implications to remediation and monitoring. Environ Pollut 153:60–70
Song Y, Deng SP, Acosta-Martínez V, Katsaliro E (2008) Characterization of redox-related soil microbial communities along a river floodplain continuum by fatty acid methyl ester (FAME) and 16S rRNA genes. Appl Soil Ecol 40:499–509
Syakti AD, Mazzella N, Nerini D, Guiliano M, Bertrand JC, Doumenq P (2006) Phospholipid fatty acids of a marine sedimentary microbial community in a laboratory microcosm: responses to petroleum hydrocarbon contamination. Org Geochem 37:1617–1628
Tayler S, May E (2000) Investigations of the localization of bacterial activity on sandstone from ancient monuments. Int Biodeter Biodegr 46:327–333
Tiessen H, Moir JO (1993) Total organic carbon. In: Carter ME (ed) Soil sampling and methods of analysis. Lewis, Ann Arbor, pp 187–211
Trevors JT, Mayfield CI, Innis WE (1982) Measurement of electron transport system (ETS) activity in soil. Microb Ecol 8:163–168
United States Army Corps of Engineers (USACE) (1999) Mahoning River Dredging Study (Ohio). Energy and Water Development Appropriations Act (Public Law 104–303)
Vestal JR, White DC (1989) Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. Biosci 39:535–541
Von Mersi V, Schinner F (1991) An improved and accurate method for determining the dehydrogenase activity of soils with iodonitrotetrazolium chloride. Biol Fertil Soils 11:216–220
Villanueva L, Navarrete A, Urmeneta J, White DC, Guerrero R (2007) Analysis of diurnal and vertical microbial diversity of a hypersaline microbial mat. Arch Microbiol 188:137–146
Wakadikar K, Sil A, Kolekar N, Tandon S, Kumar R (2011) Effect of non-aqueous drilling fluid and its synthetic base oil on soil health as indicated by its dehydrogenase activity. Environ Earth Sci 64:25–28
Whittaker P (2009) Comparison of Yersinia pestis to other closely related Yersinia species using fatty acid profiles. Food Chem 116:629–632
Williams MA, Rice CW (2007) Seven years of enhanced water availability influences the physiological, structural, and functional attributes of a soil microbial community. Appl Soil Ecol 35:535–545
Wright D (1983) Species-energy theory, an extension of species-area theory. Oikos 41:496–506
Wu Y, Ding N, Wang G, Xu J, Wu J, Brookes PC (2009) Effects of different soil weights, storage times and extraction methods on soil phospholipid fatty acid analyses. Geoderma 150:171–178
Acknowledgments
This work was supported by a Presidential Academic Center for Excellence in Research grant from Youngstown State University. The authors are grateful for useful discussions about statistics from T Diggins from the Department of Biological Sciences, Youngstown State University and J Kearns from the Department of Mathematics and Statistics, Youngstown State University and for editorial suggestions by GP Johnston from the Biology Department, Kent State University.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PPT 775 kb)
Rights and permissions
About this article
Cite this article
Pratt, B., Riesen, R. & Johnston, C.G. PLFA Analyses of Microbial Communities Associated with PAH-Contaminated Riverbank Sediment. Microb Ecol 64, 680–691 (2012). https://doi.org/10.1007/s00248-012-0060-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0060-8