Abstract
The research field of animal and plant symbioses is advancing from studying interactions between two species to whole communities of associates. High-throughput sequencing of microbial communities supports multiplexed sampling for statistically robust tests of hypotheses about symbiotic associations. We focus on ambrosia beetles, the increasingly damaging insects primarily associated with fungal symbionts, which have also been reported to support bacteria. To analyze the diversity, composition, and specificity of the beetles’ prokaryotic associates, we combine global sampling, insect anatomy, 454 sequencing of bacterial rDNA, and multivariate statistics to analyze prokaryotic communities in ambrosia beetle mycangia, organs mostly known for transporting symbiotic fungi. We analyze six beetle species that represent three types of mycangia and include several globally distributed species, some with major economic importance (Dendroctonus frontalis, Xyleborus affinis, Xyleborus bispinatus–ferrugineus, Xyleborus glabratus, Xylosandrus crassiusculus, and Xylosandrus germanus). Ninety-six beetle mycangia yielded 1,546 bacterial phylotypes. Several phylotypes appear to form the core microbiome of the mycangium. Three Mycoplasma (originally thought restricted to vertebrates), two Burkholderiales, and two Pseudomonadales are repeatedly present worldwide in multiple beetle species. However, no bacterial phylotypes were universally present, suggesting that ambrosia beetles are not obligately dependent on bacterial symbionts. The composition of bacterial communities is structured by the host beetle species more than by the locality of origin, which suggests that more bacteria are vertically transmitted than acquired from the environment. The invasive X. glabratus and the globally distributed X. crassiusculus have unique sets of bacteria, different from species native to North America. We conclude that the mycangium hosts in multiple vertically transmitted bacteria such as Mycoplasma, most of which are likely facultative commensals or parasites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The study of symbioses, whether between humans and their microbiome or insects and their symbionts, has begun to see a shift from research focusing on pairs of species toward the study of symbiotic communities. This is in part because new methods of environmental DNA sampling are revealing many previously unnoticed bacterial and fungal associates. In some symbiotic communities, both coevolved symbionts and incidental associates may play important but different roles. One such example is the ambrosia symbiosis between wood-boring beetles and diverse fungal and microbial associates. The beetles are often cited as a textbook example of tree disease vectors [1], yet in some cases, the disease agents may be incidental opportunists, rather than the primary symbionts of the beetles [2, 3]. Such complexity of interactions highlights the need to survey the full diversity of microbial associates of ambrosia beetles. Historically, studies of the ambrosia symbiosis have focused on insect interactions with the main fungal symbiont, and few addressed the role of other fungi, and even fewer studies exist on the presence and role of bacteria in this system. Here, we expand the focus of ambrosia symbiosis research to address the prokaryotic component. We combine the approaches of culture-independent microbiology, insect anatomy, and multivariate statistics to analyze bacterial communities inside one of the most frequently evolved organs mediating a symbiosis—the mycangia of ambrosia beetles.
The ambrosia symbiosis of beetles and fungi is one of the most evolutionarily successful and ecologically important insect mutualisms [4], with increasing global economic impact [3]. The symbiosis evolved in at least 11 independent bark beetle ancestors (Curculionidae, Scolytinae, and Platypodinae) and involves over 3,500 beetle species and an unknown but large number of symbiotic wood-decay fungi [4–6, 42, 43]. The fungi are carried in specialized membranous invaginations equipped with secretory glands—mycangia [7, 8]. As most of the research on ambrosia symbiosis focused on mycangial fungi, little is known about bacteria in mycangia. Even the question of whether bacteria are present in beetle mycangia is poorly resolved, and whether the bacterial community is shaped by the type of mycangium, beetle evolutionary origin, or geographic origin has never been tested.
Presence of bacteria in mycangia has been suggested by previous studies [12, 16, 18], but not explicitly tested, as bacterial associates of beetles are typically studied from crushed whole beetles or from external beetle surfaces, and the isolated bacteria might not have been inside mycangia. In this work, we targeted bacteria specifically within beetle mycangia. This often large organ [9] is the functional pivot of the ambrosia symbiosis [5]. Our current understanding is that mycangia are the products of millions of years of mutualistic coevolution between the beetle and fungal symbionts. The mycangium provides an environment in which exposure to UV light, abrasion, and tree defensive chemicals are reduced, and where symbionts are provided nutrients [10]. At the same time, it also excludes most fungi while supporting the few coevolved symbionts [11]. This selectiveness may hypothetically also affect bacteria. Bacteria are abundant and diverse on the body surface and within galleries of ambrosia and bark beetles [12–16], but whether they are also present within the mycangium, or if the mycangium is kept bacteria-free, has been only marginally explored [12]. It has been noted that bacteria are present in the ambrosia beetle niche [17, 18], but their abundance, diversity, composition, and transmission in mycangia have not been analyzed.
We used a high-throughput, culture-independent approach to search for and, if present, identify prokaryotes directly from the mycangia of five species of fungus-farming beetles. In doing so, we addressed four questions:
-
1.
Do ambrosia beetle mycangia, organs originally evolved to host fungi, also contain a significant diversity of bacteria?
-
2.
Is there a “core microbiome” associated with ambrosia beetle mycangia? One hypothesis is that mycangia of all ambrosia beetles, regardless of species, may be dominated by a suite of microbial members shared among samples, the “taxonomic core microbiome” [19]. Alternatively, associations may vary among different beetle species or individuals.
-
3.
If bacteria are present in mycangia and their communities vary among beetles, to what extent is the community composition determined by the geographic origin of the beetle, and to what extent is it the host beetle species? If bacterial communities are more specific to localities than to beetle species, they are likely facultative opportunists from the environment. On the other hand, if bacteria in mycangia display specificity to beetle species regardless of locality, they are potentially symbionts transmitted vertically across beetle generations.
-
4.
If bacterial communities are specific to beetle species, are they also more similar between closely related beetles then between unrelated beetles? Congruence between bacterial community similarity and beetle phylogeny is expected if bacteria are exclusively transmitted vertically within beetle species and families.
By sampling multiple species from nine localities from around the world (not all species were collected everywhere), we are not only assembling the broadest sample of ambrosia beetle symbionts ever collected but also creating a baseline dataset for further studies on global host–symbiont dynamics. This is important for highly invasive groups such as the ambrosia beetles [3]. Representatives of this symbiotic complex are usually not economically important in their native regions but can be devastating in invaded regions [3, 20]. It has not yet been tested whether the increased impact is caused by the change of beetle behavior in the invaded regions, or by acquisition of new symbionts (whether fungal or bacterial). By analyzing several species across their global distribution, we are implicitly testing a key assumption of the latter hypothesis: Is there evidence for acquisitions of new symbionts in newly colonized regions?
Methods
To assure sufficient statistical power of our tests, our sampling included six species of fungus-farming beetles, each represented by eight to 28 different individuals, from nine localities around the world (for details, see Table 1). To include phylogenetically diverse beetle representatives, we included species from three clades of ambrosia beetles, each with its own type of mycangium. The first type is a prothoracic mycangium in Dendroctonus frontalis, which has a paired tubular mycangium on the inside of prothoracic pleural plates; the mycangium opens near the procoxa [10]. Dendroctonus represents an independently evolved clade of fungus-farming bark beetles. These beetles inhabit tree phloem rather than xylem, and the adults feed primarily on phloem. However, the larvae depend almost entirely on coevolved fungal symbionts for nutrition, thus we include it among ambrosia beetles in this study. The second type is a mandibular mycangium, the ancestral and most widely distributed form of mycangium in the ambrosia beetle tribe Xyleborini, and characteristic of the genus Xyleborus [21]. Xyleborini are one of the largest and most economically important tribes of ambrosia beetles [3, 20]. The opening of the mandibular mycangium is in the oral cavity. The third type, representing a different clade within the Xyleborini, is the mesonotal mycangium, a large internal organ with opening between the pronotum and the elytral basis [7]. It appears to have evolved only once within the Xyleborini and coincided with a radiation of a successful clade with many genera, including Xylosandrus [21].
Beetles were obtained either by rearing from colonized logs [22] or from cup traps baited with ethanol. Beetles were preserved in 95 % ethanol and stored at −80°C. To extract bacteria from mycangia, we surface-cleaned each beetle by vortexing in water and ethanol and excised the part of each beetle that contained the mycangium. In Xyleborus spp., we sliced off a part of the head between the frons and the joints of mandibles. The taxonomic distinction between Xyleborus ferrugineus and Xyleborus bispinatus is uncertain [23], thus we treated representatives of the complex as one species. In Xylosandrus, we excised the content of the mesonotal mycangium. In Dendroctonus, we exposed both mycangia on the inner sides of the thorax and squeezed the content out with a sterile pin. The environmental control composed of two galleries of Xylosandrus germanus were collected during the same collecting event as the X. germanus beetles. Surface of the gallery containing the fungal garden was scraped into a vial and processed in the same way as the beetle mycangia.
The extracted mycangium was added to 10 μL of Ex-n-Amp™ extraction solution (Sigma-Aldrich), macerated with a pestle, and lysed at 96°C for 10 min. The reaction was deactivated with 10 μL of 3 % BSA; 1 μL of the clear supernatant was used in the polymerase chain reaction (PCR). Amplicons were generated using a universal bacterial/archeal primer pair 515 F and 806R (5′-GTGCCAGCMGCCGCGGTAA-3′ and 5′-GGACTACVSGGGTATCTAAT-3′ [24]). The primer 515 F was appended with a TC linker and a Roche 454 B pyrosequencing adapter; the 806R primer was appended with a 12-bp sample-specific barcode sequence, a CA linker, and a Roche 454 A sequencing adapter. The sample-specific, error-correcting barcode allowed for pooling all amplicons into a single pyrosequencing run. All samples were amplified in triplicated 25-μL reactions using 5 Prime HotMaster polymerase mix (5 Prime, Inc.) under the following cycling conditions—95°C, 1 min, 33 × (95°C, 30 s; 50°C, 1 min; 72°C, 1 min). Amplicons were purified using the UltraClean PCR Clean-up kit (MoBio). Concentration of each amplicon was determined using the Quant-iT™ PicoGreen® dsDNA kit (Invitrogen), and equimolar aliquots of all samples were pooled. Pyrosequencing was carried out on a Roche Genome Sequencer FLX system at Engencore, University of South Carolina, USA.
The sequencing output was processed using the package QIIME [25]. All sequences are available upon request. The output was filtered to contain only sequences with lengths >200 and <1,000 bp with an average quality score >25 and no ambiguous characters. Sequences were assigned to samples according to the 12-bp barcode. Sequences that were ≥97 % similar were grouped into phylotypes using the UCLUST method [25], and the taxonomic identity of each phylotype was determined using the RDP Classifier [26]. Phylotypes occurring in >30 % samples that were unclassified by RDP were identified individually in NCBI BLAST. To confirm the genus identity of the three common Mycoplasma phylotypes and to rule out possible misclassification in RDP and BLAST, we performed a phylogenetic analysis of the placement of our phylotypes in Mycoplasmataceae. We downloaded 16 S sequences of identified strains of Mycoplasma spp. and representatives of other genera within this family from NCBI-GenBank, aligned them with our sequences using MUSCLE [27] (270 bp alignment), and inferred a maximum likelihood phylogeny based on the Tamura-Nei model in MEGA5 [28].
For downstream analyses, a distance matrix was derived using UNIFRAC [30] which takes into account a phylogenetic structure of the community. Representative sequences from all operational taxonomic units were aligned with MUSCLE [27], and the phylogenetic tree and unifrac distance matrix were produced in QIIME. In order to avoid artifacts of PCR, sequencing, and sampling biases [31], we do not use bacterial abundance (read counts) in our analyses; instead, prevalence of a phylotype is calculated as the number of beetle samples yielding one or more reads. Likewise, sample similarities in multivariate analyses were not weighted by abundance. Prior to the analysis, samples were rarefied to 41 reads. Less-strict rarefaction levels were also tested (122 and 330 reads per sample); they did not provide greater explanatory power but resulted in exclusion of most low-yield mesonotal samples. The structure of the bacterial community was visualized using principal coordinates analysis based on the UniFrac distances.
To compare the effect of beetle species and locality on the composition of bacterial communities, we performed a permutational multivariate analysis of variance of pairwise distances between samples (the Adonis function in the Vegan package in R [29]). The factors beetle species, locality, and their interaction were considered fixed factors in the model. Mycangium type was included as a stratifying variable, such that permutations were constrained within mycangium type (the significance of mycangium type was not directly tested, as only one type was sufficiently replicated). To examine pair-wise differences between beetle species in their composition of bacterial communities, we performed Tukey’s honestly significant difference (HSD) post hoc tests on a model of bacterial community composition as a function of beetle species identity (using an analysis of multivariate homogeneity of group variances; data were pooled across beetle locality and mycangium type).
To test whether related beetle species have more similar bacterial communities than unrelated beetles, we tested the congruence between a beetle phylogeny and a dendrogram of similarity between bacterial communities summed within species. The beetle phylogeny was derived from concatenated sequences of cytochrome-oxidase I mtDNA and 28 S rDNA of each species from our previous datasets [32] and from NCBI Genbank, using maximum likelihood with the Tamura-Nei model of evolution in MEGA [28]. Records of bacterial phylotypes were summed up within beetle species, distance matrix obtained with unweighted UniFrac, and summarized in a dendrogram using the UPGMA algorithm in QIIME. Jack-knife support for nodes in the dendrogram were derived from repeating this process on successively rarefied matrices from 1,666 reads (the lowest read count, Xylosandrus crassiusculus) to 833 reads (50 % of data) per beetle species. The significance of congruence between the two matrices was assessed using Mantel test in PopTools [33] with 1,000 iterations. X. germanus was not included in most tests, as it did not yield any bacterial reads.
Results
We successfully amplified bacterial 16 S in 66 out of the initial 96 beetles. In the total of 29,842 reads of sufficient quality, we distinguished 1,546 bacterial phylotypes using a 97 % similarity threshold (see the complete dataset in the Electronic supplementary material).
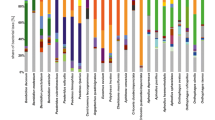
Several phylotypes occurred in many beetle species and were broadly distributed geographically (Table 2). Several phylotypes of Mycoplasma, Burkholderiales, and Pseudomonadales were recovered from more than half of the amplified samples. The genus Mycoplasma was represented by three frequently encountered phylotypes, including the most widespread phylotype (present in 75 % of samples). The placement of the three phylotypes in Mycoplasma, as opposed to placement in other Mycoplasmataceae more often reported from insect hosts, was confirmed using a phylogenetic analysis of the bacterial family (Fig. 1). The second and third most prevalent phylotypes belonged to unidentified Burkholderiales (probably Comamonadaceae, no closer match was available in RDP and BLAST) found in 69 % and 61 % of beetles, respectively. The last two phylotypes present in more than half of the samples were Pseudomonadales: one Acinetobacter and one unidentified Pseudomonadaceae (prevalence 56 % and 48 %, respectively). Several other phylotypes were found in fewer species but still prevalent within certain beetle species (Table 2). Particularly notable is the uniform microbiota of Xyleborus glabratus, where four bacterial phylotypes were shared by all beetle individuals and the same phylotypes were infrequent or absent in other beetle species.
The three most widespread Mycoplasmataceae from ambrosia beetle mycangia are more closely related to Mycoplasma spp. from vertebrates than to confamilial genera more often reported from invertebrates. They are also not monophyletic, which suggests repeated colonization of ambrosia beetles by this group of bacteria. Maximum likelihood phylogeny was reconstructed in MEGA [28] based on the Tamura-Nei model; numbers at nodes represent bootstrap support. Sequences of identified Mycoplasmataceae were obtained from NCBI-Genbank. The analysis is strictly for taxonomic purposes and did not attempt to reconstruct the actual evolution of the clade
The bacterial community as a whole differed strongly among beetle species (Fig. 2; F5,47 = 4.298, R 2 = 0.26; p < 0.001) and less strongly but still significantly among regions (F7,47 = 2.13, R 2 = 0.18; p < 0.001). The prokaryotic communities of X. crassiusculus and X. glabratus are significantly different from several other beetle species (Tukey’s HSD post hoc test; Table 3). Similarities between bacterial communities did not correspond to beetle phylogenetic relationships (Mantel test p = 0.149, Fig. 3). Interestingly, both species with mesonotal mycangia, X. crassiusculus and X. germanus, repeatedly yielded very poor amplicons of bacterial 16 S (average numbers of reads, 60 and 0, respectively). The two environmental samples from galleries of X. germanus yielded a bacterial community as diverse as samples from most mycangia, but the community was significantly different from all mycangial communities.
Principal coordinates analysis (PCoA) of bacterial assemblages from ambrosia beetle mycangia shows the significant specificity of bacterial community composition to certain beetle species. Clustered are samples from X. glabratus, X. crassiusculus, D. frontalis, and samples from the two galleries of X. germanus. Not clustered are the widely dispersed samples from X. affinis and X. bispinatus/ferrugineus. First two axes are shown, explaining 9.04 % and 7.41 % of variation. The PCoA is based on UniFrac distances unweighted by phylotype abundances
Discussion
Although mycangia are usually considered to have evolved to house and disperse the fungi on which ambrosia beetles depend, they also host several regularly occurring bacterial phylotypes. Representatives of Mycoplasma, Burkholderiales, and Pseudomonadales in particular are widespread in mycangia of multiple beetle species from diverse parts of the world. These appear to be true inhabitants of beetle mycangia, rather than environmental or laboratory contaminants: The two control samples of fungus gardens surrounding the beetles were dominated by completely different phylotypes; for example, no Mycoplasma was present.
The two most common groups of associates of ambrosia beetles have never (Mycoplasma) or only rarely (Comamonadaceae) been recovered from insect hosts. This is the first record of Mycoplasma spp. as stable associates of insects. Our discovery is at odds with the definition of the genus as parasites of vertebrates [34] and raises questions about the broader biology of the group. The confamilial genus Entomoplasma has been isolated from insects, but the phylotypes from ambrosia beetles do not fall into this clade (Fig. 3). Importantly, the Mycoplasma spp. from ambrosia beetles are also not a monophyletic clade, suggesting that several Mycoplasma clades colonized ambrosia beetles independently.
Reports of Comamonadaceae from bodies of insects are rare. Several strains have been isolated from insects but not from groups relevant to wood-boring beetles [35–37]. Pseudomonadaceae are reported more commonly. The most relevant is a record of several phylotypes of Acinetobacter in the gut of the bark beetle Dendrocotnus valens [38]. The 16 S sequence of one phylotype corresponds to the Acinetobacter phylotype detected in our samples as the fourth most widespread phylotype (only 1.5 % divergent; other reported strains are not similar). Burkholderiaceae have been previously isolated from crushed whole bodies of another Dendroctonus, D. valens [15], and Pseudomonadaceae from Dendroctonus ponderosae [14], but, in both cases, the isolates represented different 16 S phylotypes than those reported here.
Since beetle species is a stronger predictor for the bacterial community composition than locality, we suggest that more bacterial phylotypes in beetle mycangia are vertically transmitted than sampled from the environment. Given their prevalence (Table 2) and their apparent absence outside of beetle mycangia, several of the widespread phylotypes we found have the potential to be stable associates of ambrosia beetles. Based on our present knowledge, we cannot determine the ecological role of the bacterial associates. The role could be anything from mutualists of the beetles to their parasites, or even endosymbionts of the ambrosia fungi (i.e., Burkholderiales, [40]). In most cases, the beetles seem unlikely to obligately depend on the bacteria, since even the most prevalent bacterial phylotypes in the beetles did not tend to be universally present within a host species. However, an exception in this regard may be the four species-specific associates of X. glabratus recovered from all samples of this beetle regardless of the locality. These phylotypes in particular deserve additional research to better understand their role in the ambrosia symbiotic complex. It is also worth noting that mycangia are complex organs and may not have been sampled completely. We sampled the lumen of the mycangium, which is where the main mass of symbionts resides, but we did not attempt to explore the ultrastructure of its walls reported, for example, in D. frontalis [41].
The conclusion that ambrosia beetles tend to retain species-specific associates in different regions of the world is further corroborated by the unique bacterial communities in the mycangia of X. crassiusculus and X. glabratus (Table 3). Both species are non-native invaders in North America, and their bacterial communities differ significantly from those of native xyleborine beetles. Particularly intriguing is X. glabratus, since its entire non-native population likely originated from a single recent introduction [20, 39]. The bacterial community within its mycangia is unusually uniform (several phylotypes occur in every sample) and dissimilar from bacterial communities in native North American species of Xyleborus collected from the same trees. This invasive beetle thus does not appear to have adopted the bacteria of native beetles with which it is now sympatric. An interesting question we cannot resolve here is whether bacterial communities of X. glabratus mycangium have changed relative to its ancestors in its native range.
We have repeatedly failed to amplify bacterial 16 S rDNA from mesonotal mycangia of both species of the genus Xylosandrus. We are unable to distinguish whether this reflects a true absence of bacteria in the mycangia, or the presence of strong PCR inhibitors. It is however not a result of low quality of the samples. The two control samples from fungal gardens surrounding two different X. germanus were collected directly from the gallery around the beetles and processed identically, yet they yielded rich communities of bacteria.
None of the common phylotypes detected here have been previously reported from bark and ambrosia beetles. One reason may be that the majority of previous studies used culturing approaches. Here, we show that the most regular associates belong to groups that are difficult or impossible to culture, such as Mycoplasma or Rickettsia [34]. Lastly, we also show that focus on specific organs, such as the mycangium, allows refined inference of symbiotic associations, and we recommend that future research on insect symbioses takes insect anatomy into account.
To summarize our findings on the community ecology of the mycangial microbiome, it appears that (1) bacterial communities are more species-specific than locality-specific, however, (2) the core microbiome is nevertheless shared across beetle species, and (3) the similarity of bacterial communities does not reflect phylogenetic relatedness of their host beetles. This suggests that, although beetles tend to retain distinct communities even in newly colonized regions and in the presence of other ambrosia beetle species, their mycangial associates do experience horizontal cross-infection with a large amount of stochasticity and do not conform to a scenario of long-term co-cladogenesis.
References
Ciesla WM (2011) Forest entomology. Wiley-Blackwell
Six DL, Wingfield MJ (2011) The role of phytopathogenicity in bark beetle–fungus symbioses: a challenge to the classic paradigm. Annu Rev Entomol 56:255–272
Hulcr J, Dunn RR (2011) The sudden emergence of pathogenicity in insect–fungus symbiosis threatens naïve forest ecosystems. P Roy Soc London B: Biological Sciences 278(1720):2866–2873
Farrell BD, Sequeira ASO, Meara BC, Normark BB, Chung JH, Jordal BH (2001) The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae). Evolution 55:2011–2027
Beaver RA (1989) Insect–fungus relationship in the bark and ambrosia beetles. In: Wilding N, Collins NM, Hammond PM, Webber JF (eds) Insect–fungus interactions. Academic Press, London, pp 121–143
Mueller UG, Gerardo NM, Aanen DK, Six DL, Schultz TR (2005) The evolution of agriculture in insects. Annu Rev Ecol Syst 36:563–595
Francke-Grosmann H (1967) Ectosymbiosis in wood-inhabiting insects. In: Henry SM (ed) Symbiosis, vol 2—associations of invertebrates, birds, ruminants and other biota. Academic Press, New York, pp 141–206
Six DL (2003) Bark beetle–fungus symbioses. In: Bourtzis K, Miller TA (eds) Insect symbiosis. CRC Press, New York, pp 97–114
Stone WD, Nebeker TE, Monroe WA, MacGown JA (2007) Ultrastructure of the mesonotal mycangium of Xylosandrus mutilatus (Coleoptera: Curculionidae). Can J Zoolog 85(2):232–238
Happ GM, Happ CM, Barras SJ (1971) Fine structure of the prothoracic mycangium, a chamber for the culture of symbiotic fungi, in the southern pine beetle, Dendroctonus frontalis. Tissue Cell 3(2):295
Kajimura H, Hijii N (1992) Dynamics of the fungal symbionts in the gallery system and the mycangia of the ambrosia beetle, Xylosandrus mutilatus (Blandford) (Coleoptera: Scolytidae) in relation to its life history. Ecol Res 7:107–117
Hulcr J, Adams AS, Raffa KF, Hofstetter RW, Klepzig KD, Currie CR (2011) Presence and diversity of Streptomyces in Dendroctonus and sympatric bark beetle galleries across North America. Microb Ecol 61:759–768
Cardoza YJ, Klepzig KD, Raffa KF (2006) Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol Entomol 31(6):636–645
Adams AS, Currie CR, Cardoza Y, Klepzig KD, Raffa KF (2009) Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can J Forest Res 39(6):1133–1147
Adams AS, Adams SM, Currie CR, Gillette NE, Raffa KF (2010) Geographic variation in bacterial communities associated with the Red Turpentine Beetle (Coleoptera: Curculionidae). Environ Entomol 39(2):406–414
Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR (2008) Bacterial protection of beetle–fungus mutualism. Science 322(5898):63
Norris DM (1979) The mutualistic fungi of Xyleborini beetles. In: Batra LR (ed) Insect–fungus symbiosis. Allanheld, Osmund & Co, Montclair, pp 53–63
Grubbs KJ, Biedermann PHW, Suen G, Adams SM, Moeller JA, Klassen JL, Goodwin LA, Woyke T, Munk AC, Bruce D, Detter C, Tapia R, Han CS, Currie CR (2011) Genome sequence of Streptomyces griseus strain XylebKG-1, an ambrosia beetle-associated Actinomycete. J Bacteriol 193(11):2890–2891. doi:10.1128/jb.00330-11
Shade A, Handelsman J (2011) Beyond the Venn diagram: the hunt for a core microbiome. Environ Microb 14(1):4–12
Fraedrich SW, Harrington TC, Rabaglia RJ, Ulyshen MD, Mayfield AE, Hanula JL, Eickwort JM, Miller DR (2008) A fungal symbiont of the redbay ambrosia beetle causes a lethal wilt in redbay and other Lauraceae in the southeastern United States. Plant Dis 92:215–224
Cognato AI, Hulcr J, Dole SA, Jordal BH (2011) Phylogeny of haplo–diploid, fungus-growing ambrosia beetles (Curculionidae: Scolytinae: Xyleborini) inferred from molecular and morphological data. Zool Scr 40(2):174
Hulcr J, Mogia M, Isua B, Novotny V (2007) Host specificity of ambrosia and bark beetles (Col., Curculionidae: Scolytinae and Platypodinae) in a New Guinea rain forest. Ecol Entomol 32:762–772
Kirkendall LR, Jordal BH (2006) The bark and ambrosia beetles (Curculionidae, Scolytinae) of Cocos Island, Costa Rica and the role of mating systems in island zoogeography. Biol J Linn Soc 89:729–743
Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N (2011) Examining the global distribution of dominant archaeal populations in soil. ISME J 5(5):908
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2011) Vegan: community ecology R package, version 1.17-10. URL: http://CRANR-projectorg/package=vegan
Lozupone C, Knight R (2005) UNIFRAC: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71(12):8228–8235
Lozupone C, Hamady M, Kelley S, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73(5):1576–1585
Hulcr J, Cognato AI (2010) Repeated evolution of crop theft in fungus-farming ambrosia beetles. Evolution 64(11):3205–3212
Hood GM (2006) PopTools version 2.7.2. URL: http://www.cse.csiro.au/poptools
Razin S, Herrmann R (2002) Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic and Plenum Publishers, London, UK
Bextine B, Lauzon C, Potter S, Lampe D, Miller TA (2004) Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Curr Microbiol 48:327–331
Le Guyader A, Rivault C, Chaperon J (1989) Microbial organisms carried by brown banded cockroaches in relation to their spatial distribution in a hospital. Epidemiol Infect 12(3):485–492
Zouache K, Rharaimalala FN, Raquin V, Tran-Van V, Raveloson LH, Ravelonandro P, Mavingui P (2010) Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol 75(3):377–389
Morales-Jiménez J, Zuniga G, Villa-Tanaca L, Hernández-Rodríguez C (2009) Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb Ecol 58(4):879–891
Rabaglia RJ, Dole S, Cognato AI (2006) Review of American Xyleborina (Coleoptera: Curculionidae: Scolytinae) occurring North of Mexico, with an illustrated key. Ann Entomol Soc Amer 99(6):1034–1056
Hoffman MT, Arnold AE (2010) Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microb 76:4063–4075
Yuceer C, Hsu CY, Erbilgin N, Klepzig KD (2011) Ultrastructure of the mycangium of the southern pine beetle, Dendroctonus frontalis (Coleoptera: Curculionidae, Scolytinae): complex morphology for complex interactions. Acta Zool 92:216–224
Hulcr J, Kolarik M, Kirkendall LR (2007) A new record of fungus–beetle symbiosis in Scolytodes bark beetles (Scolytinae, Curculionidae, Coleoptera). Symbiosis 43:151–159
Kolarik M, Kirkendall LR (2010) Evidence for a new lineage of primary ambrosia fungi in Geosmithia Pitt (Ascomycota: Hypocreales). Fungal Bio 114:676–689
Acknowledgement
A number of collaborators contributed beetle samples: Vojtech Novotny, Rich Hofstetter, Sarah M. Smith, Anthony I. Cognato, Shannon Pelini, Hisashi Kajimura, and Kazuki Tsuji. Trudy McKay allowed us to use reagents in her lab. JH was supported by US Forest Service Cooperative Agreement and by NSF-CAREER grant 09533390. RRD was supported by DOE-PER grant DE-FG02-08ER64510, a NASA award NNX09AK22G, and an NSF-CAREER grant 09533390. LLS was supported by a USDA-SCRI grant.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Overview of bacterial phylotypes detected in mycangia of ambrosia beetles, including the taxonomic classification of each phylotype.
ESM 1
(XLSX 339 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hulcr, J., Rountree, N.R., Diamond, S.E. et al. Mycangia of Ambrosia Beetles Host Communities of Bacteria. Microb Ecol 64, 784–793 (2012). https://doi.org/10.1007/s00248-012-0055-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0055-5