Abstract
The quality of torrefied grass fibers (TGF) as a new potting soil ingredient was tested in a greenhouse experiment. TGF was colonized with previously selected microorganisms. Four colonization treatments were compared: (1) no inoculants, (2) the fungus Coniochaeta ligniaria F/TGF15 alone, (3) the fungus followed by inoculation with two selected bacteria, and (4) the fungus with seven selected bacteria. Cultivation-based and DNA-based methods, i.e., PCR-DGGE and BOX-PCR, were applied to assess the bacterial and fungal communities established in the TGF. Although colonization was not performed under sterile conditions, all inoculated strains were recovered from TGF up to 26 days incubation. Stable fungal and bacterial populations of 108 and 109 CFU/g TGF, respectively, were reached. As a side effect of the torrefaction process that aimed at the chemical stabilization of grass fibers, potentially phytotoxic compounds were generated. These phytotoxic compounds were cold-extracted from the fibers and analyzed by gas chromatography mass spectrometry. Four of 15 target compounds that had previously been found in the extract of TGF were encountered, namely phenol, 2-methoxyphenol, benzopyran-2-one, and tetrahydro-5,6,7,7a-benzofuranone. The concentration of these compounds decreased significantly during incubation. The colonized TGF was mixed with peat (P) in a range of 100%:0%, 50%:50%, 20%:80%, and 0%:100% TGF/P (w/w), respectively, to assess suitability for plant growth. Germination of tomato seeds was assessed three times, i.e., with inoculated TGF that had been incubated for 12, 21, and 26 days. In these tests, 90–100% of the seeds germinated in 50%:50% and 20%:80% TGF/P, whereas on average only 50% of the seeds germinated in pure TGF. Germination was not improved by the microbial inoculants. However, plant fresh weight as well as leaf area of 28-day-old tomato plants were significantly increased in all treatments where C. ligniaria F/TGF15 was inoculated compared to the control treatment without microbial inoculants. Colonization with C. ligniaria also protected the substrate from uncontrolled colonization by other fungi. The excellent colonization of TGF by the selected plant-health promoting bacteria in combination with the fungus C. ligniaria offers the possibility to create disease suppressive substrate, meanwhile replacing 20% to 50% of peat in potting soil by TGF.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thermally treated (torrefied) grass fibers (TGF) are a viable substitute for peat in potting media [21]. TGF have good water holding capacity, and grass is a renewable substrate. Torrefaction was applied in order to transform grass fibers into a (chemically) more stable product [21]. After torrefaction, TGF contains abundant acetic acid as well as formic acid, which can serve as nutrient and energy sources for particular microorganisms [4, 6, 8, 18, 24]. Phytotoxic compounds such as phenol, phenolic compounds [15, 19, 21, 23], and 2-furalaldehyde (furfural) [2, 9, 10, 14] are produced as side compounds during the heating process, but they may serve as nutrient sources for specific microorganisms.
A proper microbial colonization of TGF, which is sterile after the heating process, is crucial to obtain a stable microbial community and reduce the phytotoxicity of the substrate. Meanwhile, this is a perfect opportunity to inoculate the substrate with plant-beneficial microorganisms. Agricultural soil, compost, as well as potting soil, have often been inoculated with plant-beneficial bacteria or fungi to control plant diseases. The survival of such inoculated organisms has generally been found to be low; mostly less than 1% survival was noted following incubation [1, 3, 9, 11, 12]. Also, colonization of compost by beneficial microorganisms was often disappointing [13, 16]. A main factor is the lack of competitiveness of the introduced microorganisms versus the indigenous microbiota. In the case of TGF, the introduced microorganisms have several advantages: (1) there is no indigenous microbiota immediately after the torrefaction process and (2) bacteria and fungi with the ability to grow on the specific (toxic) compounds present in TGF will be in favor.
In our previous work, it was found that the ascomycete Coniochaeta ligniaria F/TGF15 together with the bacterial strains Pseudomonas putida 15/TGE5, Serratia plymuthica 23/TGE5, Pseudomonas corrugata 31/TGE5, Methylobacterium radiotolerans 56/TGF10, Leifsonia xyli subsp. xyli 66/TGF15, Mycobacterium anthracenicum 70/TGF15, and Agromyces aurantiacus 95/TGF15 could utilize some of the toxic compounds present on TGF and use these as carbon sources [22]. This combination of microorganisms was selected on the basis of successful colonization and persistence on TGF and reduction of phytotoxicity in TGF extracts [20]. S. plymuthica and P. corrugata are of special interest since they could inhibit several plant pathogenic fungi. An initial colonization of TGF by C. ligniaria F/TGF15 was crucial to allow establishment of the selected bacteria on the TGF [20] C. ligniaria F/TGF15, thus, paved the way for bacterial growth by removing bacteriostatic or bactericidal factors from TGF. Up to now, colonization in TGF was studied under otherwise sterile conditions in small systems, whereas in practice, TGF will be applied for plant growth in larger systems. It is difficult to keep such large volumes sterile; during storage and incubation, the substrate will be in contact with (non-sterile) air. Information about the colonization of TGF by the selected strains under non-sterile conditions is not available yet.
The current experiment aimed to test the ability of the previously selected ascomycete C. ligniaria F/TGF15 and the plant-beneficial bacteria P. putida 15/TGE5, S. plymuthica 23/TGE5, P. corrugata 31/TGE5, M. radiotolerans 56/TGF10, L. xyli subsp. xyli 66/TGF15, M. anthracenicum 70/TGF15, and A. aurantiacus 95/TGF15 to colonize TGF under non-sterile conditions. TGF with different inoculation treatments was applied as a potting substrate, alone or mixed with peat, and evaluated in the greenhouse to test its suitability as a matrix for plant growth. Cultivation-based and DNA-based methods, i.e., PCR-DGGE and BOX-PCR, were applied to analyze the microbial and fungal communities in the matrix. The quality of TGF as a potting substrate was assessed by measuring the germination of tomato seeds, as well as plant weight and leaf area. In addition, the presence of phytotoxic compounds in the TGF substrate was assessed with gas chromatography mass spectrometry (GCMS).
Methods
Preparation of TGF
Grass material was collected in August 2004 from unfertilized, seminatural grassland on a peaty clay soil near Wageningen, the Netherlands. The biomass was harvested as a first cut and consisted of a mixture of more than 30 species (mainly grasses, rushes, sedges, and herbs) with a dry matter content of 37.5% w/w. Dry matter content of the plant biomass was measured after drying aliquots at 105°C. The harvested material was dried at 105°C, ground in a hammer mill to pieces <1 mm, and stored until use in torrefaction experiments.
The ground material was torrefied in a preheated Carbolite muffle furnace (Model CWF 1100) at a temperature of 240°C during 1 h. To ensure the occurrence of anoxic conditions, ground biomass was placed in aluminum boxes (approximately 40 g dry matter in a 200-ml box). The boxes were closed with their lid leaving a narrow split for gas exchange. The boxes were placed upside down in the oven to promote homogeneous heating of the contents. Boxes were weighed before and after heating to determine weight loss of the samples. The loss of dry weight due to torrefaction was 11% w/w. The pH of the TGF was 5.2. TGF was stored air-dry at room temperature in a closed but non-sterile plastic container.
Preparation of a Fungal Inoculum
The fungal strain C. ligniaria F/TGF15 was grown on potato dextrose agar (39 g/l, Oxoid, London, UK) for 10–12 days at 20°C in the dark. Then, 10 ml sterilized tap water was added per plate and fungal mycelium and spores were scraped off with a spatula. The optical density (OD600) of the suspension was adjusted to 0.1 with sterilized tap water and measured using a Beckman DU® 530 Life Science UV/Vis spectrophotometer. The suspension was further diluted 1,000-fold to obtain approximately 105 fungal propagules per ml suspension. Inoculum concentrations were enumerated by plating on R2A (Difco, Detroit, MI, USA).
Preparation of a Bacterial Inoculum
Freshly grown bacterial strains that have been previously isolated from a subsequent enrichment in TGF and its extract (TGE) [21], namely P. putida 15/TGE5, S. plymuthica 23/TGE5, P. corrugata 31/TGE5, M. radiotolerans 56/TGF10, L. xyli subsp. xyli 66/TGF15, M. anthracenicum 70/TGF15, and A. aurantiacus 95/TGF15 were harvested from R2A (Difco, Detroit, MI, USA) by adding 5 ml mineral salt medium (MSM: K2HPO4 1.4 g/l, KH2PO4 1.4 g/l, MgSO4.7H2O 0.1 g/l, (NH4)2SO4 1.0 g/l, MnSO4.7H2O 0.001 g/l, and FeSO4.7H2O 0.01 g/l) to the Petri dishes. The optical density (OD600) of the suspensions was adjusted to 0.1 with MSM and 1,000-fold dilutions were made to obtain suspensions with approximately 105 CFU/ml. The inoculum concentrations were checked by plating on R2A.The bacterial consortium with seven strains in roughly equal CFU numbers was mixed with the TGF.
Colonization of TGF
In total, 12 spawn bags (30 × 60 cm) containing a 160-mm filter with 100-μm perforations (Van Leer Packaging, Pont-Audemer Cedex, France) were filled with 420 g air-dry TGF. Four treatments, each with three replicates, were prepared: C, uninoculated control, F, fungus alone, F + 2B, the fungus plus bacterial strains S. plymuthica 23/TGE5 and P. corrugata 31/TGE5, and F + 7B, the fungus plus seven bacterial strains, i.e., P. putida 15/TGE5, S. plymuthica 23/TGE5, P. corrugata 31/TGE5, M. radiotolerans 56/TGF10, L. xyli subsp. xyli 66/TGF15, M. anthracenicum 70/TGF15, and A. aurantiacus 95/TGF15. The C. ligniaria F/TGF15 inoculum was added meanwhile moistening TGF up to 50% of its water holding capacity. Fungal spores (5 × 104/g TGF) were added for treatments F, F + 2B, and F + 7B. In the control treatment, an equal volume of sterilized tap water was added. The treated TGF was mixed well and incubated for 5 days at 25°C in the dark. Subsequently, the bacterial inocula were added to treatments F + 2B and F + 7B using 105 cells of the bacterial mixtures per g dry TGF. In order to moisten all treatments equally, 0.6-ml sterilized MSM was added per g TGF to the control and to treatment F. All treatments were moistened to 60% of the TGF water holding capacity, mixed well, and incubated for another 7 days at 25°C in the dark.
Bacterial and fungal populations in the different treatments were analyzed over time by plate counts and PCR-DGGE. The pH and presence of selected compounds (GC analyses) were measured 12 and 26 days after the first inoculation. The four TGF treatments incubated for 12 days were used, alone or mixed with peat, to test tomato seed germination and plant growth. The seed germination test was repeated when the TGF treatments were incubated during 21 and 26 days.
Assessment of Microbial Colonization
Bacterial and fungal populations in the four TGF treatments, each with three replicates, were enumerated by plate counts. A sample of 5 g moist TGF was shaken vigorously in 95 ml 0.1% sodium pyrophosphate and 10 g gravel. Tenfold dilution series were prepared in 0.85% NaCl and plated on R2A medium. Results are presented as logarithmic numbers of CFU calculated per g dry TGF. The introduced fungal strain C. ligniaria F/TGF15 was recognized by its typical morphological characteristics. The plate readings were done on days 2, 4, 6, 8, 10, 12, 14, 16, and 18 for different dilution series. The presence of the seven inoculated bacterial strains was detected by performing BOX-PCR on 100 randomly selected colonies from the F + 7B treatment after 26 days incubation.
BOX-PCR fingerprints of the colonies were performed by PCR with primer BOX-261 (5′-CTACGGCAAGGCGACGCTGACG-3′). Briefly, 1 μl of cell lysate was received as explained previously [21] and was added to 24 μl of a mix containing 15 μl dNTP (Boehringer, Almere, the Netherlands), 2.5 μl buffer super Taq, 0.25 μl primer, 3.75 μl MgCl2 (Perkin-Elmer, Nieuwersluis, the Netherlands), 0.25 μl superTaq, and 2.75 μl MQ. Thermal cycling was as follows: initial denaturation at 95°C for 7 min, followed by 90°C for 30 s, 95°C for 1 min, 52°C for 1 min, 65°C for 8 min, 29 cycles of 90°C for 30 s, and 65°C for 16 min. The PCR products were analyzed by running 25 µl aliquots of the reaction mixtures in 1.5% agarose gel in 0.5 × TBE buffer for 3 h, 25 mV. Strains that were not successfully amplified with the BOX-261 primer were subjected to a second PCR reaction in which a diluted volume of crude cell lysate was added.
Bacterial and Fungal PCR-DGGE Analyses
Total community DNA of the four treatments were extracted and purified with the UltraClean™ soil DNA kit (MoBio Laboratories, BIOzymTC, Landgraaf, the Netherlands) according to the protocol of the supplier, except that the cells were disrupted by bead beating two times 30 s in a Braun’s cell homogenizer (Braun, Melsungen, Germany) at maximum speed. For disruption, glass beads (50 mg, 0.11 mm dia) were added to 0.5 g of TGF. The bead beating step was included to ensure maximal cell lysis without severe shearing of the DNA. DNA quality and quantity were assessed by electrophoresis in 1% agarose gels in a 0.5 × TBE buffer [17] by comparison to a standard 1-kb ladder (Invitrogen, Cat.15615-024, Carlsbad, USA). DNA size was 10–40 kb, on average.
Amplification of 16 S ribosomal (r) RNA genes was done by using PCR in 50-µl reaction volumes as described before. For DGGE, a GC-clamped version of U968, i.e. U968-GC (5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGAACGCGAA GAACCTTAC-3′) was used. Thermal cycling was as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 65°C for 90 s, 72°C for 2 min, final extension at 72°C for 10 min. The PCR products (expected sizes about 450 bp) were analyzed by running 5 µl aliquots of the reaction mixtures in 1% agarose gels.
For fungi, a nested PCR of the ITS region was performed. The first PCR was carried out in 25-µl reaction volumes containing 0.4 µM of each primer, 0.5 mM MgCl2 (Perkin-Elmer, Nieuwersluis, the Netherlands ) and 200 µM of each dNTP (Boehringer, Almere, the Netherlands) and 0.25 µg of T4 gene 32 protein (Boehringer, Mannheim, Germany) using 3.5 U expand HF polymerase in HF buffer 2. The primers used were ITS4, ITS primer in 28 S (TCCTCCGCTTATTGATATGC) and EF4, ITS primer, located in 18S rDNA (GGAAGG GRTGTATTTATTAG). The PCR product was used as a template for PCR-ITS2. The second PCR was carried out in 50-µl reaction volumes containing 0.4 µM of each primer, 0.5 mM MgCl2 (Perkin-Elmer, Nieuwersluis, the Netherlands) and 200 µM of each dNTP (Boehringer, Almere, the Netherlands) and 5 U Super Taq polymerase in super Taq Buffer. The primers used were ITS2 primer in 5.8 S (GCTGCGTTCTTCATCGATGC) and ITS1F primer +GC-clamped version in 18S (CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGG GCTTGGTCATTTAGAGGAAGTAA).
Bacterial DGGE analyses were done using 6% polyacrylamide gels (ratio of acrylamide to bisacrylamide 37:1) with a gradient of 45% to 65% denaturants (100% denaturant was defined as 7 M urea plus 40% formamide). The gradient of fungal DGGE was 30% to 80% denaturants. The gels were run at 60°C (100 V) for 16 h in a Phor-U2 apparatus (Ingeny International, Goes, the Netherlands) and stained with SYBR gold (Molecular Probes, Leiden, the Netherlands).
GCMS Analyses and pH
Extracts of the four different TGF treatments, each with three replicates, were prepared by the following protocol: 4.3 g of torrefied grass fiber was cold-extracted at room temperature with 113 ml distilled water for 2 h using a magnetic stirrer at 700 rpm. Following the cold extraction, the suspension was paper-filtered to remove grass fiber particles. The extract was then filter-sterilized (Millex GV bacterial filter Unit 0.22 µm Millipore) and GCMS analyses were done. Two ml filtrate and 2 ml of dichloromethane were mixed and the mixture was vortexed for about 10 sec, until a homogeneous solution was obtained. After centrifugation (1,200 × g, 5 min), the upper inorganic layer was discarded and the organic layer was transferred to a clean vial and passed over a short column (Pasteur capillary pipette filled with siliconized glass wool and anhydrous Na2SO4). After this, the clean filtrate was collected and stored at -20°C until measurements were performed. Samples were concentrated 5-fold under a gentle flow of nitrogen gas.
Two µl of the filtered organic phase were injected in splitless mode into the injection port of a gas chromatograph (5888 series II, Hewlett-Packard GMI, USA) coupled to a mass spectrometer (model 5972A, Hewlett-Packard GMI, USA) with a Zebron ZB-5 ms column (30 m × 0.25 mm I.D. × 0.25 µm film thickness) (PheNMenex, USA). The carrier gas was He (1 ml min-1). The oven was programmed at an initial temperature of 45°C for 1 min, with a ramp of 10°C per min to 310°C, and final time of 8.5 min. The injection temperature was 250°C and the detection temperature was 290°C. Peaks were identified by comparison of the spectra to NIST databases as well as to reference compounds for phenol and 2-methoxyphenol. The matching factors and probability by comparing to NIST for phenol, 2-methoxyphenol, benzopyran-2-one and 5,6,6,7a tetrahydro-benzofuranonare are: match 816 prob 52%, match 761 prob 55%, match 920 prob 83%, match 880 prob 75% respectively. The matching factors for phenol and 2-methoxyphenol were the lowest. However, we confirmed spectra and retention times for these compounds by injecting the reference compounds: phenol (QBiogene, USA), 2-methoxyphenol, (Alfa Aesar GmbH & Co.KG, Germany).
The pH of the four colonization treatments in TGF after 12 and 26 days incubation was measured in the solid substrate by placing an electrode directly into the moist TGF.
Plant Assay
TGF of the four colonization treatments after 12 days incubation were mixed in different ratio’s with a non-fertilized horizontally harvested peat (P) from the Baltic region (Lentse Potgrond, Cuijk, the Netherlands). The pH of the peat was 3.5. Mixtures of 100%:0% 50%:50% and 20%:80% and 0%:100% TGF/P were used. Plastic pots 6 × 6 × 6 cm were filled with 200 g of the mixed substrates. Nine tomato seeds (cv Pronto, De Ruiter, the Netherlands) were sown per pot. All treatments had six replicate pots in a block design. The pots were watered after sowing and covered with plastic for a week. Thereafter, water was given two to three times a week and the nutrient solution once a week. This nutrient solution contained 0.5 NH4, 7.2 K, 0.5 Na, 3.1 Ca,1.6 Mg, 9.4 NO3, 0.3 Cl, 2.8 S, 0.5 HCO3, 1.34 P, 0.01 Si [mmol/l], and 35 Fe, 3.5 Mn, 4.3 Zn, 25 B, 0.7 Cu and 0.6 Mo [μmol/l], pH 5.9, EC 2.0 [mS/cm]. The plants were cultivated in the greenhouse at 20°C with 16 h light/8 h dark and a 70% relative air humidity. Seed germination was scored 7 and 14 days after sowing.
After assessment of germination, one healthy plant per pot was maintained and the surplus plants were removed. The single plant per pot was harvested 14 days later; plant fresh weight (sprout), and area of detached leaves per plant were measured.
The germination assay was repeated after TGF treatments had been incubated for 21 and 26 days.
Statistical Analysis
Analyses of variance were performed for all measurements with the statistical program Genstat 10 (Rothamsted Experimental Station, Harpenden, UK). Least significant differences (LSD) were calculated at a significant level of P = 0.05. CFU were analyzed after logarithmic transformation and GCMS data were square root performed to balance the residual values.
Results
Bacterial Populations
Bacterial populations in the treatments F + 2B and F + 7B increased from the introduced 105 CFU/g TGF up to 108 CFU/g after 12 days and nearly 109 CFU/g after 26 and 40 days of incubation (Fig. 1). In the treatment F + 2B, two colony types dominated, i.e., red colonies corresponding to S. plymuthica 23/TGE5 and white slimy colonies corresponding to P. corrugata 31/TGE5. The percentages of S. plymuthica 23/TGE5 CFU were 37%, 31%, and 12% in the treatment F + 2B after 12, 26, and 40 days, whereas P. corrugata 31/TGE5 encompassed the remainder. S. plymuthica 23/TGE5 CFU made up 14%, 17%, and 11% in the F + 7B treatment after 12, 26, and 40 days incubation, respectively. In the latter treatment after 26 days, all seven inoculated bacterial strains were found back by BOX-PCR using 100 randomly selected colonies. The occurrence of strains was: 15% P. putida 15/TGE5, 17% S. plymuthica 23/TGE5, 18% P. corrugata 31/TGE5, 8% M. radiotolerans 56/TGF10, 12% L. xyli subsp. xyli 66/TGF15, 12% M. anthracenicum 70/TGF15, and 13% A. aurantiacus 95/TGF15. Only 5% of the colonies did not correspond to the seven introduced strains, showing a clear dominance of these.
Total culturable bacterial and fungal population in TGF on 12, 26, and 40 days after fungal inoculation. Treatments are: C control without inoculants; F C. ligniaria F/TGF15; F + 2B C. ligniaria F/TGF15 and, 5 days later, P. putida 15/TGE5 and S. plymuthica 23/TGE5; F + 7B C. ligniaria F/TGF15 and, 5 days later, seven selected bacteria. Least significant difference at P = 0.05 is 0.47 for bacteria and 0.56 for fungi. Bars with the same letter (within each graph) are not significantly different
The treatments C and F which had not been inoculated with any bacterial strain contained or acquired indigenous bacterial populations. The populations in treatment C and F increased up to 5 × 108 and 109 CFU/g after 26 and 40 days, respectively (Fig. 1).
The DGGE fingerprints of treatment F + 2B on day 12 showed a dominant band corresponding with the introduced S. plymuthica 23/TGE5 and a double band representing P. corrugata 31/TGE5 (Fig. 2). Treatment F + 7B contained, additionally, the bands of L. xyli subsp. xyli 66/TGF15 and M. anthracenicum 70/TGF15 and probably of A. aurantiacus 95/TGF15. P. putida 15/TGE5 and M. radiotolerans 56/TGF10 were not detectable on gel since their bands overlap with those of P. corrugata 31/TGE5. The introduced S. plymuthica 23/TGE5, P. corrugata 31/TGE5, L. xyli subsp. xyli 66/TGF15, M. anthracenicum 70/TGF15, and A. aurantiacus 95/TGF15 were also detectable in the DGGE patterns at 26 and 40 days of incubation (data not shown).
Bacterial (upper figure) and fungal (lower figure) DGGE fingerprints of TGF on day 12 in triplicate samples. Treatments are: C control without inoculants; F C. ligniaria F/TGF15; F + 2B C. ligniaria F/TGF15 and, 5 days later, P. putida 15/TGE5 and S. plymuthica 23/TGE5; F + 7B C. ligniaria F/TGF15 and, 5 days later, seven selected bacteria. Numbers 15, 23, 31, 56, 66, 70, and 95 correspond with the band(s) of P. putida 15/TGE5, S. plymuthica 23/TGE5, P. corrugata 31/TGE5, M. radiotolerans 56/TGF10, L. xyli subsp. xyli 66/TGF15, M. anthracenicum 70/TGF15, and A. aurantiacus 95/TGF15, respectively. M is the fungal marker, and X is the fungal strain C. ligniaria F/TGF15 with its band F
From control treatment C, in which bacteria had not been introduced and CFU numbers were low at day 12, no bacterial DNA could be obtained on day 12. Thus, patterns could not be included in the DGGE fingerprint (Fig. 2). Also, treatment F did not receive bacterial inoculum. Nevertheless, several bands were present in this treatment at day 12, but they were different from those of the introduced strains (Fig. 2). The presence of bacterial bands in treatment F corresponded with the high CFU numbers on day 12 (Fig. 1).
Fungal Populations
Total fungal populations were 107 to 108 CFU/g TGF on days 12, 26, and 40 (Fig. 1). Colonies of the fungus C. ligniaria F/TGF15 dominated (>95%) in all treatments where it was introduced (data not shown). C. ligniaria F/TGF15 was also clearly detected in the fungal DGGE patterns of treatments F, F + 2B, and F + 7B on days 12 (Fig. 2), 26, and 40. The numbers of other bands in the DGGE patterns from these treatments was limited, i.e., ranged from four (day 12) to seven (days 26 and 40). In treatment C, where C. ligniaria F/TGF15 was not introduced, other fungal colonies were present.
The bags of control treatment C were visually contaminated with an unidentified pink-orange colored fungus. This was not the case with treatments F, F + 2B, and F + 7B.
GCMS Analyses
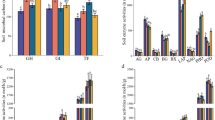
In total, four compounds were detected in TGF with GCMS analyses (Fig. 3) out of 15 target compounds which were previously found in the extract of TGF [22]. These four compounds were phenol, 2-methoxyphenol, benzopyran-2-one, and tetrahydro-5,6,7,7a-benzofuranone. In general, large variation between the levels of these compounds in independent samples was detected. On day 12, the level of phenol was significantly decreased in treatment F + 7B compared to the other treatments. Also for 2-methoxyphenol and benzopyran-2-one levels were lowest in treatment F + 7B, but levels were not significantly different from the control treatment C. Remarkably, tetrahydro-5,6,7,7a-benzofuranone had increased in the treatments F, F + 2B, and F + 7B compared to the control treatment C. In general, compound levels decreased significantly during the incubation period from days 12 to 26 for all treatments (Fig. 3).
Presence of phenol, 2-methoxyphenol, benzopyran-2-one, and 5,6,7,7a tetrahydro-benzofuranon in TGF 12 and 26 days after the inoculation treatments measured with GCMS. Treatments are: C control without inoculants; F C. ligniaria F/TGF15; F + 2B C. ligniaria F/TGF15 and, 5 days later, P. putida 15/TGE5 and S. plymuthica 23/TGE5; F + 7B C. ligniaria F/TGF15 and, 5 days later, seven selected bacteria. Bars with the same letter (within each graph) are not significantly different
pH
The pH values in TGF increased remarkably in all treatments colonized with the fungus C. ligniaria F/TGF15; from 5.2 in the fresh TGF up to 7.6 after colonization (Table 1). The pH in the control treatment without C. ligniaria F/TGF15 was 6.5 and 6.8 on days 12 and 26, respectively. Especially in the control treatment, the variation between the replicates was large.
Plant Assay
Seed germination was assessed 7 and 14 days after sowing into the pots containing the differently treated potting mixes. The results of 12 days colonized TGF are shown in Fig. 4. Germination of seeds in pure TGF (100%) was 0 after 7 days and 30–57% after 14 days. Germination in 50% TGF was 7–15% after 7 days but increased up to 89–96% after 14 days. Eighty percent of the seeds had germinated after 7 days in 20% TGF; hence, this matrix was the best substrate for tomato seed germination among the tested TGF-peat mixtures. The pure peat (0% TGF) with a very low pH value showed high germination rates, i.e. 70–80% on days 7 and 14. None of the inoculants significantly improved seed germination in any of the TGF-peat mixtures.
Percentage of germinated tomato seeds in different TGF-peat mixtures 7 and 14 days after sowing. Treatments are: C control without inoculants; F C. ligniaria F/TGF15; F + 2B C. ligniaria F/TGF15 and, 5 days later, P. putida 15/TGE5 and S. plymuthica 23/TGE5; F + 7B C. ligniaria F/TGF15 and, 5 days later, seven selected bacteria. Least significant differences at P = 0.05 are 18.6 and 19.2, 7 and 14 days after sowing, respectively. Bars with the same letter (within each graph) are not significantly different
The germination experiment was repeated twice showing similar results. In all cases, 100% TGF showed the lowest germination percentage and 20% TGF the highest (data not shown). There was no improvement in germination by the inoculation of the selected microorganisms in any of the experiments (Table 2). In fact, in some cases, the control treatment C showed higher germination percentage than the inoculated treatments.
Values of plant weight are shown in Fig. 5. Plant weight in treatment C was low when using 100% TGF, higher with 50%, and highest with 20% TGF. Plant weight was extremely low (0.1 g) in the pure peat (0% TGF) with low pH. Significant improvement was found in all treatments in which the fungus C. ligniaria F/TGF15 was used. Especially in the mixture of 50% TGF, plant weight increased drastically in all treatments with the fungus as compared to that in the control. When the fungus was present, the mixture with 50% TGF was as good as the mixture with 20% TGF. The co-presence of bacterial inoculants had some effect in 100% TGF, showing a slight increase (not significant) in plant weight in treatment F + 2B and F + 7B compared to treatment C and F.
Plant fresh weight (g) and leaf area (cm2/plant) of tomato plants 28 days after sowing. Treatments are: C control without inoculants; F C. ligniaria F/TGF15; F + 2B C. ligniaria F/TGF15 and, 5 days later, P. putida 15/TGE5 and S. plymuthica 23/TGE5; F + 7B, C. ligniaria F/TGF15 and, 5 days later, seven selected bacteria. Least significant differences at P = 0.05 are 0.40 and 15.8 for plant weight and leaf area, respectively. Bars with the same letter (within each graph) are not significantly different
The results of the plant weight were confirmed by the leaf area measurements (Fig. 5). Again, C. ligniaria F/TGF15 (treatment F) either or not in combination with two (treatment F + 2B) or seven bacterial inoculants (treatment F + 7B) incited a larger leaf area per plant than that observed in the control treatment C. A slight but not significant improvement occurred in 100% TGF with treatment F + 2B and F + 7B compared to treatment C and F.
Discussion
This study is a continuation of our previous research where a microbial community including C. ligniaria F/TGF15 and several selected beneficial bacteria were established on TGF [20]. The microbial consortium was selected on the basis of several parameters: (1) successful colonization and persistence on TGF, (2) reduction of phytotoxicity in TGF extracts, and (3) the possession of plant-beneficial properties. In the current study, the practical application of TGF was evaluated by assessing the colonization of the microbial consortium under non-sterile conditions and by testing the novel substrate in a greenhouse experiment with tomato seedlings. Regular potting soil is composed of different types of peat mixed with other organic compounds, fertilizer, and lime. In this study, different mixtures of the novel substrate TGF were prepared with pure peat, in order to test the proportion of peat which can be substituted by TGF.
According to the CFU enumerations, the bacterial consortia of respectively two or seven bacterial species in combination with the fungus C. ligniaria F/TGF15 were relatively stable in the matrix during a period of 40 days under non-sterile conditions. Populations reached nearly 109 CFU/g TGF. Moreover, the introduced bacterial consortia dominated and all introduced bacteria were found back with BOX-PCR among the isolated colonies after 26 days, being 95% of the total culturable bacterial population. Also the inoculated fungus C. ligniaria F/TGF15 proliferated well, up to 107–108 CFU/g TGF and was dominantly present after its inoculation. In general, survival of inoculants in agricultural soil or peat is poor; fungal or bacterial inoculants added to non-sterile soil or peat often do not reach more than 1% of the total population [1, 9, 11, 16]. Our results showed that on the one hand TGF is a rich substrate for microorganisms (high population levels); on the other hand, the inoculated consortium proliferated and was dominantly present. This proves the advantages of the applied strategy concerning: (1) the absence of indigenous microbiota immediately after the torrefaction process and (2) introducing microorganism with the ability to grow on the specific (toxic) compounds present in TGF.
Colonization of fresh organic substrates by undesired fungi is regularly occurring in practice and decreases the value of the substrate, especially if fungal mycelium is visually present [5, 7]. In the current experiment where the bags with TGF were exposed to open air, an unknown pink-orange colored fungus contaminated all bags of the control treatment without microbial inoculants. Reassuringly, the pink-orange fungus was not found in treatments in which C. ligniaria F/TGF15 had been used. Thus, controlled inoculation of TGF with C. ligniaria F/TGF15 has the advantage that the substrate is prevented from undesirable and uncontrolled invasion by other fungi.
GCMS analyses revealed the presence of four compounds, phenol, 2-methoxyphenol, 2-one-benzopyran, and tetrahydro-5,6,7,7a-benzofuranone, in the TGF. A standard curve was prepared for 2-methoxyphenol [22], indicating that approximately 3 mg of this compound was present per kg dry TGF on day 12, which was roughly 30% of the measured values in freshly torrefied grass. The concentrations of all compounds decreased during the incubation period between 12 and 26 days. However, there was, in general, no clear reduction in the concentration of the compounds due to the introduced fungus C. ligniaria F/TGF15 and bacteria. In the previous research, the fungus effectively removed several compounds, including phenol, from the extract of TGF [22]. The difference between the current and previous research was that, respectively, TGF and the extract of TGF were assessed. The discrepancy could be due to the fact that only part of the compounds (quantitatively and qualitatively) in TGF dissolve in the extract or to the fact that solved compounds are better available for the microorganisms.
Although limited levels of toxic compounds were measured with GCMS, the pure TGF was apparently still too phytotoxic, with on average 50% germination after 14 days. Germination in the mix with 50% TGF was delayed; however, it became substantial (90% germination) over time. The potting mix containing 20% TGF was the substrate that allowed the highest germination rate within 7 days. Unexpectedly, the seed germination did not improve after longer incubations of the TGF substrate, although the compounds measured with GCMS analyses decreased over time.
Seed germination was not improved by any of the bacterial inoculants or by the fungus. These inoculants had been selected, since previous studies showed that C. ligniaria F/TGF15 and the selected bacteria could reduce the toxicity and improve seed germination in extracts of TGF [22]. The fungus and several of the bacterial strains could grow on single model toxic compounds and reduce the presence of these compounds in the TGF extract [22]. The difference in results between the extract of TGF (previous results) and the solid TGF substrate again indicates that the solid substrate might contain more or other toxic compounds than its extract.
Interestingly, plant growth was significantly improved by C. ligniaria F/TGF15 in the 20% and 50% TGF mixtures. Leaf area as well as plant weight were significantly enhanced when the fungus was introduced into TGF. As a result, the mixtures with 20% as well as 50% TGF with the fungus showed the best plant growth. Since this was tested only in one experiment, more repeats in different conditions and crops should be performed to conclude about the benefit of the fungus for horticultural practice.
C. ligniaria F/TGF15 also influenced the pH of TGF upon colonization; pH increased up to 7.6. The pH in the TGF control treatment without fungus was, on average, 6.6 and was always lower than in the presence of C. ligniaria F/TGF15. This pH increase can be explained by utilization of the acid compounds in TGF by C. ligniaria F/TGF15. In the current experiment where TGF was mixed with peat of a low pH, this could have a positive influence on plant growth parameters.
Although there was no effect of the bacteria on germination and plant growth, they colonized TGF as a co-inoculant of the fungus up to unexpected high levels. The plant-beneficial and pathogen-inhibiting bacteria among the inoculated consortium proliferated and formed a dominant population in TGF up to at least 26 days. In several other studies, colonization of compost or soil by beneficial microorganisms was found to be disappointing, since the introduced microorganisms did not survive sufficiently [1, 9, 11, 13, 16]. Thus, the successfully colonized TGF offers a unique opportunity for disease suppression. Data on this aspect are not available yet, since the substrate mix was free of pathogens.
With the current greenhouse experiment we showed that TGF can form an acceptable component of potting soil. It can be applied to up to 20% in a potting mix if tomato plants have to be grown from seeds or up to 50% if germinated seeds are used. The ascomycete C. ligniaria F/TGF15 improved the quality of TGF as a partial peat replacement in potting soil and prevented the substrate from run-away colonization by other fungi. TGF was well colonized by the potential plant-growth-promoting bacteria P. putida 15/TGE5, S. plymuthica 23/TGE5 and P. corrugata 31/TGE5. The colonization of the microbiologically virgin TGF with a plant-health-improving microbial consortium offers additional advantages to create a disease-suppressive substrate, meanwhile replacing peat in potting soil.
References
Amarger N (2002) Genetically modified bacteria in agriculture. Biochimie 84:1061–1072
Boopathy R (2002) Methanogenesis from furfural by defined mixed cultures. Curr Microb 44:406–410
Bruck DJ, Solter LF, Lake A (2008) Effects of a novel microsporidium on the black vine weevil, Otiorhynchus sulcatus (F.) (Coleoptera: Curculionidae). J Invert Pathol 98:351–355
Canibe N, Miettinen H, Jensen BB (2008) Effect of adding Lactobacillus plantarum or a formic acid containing-product to fermented liquid feed on gastrointestinal ecology and growth performance of piglets. Livestock Sci 114:251–262
Carlile WR (2001) Problems of Saprophytic fungi in peat free growing media. Acta Hort 548:243–246
Dias M, Salvado JC, Monperrus M, Caumette P, Amouroux D, Duran R, Guyoneaud R (2008) Characterization of Desulfomicrobium salsuginis sp. nov. and Desulfomicrobium aestuarii sp. nov., two new sulfate-reducing bacteria isolated from the Adour estuary (French Atlantic coast) with specific mercury methylation potentials. Syst Appl Microbiol 31:30–37
Döbbeler P (2004) Octosporella erythrostigma (Pezizales) and Pithyella frullaniae (Helotiales), two remarkable ascomycetes on Frullania dilatata. Feddes Repertorium 115:5–14
Gössner AS, Küsel K, Schulz D, Trenz S, Acker G, Lovell CR, Drake HL (2006) Trophic interaction of the aerotolerant anaerobe Clostridium intestinale and the acetogen Sporomusa rhizae sp. nov. isolated from roots of the black needlerush Juncus roemerianus. Microbiol 152:1209–1219
Götz M, Gomes NCM, Dratwinski A, Costa R, Berg G, Peixoto R, Mendonça-Hagler L, Smalla K (2006) Survival of gfp-tagged antagonistic bacteria in the rhizosphere of tomato plants and their effects on the indigenous bacterial community. FEMS Microb Ecol 56:207–218
Gutiérrez T, Buszko M, Ingram L, Preston J (2002) Reduction of furfural to furfuryl alcohol by ethanologenic strains of bacteria and its effect on ethanol production from xylose. Appl Biochem Biotechnol 98–100:327–340
Johansen A, Knudsen IMB, Binnerup SJ, Winding A, Johansen JE, Jensen LE, Andersen KS, Svenning MM, Bonde TA (2005) Non-target effects of the microbial control agents Pseudomonas fluorescens DR54 and Clonostachys rosea IK726 in soils cropped with barley followed by sugar beet: a greenhouse assessment. Soil Biol Biochem 37:2225–2239
Kokalis-Burelle N, Kloepper JW, Reddy MS (2006) Plant growth-promoting rhizobacteria as transplant amendments and their effects on indigenous rhizosphere microorganisms. Appl Soil Ecol 31:91–100
Larkin RP (2008) Relative effects of biological amendments and crop rotations on soil microbial communities and soilborne diseases of potato. Soil Biol Biochem 40:1341–1351
López MJ, Nichols NN, Dien BS, Moreno J, Bothast RJ (2004) Isolation of microorganisms for biological detoxification of lignocellulosic hydrolysates. Appl Microb Biotech 64:125–131
Orozco AL, Pérez MI, Guevara O, Rodríguez J, Hernández M, González-Vila FJ, Polvillo O, Arias ME (2008) Biotechnological enhancement of coffee pulp residues by solid-state fermentation with Streptomyces. Py-GC/MS analysis. J Anal Appl Pyro lysis 81:247–252
Postma J, Montanari M, van den Boogert PHJF (2003) Microbial enrichment to enhance the disease suppressive activity of compost. Eur J Soil Biol 39:157–163
Sambrook J, Fritsch E, Maniatis T (1998) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, NY, USA
Shinagawa E, Toyama H, Matsushita K, Tuitemwong P, Theeragool G, Adachi O (2008) Formaldehyde elimination with formaldehyde and formate oxidase in membrane of acetic acid bacteria. J Biosci Bioeng 105:292–295
Trautwein K, Kuhner S, Wohlbrand L, Halder T, Kuchta K, Steinbuchel A, Rabus R (2008) Solvent stress response of the denitrifying bacterium “Aromatoleum aromaticum” strain EbN1. Appl Environ Microbiol 74:2267–2274
Trifonova R, Babini V, Postma J, Ketelaars JJMH, van Elsas JD (2009) Colonization of torrefied grass fibers by plant-beneficial microorganisms. Appl Soil Ecol 41:98–106
Trifonova R, Postma J, Ketelaars JJMH, van Elsas JD (2008) Thermally treated grass fibres as colonisable substrate for beneficial bacterial inoculum. Microb Ecol 56:561–571
Trifonova R, Postma J, Verstappen FWA, Bouwmeester HJ, Ketelaars JJMH, van Elsas JD (2008) Removal of phytotoxic compounds from torrefied grass fibres by plant-beneficial microorganisms. FEMS Microbiol Ecol 66:158–168
Vasiliadou IA, Tziotzios G, Vayenas DV (2008) A kinetic study of combined aerobic biological phenol and nitrate removal in batch suspended growth cultures. Int Biodeterior Biodegrad 61:261–271
Zaitsev G, Uotila J, Häggblom M (2007) Biodegradation of methyl tert-butyl ether by cold-adapted mixed and pure bacterial cultures. Appl Microbiol Biotechnol 74:1092–1102
Acknowledgements
This research was supported by the Dutch Ministry of Agriculture, Nature, and Food Quality. We thank Herman Meurs for his technical help and Francel W.A. Verstappen for carrying out the GCMS analyses.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Trifonova, R., Postma, J., Schilder, M.T. et al. Microbial Enrichment of a Novel Growing Substrate and its Effect on Plant Growth. Microb Ecol 58, 632–641 (2009). https://doi.org/10.1007/s00248-009-9518-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9518-8