Abstract
The coastal marine environment of the Northwest Atlantic contains strong environmental gradients that create distinct marine biogeographic provinces by limiting dispersal, recruitment, and survival. This region has also been subjected to numerous Pleistocene glacial cycles, resulting in repeated extirpations and recolonizations in northern populations of marine organisms. In this study, we examined patterns of genetic structure and historical demography in the Atlantic silverside, Menidia menidia, an annual marine fish with high dispersal potential but with well-documented patterns of clinal phenotypic adaptation along the environmental gradients of the Northwest Atlantic. Contrary to previous studies indicating genetic homogeneity that should preclude regional adaptation, results demonstrate subtle but significant (F ST = 0.07; P < 0.0001) genetic structure among three phylogeographic regions that partially correspond with biogeographic provinces, suggesting regional limits to gene flow. Tests for non-equilibrium population dynamics and latitudinal patterns in genetic diversity indicate northward population expansion from a single southern refugium following the last glacial maximum, suggesting that phylogeographic and phenotypic patterns have relatively recent origins. The recovery of phylogeographic structure and the partial correspondence of these regions to recognized biogeographic provinces suggest that the environmental gradients that shape biogeographic patterns in the Northwest Atlantic may also limit gene flow in M. menidia, creating phylogeographic structure and contributing to the creation of latitudinal phenotypic clines in this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Discontinuities in environmental conditions can form barriers to species distributions, creating distinct biogeographic provinces. However, these same environmental gradients may also act as filters to gene flow, creating population genetic structure in wide-ranging species that span biogeographic boundaries. The shallow marine environments of temperate North America show many examples of concordance between well-established marine biogeographic zones and intraspecific phylogeographic structure (see reviews by Dawson 2001; Wares and Cunningham 2001). However, these reviews also show many examples where intraspecific phylogeographic patterns do not conform to contemporary biogeographic provinces, highlighting the need to examine both historical biogeographic and oceanographic processes to understand the processes shaping species distributions and contemporary population structure. Understanding the processes driving such microevolutionary patterns may help our understanding of the processes shaping the broader biogeographic patterns (Avise et al. 1987).
In the Northwest Atlantic, the collision of warm, high-salinity Gulf Stream waters with cold, less-saline waters of the Labrador Current creates pronounced latitudinal gradients in both salinity and temperature (Bower et al. 1985), the latter of which is further magnified by latitudinal variation in ambient air temperature. While these gradients are mostly continuous across this region, there are specific areas where more pronounced environmental transitions enforce the northern or southern limits of species ranges, resulting in several well-established biogeographic provinces. Cape Hatteras separates the warmer temperate waters of the Carolinian province to the south from the cooler temperate waters of the Virginian province to the north. Cape Cod separates the Virginian and Acadian provinces, the former of which experiences extreme seasonal temperature fluctuations and the latter of which is cold year round (Briggs 1974; Engle and Summers 1999). Additionally, Nova Scotia separates warmer shallow waters in the Gulf of St. Lawrence from the cold waters of the Labrador Current, providing refugia for many marine coastal species (Bousfield and Thomas 1975). The boundaries between these biogeographic provinces are filters to gene flow in many western North Atlantic species, including amphipods (Jones 2005; Kelly et al. 2006), polychaetes (Jennings et al. 2009), and lobster (Kenchington et al. 2009) through limits to larval dispersal, recruitment, adult survival, or adult habitat choice. However, despite the apparent strength of these environmental filters, they are largely invisible to others species such as weakfish (Cordes and Graves 2003), bivalves (Dahlgren et al. 2000; Hare and Weinberg 2005; Strasser and Barber 2008), and other gastropods (Collin 2001) (see Wares 2002 for review).
One potential explanation for lack of structure in some species is that during the Pliocene and Pleistocene, glaciers covered much of the New England and Canadian coastline (Pratt and Schlee 1969; Shackleton et al. 1984; Pielou 1991). These glaciers, combined with the resulting 130-m decrease in sea level (Porter 1989), exposed the shallow continental shelf of eastern North America (Fig. 1) and forced many marine species out of the high latitudes and into southern refugia during glacial periods from which modern populations were recolonized (Bucklin and Wiebe 1998; Wares and Cunningham 2001; Wares 2002). Recently recolonized northern latitude populations often show signals of rapid population expansion and reduction of genetic diversity (see Hewitt 1996 for review) that could mask filters to gene flow across biogeographic boundaries. In contrast, while recolonization of northern latitudes would be facilitated by northward advection of pelagic marine larvae on the strong currents of the Gulf Stream, this process would also likely result of the transport of larvae adapted to warmer biogeographic regions of the south into cooler biogeographic regions of the north. Advection of larvae into environments to which they may be poorly suited could result in limited realized recruitment and gene flow across the environmental gradients (Rocha et al. 2005), shaping the regional biogeographic provinces of the Northwest Atlantic.
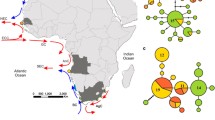
Collection sites for M. menidia and geological features of the western North Atlantic. Site numbers correspond to those in Table 1. Approximate coastline for the LGM corresponds to the 120-m isobath (Rohling et al. 1998), which is also the maximum depth limit for winter populations of M. menidia (Conover 1998). The 40-m isobath corresponds to the typical depth limit for winter populations (Conover 1998). Maximum ice extent at the LGM is approximate (after Shaw et al. 2002). Biogeographic provinces are from Briggs (1974). GSL Gulf of St. Lawrence, GOM Gulf of Maine, CC Cape Cod, CH Cape Hatteras, NF Cape Canaveral in northern Florida
The Atlantic silverside, Menidia menidia, is a broadly distributed fish that inhabits the near-shore zone of bays and estuaries along almost the entire east coast of North America from northeastern Florida to the Gulf of St. Lawrence. It is principally an annual species (Conover and Ross 1982) that spawns in salt marshes in the spring, where eggs hatch and juveniles grow to adult size. At high latitudes, M. menidia migrate offshore to the inner continental shelf to overwinter in deeper water (Conover and Murawski 1982; Conover and Ross 1982), while at lower latitudes, fish move into the deeper waters of the lower estuary but do not migrate offshore (Cain and Dean 1976; Shenker and Dean 1979) before returning to the salt marshes to spawn in spring. In the absence of physical, ecological, or behavioral limits to dispersal, the annual life history should promote widespread mixing and genetic homogeneity, a prediction supported by allozyme studies (Johnson 1975; Conover 1998) that indicate M. menidia has no significant population genetic structure across its range.
In response to the pronounced environmental gradients of the western North Atlantic, however, M. menidia has evolved strong and well-documented clinal patterns of countergradient and cogradient variation in a suite of physiological, behavioral, and morphological traits (Fig. 2) including growth rate, growth efficiency, foraging behavior, and vertebral number (Conover and Present 1990; Present and Conover 1992; Billerbeck et al. 1997; Yamahira and Conover 2002; Conover et al. 2005; Yamahira et al. 2006). Studies along the coast show that most of these traits are highly correlated (r = 0.80–0.99) with latitude of origin (Fig. 2) and are highly heritable (Conover 1998; Conover et al. 2005). The presence of strong heritable phenotypic clines seems incompatible with the life history of M. menidia. High dispersal and gene flow should swamp locally adapted gene complexes, limiting the ability of natural selection to form adaptive clines. Yet, previously reported high levels of gene flow may simply reflect stabilizing selection or the lack of power of allozymes to detect genetic differences in a species following a recent population bottleneck. Indeed, recent studies using otolith analysis have suggested M. menidia individuals are largely returning to their natal estuaries (Clarke et al. in press).
Trait variation with latitude (or length of growing season) in M. menidia (Conover 1998); a growth rate (mm/day; R 2 = 0.97, P < 0.01, y = −0.0246x + 0.4831), b environmental sex determination (level of ESD; R 2 = 0.86, P < 0.05, y = 0.0724x − 0.059), c mean vertebrae (count; R 2 = 0.98, P < 0.001, y = 0.334x + 28.016). Length of the growing season increases as latitude decreases
The presence of these phenotypic clines also seems incompatible with the geologic history of the western North Atlantic. Pleistocene glacial cycles followed by a range expansion at the end of the last glacial maximum (LGM) approximately 18,000 kya could result in reduced genetic diversity (Hewitt 1996; Schmitt 2007), providing a limited amount of genetic variation and a limited amount of time for heritable phenotypic clines to evolve. However, it has been suggested that some species may have weathered western North Atlantic glacial periods in ice-free northern refugia (Dyke and Prest 1987; Holder et al. 1999), mitigating loss of genetic diversity. Persistence in multiple refugia could allow for the preservation of genetic diversity and regional adaptation, with contemporary patterns of clinal variation resulting from the subsequent admixture of these groups. In fact, secondary contact is thought to explain similar morphological and physiological clines in the mummichog, Fundulus heteroclitus (Gonzalez Villasenor and Powers 1990; Bernardi et al. 1993; Adams et al. 2006; Durand et al. 2009), which shares an identical estuarine distribution along the western North Atlantic coast with M. menidia.
Here we use DNA sequence variation in the rapidly evolving mitochondrial control region to examine spatial and temporal patterns of genetic diversity and genetic structure in populations of M. menidia. First, we test whether the population structure of M. menidia is not as open as suggested by its life history and previous allozyme studies. In particular, we examine whether there are reductions in genetic exchange across the environmental discontinuities that shape traditional western North Atlantic biogeographic provinces. Second, we conduct a temporal analysis of genetic structure to examine the temporal stability of phylogeographic patterns and test for confounding effects resulting from sample collections that span multiple years and to test for regional fidelity of migrating fish. Lastly, because both population genetic patterns and phenotypic patterns could be influenced by population expansions from multiple refugia, we test for non-equilibrium population dynamics and latitudinal patterns in genetic diversity, signals indicative of post-glacial expansion.
Materials and methods
Sample collection and DNA sequencing
We collected 704 adult Menidia menidia using 30–100′ seine nets from twenty-five sites along its entire known range from northeastern Florida, USA, to Quebec, Canada, between May and August 2005 (n = 12 populations) and May and August 2006 (n = 13 populations). To determine the effect of combining samples across years as well as to examine for temporal stability of regional patterns, an additional 9 sites sampled in 2005 were resampled in 2006 (325 fish) for a combined total of 1,029 samples (Table 1; Fig. 1). Pectoral fins were removed and then preserved in 95% ethanol for DNA analysis. An equal sex ratio of males and females was examined from each site.
DNA was extracted from ~1-mm2 section of fin using 10% Chelex® (BioRad) solution (Walsh et al. 1991). A 340-base-pair fragment of the D-loop portion of the mitochondrial control region was amplified using the primers CRA and CRE (Lee et al. 1995). Polymerase chain reaction (PCR) amplifications of the mtDNA fragments were carried out in 25 μl reactions consisting of 0.625 units AmpliTaq® or AmpliTaq Gold® DNA polymerase (Applied Biosystems Inc., Foster City, CA), 1× GeneAmp PCR Buffer II (10 mM Tris–HCL, pH 8.3, 50 mM KCL; Applied Biosystems Inc.), 200 μM each dNTP (Promega), 2 mM MgCl2, 0.5 μM of each primer, and 1 ul of template DNA. For reactions containing AmpliTaq® DNA polymerase, manual hot-start PCRs were performed by combining template-containing and enzyme-containing fractions at 80°C before continuing to cycles. Reactions containing Amplitaq Gold® differed in that a chemical hot start was performed by incubating the entire reaction for 5 min at 95°C to activate the enzyme before continuing to cycles. Regardless of enzyme, thermal cycling was performed in a Biometra® thermocycler for 38 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 3 min.
PCR fragments were electrophoresed on 1% agarose gels at 200 volts in 1× Sodium Boric Acid buffer (Brody and Kern 2004) and visualized with Ethidium Bromide on a UV lightbox. PCR fragments were prepared for sequencing by combining 5 μl of PCR product, 0.5 units of Shrimp Alkaline Phosphatase and 5 units of Exonuclease I (Amersham), then incubating at 37°C for 30 min and 80°C for 15 min. Cleaned PCR fragments were sequenced using BigDye© (Applied Biosystems Inc., Foster City, CA) terminator chemistry and visualized on an ABI 377 or ABI 3730 following manufacturer supplied protocols. Forward and reverse sequences were proofread in Sequencher 4.5 (Gene Codes Corp., Ann Arbor, MI), and all 340-base-pair (bp) fragments were aligned by eye. Sequences of all haplotypes recovered are in GenBank (accession numbers HQ450218–HQ450288).
Genetic analysis
Tests of genetic structure
To examine genetic structure across the latitudinal range of M. menidia, unique haplotypes from all 25 sites (Table 1) were determined using Maclade 4.05 (Maddison and Maddison 2002) The relationship between these haplotypes was determined via the MINSPNET algorithm as employed Arlequin 3.0 (Excoffier et al. 2005). The resulting minimum-spanning tree was drawn by hand in Adobe Illustrator CS™. Because of the absence of clearly defined clades in this tree, the frequency of the six most common haplotypes, rare haplotypes, and singleton alleles was plotted in pie diagrams onto geography to initially explore regional subdivisions.
To more robustly investigate genetic differentiation across a latitudinal gradient, levels of population subdivision were estimated using the analysis of molecular variance (AMOVA) algorithm as implemented in Arlequin 3.0 (Excoffier et al. 2005). AMOVA were run in three ways: (1) assuming no a priori regional geographic structure; (2) enforcing regional structure corresponding to traditionally recognized marine biogeographic provinces according to Briggs (1974) for the western North Atlantic: the Carolinian province (northeast Florida to Cape Hatteras), the Virginian province (Cape Hatteras to Cape Cod), and the Acadian province (Cape Cod to the Gulf of St. Lawrence; Fig. 1); and (3) enforcing three regions as suggested by the spatial distribution of the mtDNA haplotypes: a Florida to Cape Cod region, a Gulf of Maine region (including the Bay of Fundy), and a Gulf of St. Lawrence region.
Patterns of gene flow among sampled sites were assessed by pairwise ϕST and between regions by ϕCT in Arlequin 3.0. Because of the high number of pairwise comparisons, significant values for pairwise ϕST estimates were adjusted via Bonferroni correction, and sequential Bonferroni was calculated using MacBonferroni (Watkins 2002). Additionally, we calculated a standardized F ST that takes into account within-population genetic variation (F′ST) (Hedrick 2005; Meirmans 2006). Significance levels were determined through 10,000 randomization replicates. To confirm the genetic structure tested above, ϕST and F′ST matrices were plotted on a map created via Delaunay triangulation and Voronoi tessellation using the software Barrier 2.2 (Manni et al. 2004). We applied the default settings to plot up to 10 barriers.
To test the effect of sampling intensity on population structure results, a power analysis was designed to compare AMOVA results at various levels of sampling intensity when enforcing no a priori regional geographic structure, biogeographic provinces, and phylogeographic provinces, as described above. Samples were structured by (1) all 25 sites in both 2005 and 2006 (n = 34) using all samples collected at those sites, (2) all 25 sites with no temporally sampled 2006 sites (n = 25) using all samples collected at those sites, (3) all 25 sites (n = 25) using 10 samples randomly selected per site, (4) 10 sites (selected ~300 km apart; n = 10) using all samples at those 10 sites, and (5) 10 sites (n = 10) using 10 samples randomly selected per site.
To test for isolation by distance (IBD), pairwise ϕST and F′ST values were plotted against geographic distance among sites sampled. Geographic distances were estimated using Arcview™ 3.2 (ESRI), and distance calculations consisted of the shortest path, by sea, between any two sample sites. Significance was determined using Mantel tests as implemented in Isolation By Distance (Jensen et al. 2005) with 10,000 permutations. This test was run using a global analysis of all sites sampled in 2005 and new sites in 2006, as well as with the regions found to explain the highest partitioning of variance by AMOVA (comparison number 3 above); however, these analyses could only be performed for the Florida to Cape Cod and Gulf of Maine regions as the Gulf of St. Lawrence region contained only two sampled sites.
Temporal stability analysis
As with many phylogeography studies, the time-consuming nature of collections required that sample collection spans multiple years. Because of the mostly annual life history of M. menidia, combining samples across multiple years could impact the results as the genetic composition of local populations may vary over time due to yearly recolonization patterns following offshore migration. To determine whether the recovered patterns of genetic structure may be influenced by combining samples across years in an annual species, samples were collected from nine sites in both 2005 (n = 345) and 2006 (n = 327). Patterns of geographic structure between 2005 and 2006 were assessed for each temporally sampled site by treating temporal samples as unique sites and testing for significant pairwise ϕST and ϕCT among temporal samples using Arlequin 3.0 (Excoffier et al. 2005), following methods described above. An ANCOVA was also used to test similarities in haplotypic diversity across sample years with latitude as a covariate in Statistica 6.1 (StatSoft Co.). To further explore annual differences in the distribution of genetic variation, the frequency of the six most common haplotypes (those haplotypes found in more than 10 individuals) was compared between 2005 and 2006 in each of the nine resampled sites. Frequencies of these haplotypes across years were analyzed using chi-squared analysis.
Tests of demographic expansion and selection
Due to the geologic history of the Northwest Atlantic, non-equilibrium dynamics may result from recolonization of previously glaciated regions. However, non-equilibrium dynamics may result from the effects of selection across the ecological gradients of this region. To explore the impacts of population expansion and selection on the data set, we employed a variety of methods. First, following Crandall et al. (2008), we calculated Fu’s F S (Fu 1997) and Fu and Li’s D* (Fu and Li 1993). These tests have been developed to test for departures from the neutral model due to population expansion, background selection, or positive selection, with each test being more powerful at detecting certain forces than the other. These analyses were performed in DnaSP 4.10.7 (Rozas et al. 2003) with significance of F S and D* determined by 1,000 simulations of the neutral model.
Second, because a northward expansion from a southern glacial refuge should result in a genetic diversity gradient that decreases from south to north, whereas an expansion from a northern refuge should produce the opposite trend, haplotypic, nucleotide, and theta diversity were calculated in Arlequin 3.0 (Excoffier et al. 2005) and plotted against latitude in Statistica 6.1 (StatSoft Co.) to explore statistical associations. In addition, analyses of variance (ANOVA) were used to test for differences in theta between phylogeographic regions, and a post hoc test (Tukey t-test) was performed to detect specific differences between the regions (JMP 7.0 2007). While such a diversity gradient is a prediction of recolonization, there is no expectation of latitudinal gradients in genetic diversity under selection.
Third, we specifically tested for demographic expansion by analyzing the distribution of all pairwise haplotype differences using Arlequin 3.0 (Excoffier et al. 2005). We calculated goodness of fit of the estimated distribution predicted by a sudden expansion model (10,000 simulations) and associated raggedness index (Harpending 1994) for a global data set of all sites sampled and regional data sets of the Gulf of St. Lawrence, Gulf of Maine, and Florida to Cape Cod. Mismatch distributions tend to be unimodal and smooth in populations that have undergone population expansions, while multimodal and rough distributions describe populations that have experienced long-term stability. The goodness of fit of the observed data to the predicted distribution modeled for rapid population expansion was explained using a sum of squares method (SSD). When the observed distributions fit the sudden expansion model (P ≥ 0.05), we estimated the number of generations since the expansion (t) from the peak of the distribution (τ) as t = τ/2μ where μ is the rate of mutation per gene per generation.
Results
A total of 340 base pairs of mitochondrial control region (D-Loop) data were collected from 1,029 individuals in 25 sample sites, including samples collected both in 2005 (n = 421) and in 2006 (n = 283) plus the nine sites that were resampled in 2006 (n = 325; Table 1). Sequences were easily aligned with only a single insertion/deletion observed, yielding 42 polymorphic sites and 76 haplotypes. Maximum pairwise sequence variation was 2.4% (eight steps). Genetic diversity varied greatly among sites. Haplotype diversity ranged from 0.074 to 0.8571 in sites 23 and 4, respectively, nucleotide diversity ranged from 0.0002 to 0.0047 in sites 23 and 4, respectively, and theta ranged from 0.2594 to 3.3830 in sites 23 and 4, respectively.
The minimum-spanning tree of 71 haplotypes shows a star-like phylogeny with most haplotypes differing from the most common haplotype (haplotype 1) by a single substitution (Fig. 3a), and no haplotype being more than four substitutions different from haplotype 1. Haplotype 1 is not only the most common haplotype overall, found in 66% of all sampled individuals, but also the most common haplotype within all sites sampled with the exception of the most northern site, the Magdalen Islands (Site 25), where it is matched in frequency by haplotype 2 (Fig. 3a). Of the remaining haplotypes, 27 were shared among multiple sample sites, and the remaining 43 were singletons found in only a single individual within a single site.
a Map of the western North Atlantic coastline with site numbers (corresponding to Table 1) and pie diagrams showing the relative frequency of M. menidia’s mitochondrial control region haplotypes in each of the 25 collection sites. For display purposes, only the six most common haplotypes are differentiated from the others, with the less common (gray) and singleton (white) haplotypes lumped together. The unrooted minimum-spanning tree depicts the genetic relationship of 71 unique mitochondrial control region haplotypes. The 120-m isobath, indicating the shoreline at the height of the LGM, is also shown. Colors are as in Fig. 3b. b Relative frequencies of the mitochondrial control region haplotypes from each of the nine sites temporally sampled in 2005 (pie on the left) and 2006 (pie on the right). Sample sites and data correspond to that in Table 1
Regional genetic structure
The global ϕST analysis of all fish collected in both 2005 and 2006 indicated low but significant population structure (AMOVA, ϕST = 0.04, P < 0.0001) with 95.78% of the genetic variation was within sites and only 4.23% of the variation among sites (Table 2). A second AMOVA analysis which imposed a regional genetic structure that corresponded to the main biogeographic provinces of the western North Atlantic (Briggs 1974; Engle and Summers 1999) showed that 2.17% the observed genetic variation was due to differences between these provinces (AMOVA, ϕCT = 0.02, P < 0.0001), while 2.72% was due to differences between sites within these provinces (AMOVA, ϕSC = 0.03, P < 0.0001; Table 2).
Despite the low level of population structure observed when no groupings were imposed in the AMOVA analysis, distinct regional patterns clearly emerge when haplotypes are plotted onto geography, particularly if one focuses on the three most common haplotypes (haplotypes 1–3; Fig. 3a). Although haplotype 1 was present in the highest frequencies across all sampled sites (with the exception listed above), the Gulf of St. Lawrence sites had a disproportionately high frequency of haplotype 2, while the Gulf of Maine and Bay of Fundy sites had disproportionately high frequencies of haplotype 3. While both haplotypes 2 and 3 were found in the Florida to Cape Cod region, they were generally seen in lower frequencies. An AMOVA imposing three regions corresponding to (1) Florida to Cape Cod, (2) Gulf of Maine, and (3) Gulf of St. Lawrence revealed significant population structure (AMOVA, ϕST = 0.07, P < 0.0001) with 93% of the genetic variation within sites and 6.38% of due to differences between these phylogeographic regions (AMOVA, ϕCT = 0.06, P < 0.0001), a higher value than the level of population structure due to differences between biogeographic regions reported above. The percent variation among sites within these regions was low (0.83%) but significant (AMOVA, ϕSC = 0.01, P = 0.0285; Table 2) and lower than levels found with the previous biogeographic regions. While these regional differences are relatively subtle due to low genetic variation, the percentage of variation attributed to differences between regions was depressed due to the high frequency of haplotype 1 in all sites sampled. As a heuristic to explore differences between the remaining haplotypes, we conducted a structured AMOVA after the removal of haplotype 1. Results of this analysis indicate 23.72% of the genetic variation was due to differences between regions defined by the geographic distribution of haplotypes (AMOVA, ϕCT = 0.24, P < 0.0001; ϕST = 0.24, P < 0.0001; ϕSC = 0.01, P < 0.1760), with only 0.55% of the variation among sites within regions.
Barrier 2.2 analysis identified genetic structure that was largely consistent between genetic distance measures, ϕST and F′ ST, and with phylogeographic and biogeographic regions. For F′ ST, the first-order barrier corresponded with the phylogeographic barrier between the Gulf of St. Lawrence and the Gulf of Maine, additional structure was found within the Virginian province between South Carolina and North Carolina, at Massachusetts Bay and at Cape Hatteras (presented in order, as described by Barrier 2.2). ϕST data found the same limits to gene flow but in a different order, and with additional structure between sites sampled in Maine.
Consistent with the results of the AMOVAs, analyses of pairwise ϕST and F′ST among all sites sampled in 2005 plus all new sites sampled in 2006 (no sites resampled) indicated few limits to genetic exchange except among regions (Supplementary Table 1). A total of 105 of the 300 pairwise ϕST comparisons were found to be significant (P = 0.05 − ≤0.0001). Of these, 80 were contrasts between sites north and south of Cape Cod, 12 between the Gulf of Maine and Gulf of St. Lawrence, 10 within the Florida to Cape Cod region, 2 within the Gulf of Maine, and 1 within the Gulf of St. Lawrence. Following Bonferroni correction (alpha = 0.000016), only 27 ϕST pairwise values remained significant, 21 of which were contrasts between sites north and south of Cape Cod and 6 between the Gulf of Maine and Gulf of St. Lawrence. A less conservative sequential Bonferroni correction (Rice 1989) found the same 27 pairwise ϕST to be significant. As expected, analyses of F′ST pairwise distances found similar results to ϕST (Supplementary Table 1).
A power analysis testing the effect of sampling intensity on resulting genetic structure found that significant results were recovered in all analyses, except for those with the fewest sites or fewest individuals per site (Supplementary Table 2). Regional structure (ϕCT) results did not change under decreased sampling intensity when enforcing groupings based on observed phylogeographic, although reduced sampling resulted in higher ϕCT values than with higher numbers of individuals or sites. All analyses enforcing structure corresponding to the biogeographic provinces were also significant except when 10 sites were analyzed whether including all samples or 10 samples. ϕST values were similarly significant for all comparisons except for the lowest sampling intensity of 10 individuals and 10 sites. The largest impact of sampling intensity was seen in the ability to detect structure among sites within regions (ϕSC) where only the highest sampling intensities could resolve this additional structure.
A subtle but significant relationship between genetic distance and geographic distance was indicated by a Mantel test (ϕST, R 2 = 0.203, N = 25, m = 1.18 × 10−04, P = 0.0006; F′ST, R 2 = 0.241, N = 25, m = 2.33 × 10−04, P = 0.0001) suggesting isolation by distance (IBD) across the western North Atlantic. However, because patterns of IBD can be influenced by regional genetic structure, IBD analyses were performed with sites grouped into the three regions described above. No correlation between genetic and geographic distance was observed within the Florida to Cape Cod (ϕST, R 2 = 0.007, N = 17, m = −5.46 × 10−05, P = 0.772; F′ST, R 2 = 0.0003, m = 2.22 × 10−04, P = 0.355) region, suggesting that the global pattern of IBD is likely driven by allele frequency differences between regions. However, within the Gulf of Maine, while ϕST does not show a relationship between genetic and geographic distance (N = 6, R 2 = 0.0002, m = −1.91 × 10−04, P = 0.4776), F′ST did show a significant relationship (N = 6, R 2 = 0.506, m = 5.135 × 10−05, P = 0.023).
Temporal analyses
Pairwise ϕST comparison of the 9 temporally sampled sites recovered 69 of 144 significant values; however, all of these significant values were from comparisons between regions. No significant values were observed among samples taken from a single locality in 2005 and 2006. Similar results were obtained using F′ST. Temporal stability of regional patterns is further seen in the plot of the frequencies of the six most common haplotypes, which demonstrates clear similarities within sites sampled in both 2005 and 2006 as well as the pronounced differences between the Florida to Cape Cod, Gulf of Maine, and Gulf of St. Lawrence regions (Fig. 3b). An ANCOVA further demonstrated that for any given latitude, haplotypic diversity does not change from year to year (ANCOVA, F 1,15 = 0.064, P = 0.8044). Chi-squared analysis of the frequencies of the 6 most common haplotypes found a significant difference between years only in New Jersey (X 2 = 11.874; P = 0.037), and this relationship is not significant after Bonferroni correction (α = 0.0056), suggesting haplotypes are returning at the same frequencies to most sites from year to year.
Demographic analyses
Fu’s F S values were negative at all sites, and all were highly significant except the sites in the Gulf of St. Lawrence. In contrast, only 5 sites had significant D* values: 4 to the south of Cape Cod and one in the Gulf of Maine Table 1). Global, Gulf of Maine, and Florida to Cape Cod regional analyses also found significant F S values (Table 3; P ≤ 0.05) and were negative; however, the Gulf of St. Lawrence F S value was not significant, perhaps due to the few sites sampled in this region.
Although genetic diversity was very low, genetic diversity indices showed a pronounced decrease with increasing latitude. Haplotype diversity decreased sharply with increasing latitude (R 2 = 0.44, P < 0.0001, y = −0.0279x + 1.6439; Table 1; Fig. 4), nucleotide diversity (R 2 = 0.54, P = 0.00017, y = −0.0002x + 0.01; Table 1; Fig. 4), and theta also decreased as latitude increased (R 2 = 0.59, P < 0.0001, y = −0.1303x + 6.8456; Table 1; Fig. 4). Theta (ANOVA, F = 19.30, P < 0.0001) was significantly lower in the Gulf of Maine and Gulf of St. Lawrence than in Florida to Cape Cod (Tukey t-test, P < 0.05).
Genetic diversities vs. latitude for 25 sites (no temporally resampled sites) of M. menidia with data taken from Table 1; a Haplotypic diversity vs. latitude (n = 25; R 2 = 0.44, P < 0.0001, y = −0.0279x + 1.64), b Nucleotide diversity vs. latitude (n = 25; R 2 = 0.54, P = 0.00017, y = −0.0002x + 0.01), c Theta vs. latitude (n = 25, R 2 = 0.59, P < 0.0001, y = −0.1303x + 6.8456)
Using a mutation rate (μ) for the mitochondrial control region of 2 × 10−8 (a conservative estimate) to 5 × 10−8 substitutions/site/year (Bowen et al. 2006) and an estimate of a generation time of 1 year (Conover and Ross 1982), we tested for population expansion using mismatch distribution estimates (Table 3). A global mismatch analysis of all regions together did not deviate significantly from a unimodal mean pairwise difference distribution (P = 0.99), suggesting a recent population expansion. Regional analysis of mismatch distribution estimates found that the Gulf of Maine and Florida to Cape Cod regions also conform to a model of population expansion (P = 0.95 and 0.6, respectively). However, the northern Gulf of St. Lawrence region differed significantly from the model of rapid population expansion (P = 0.029). These regional trends were reinforced with Harpending’s raggedness (r) values (Harpending 1994), which were significantly different from ragged distribution in Gulf of Maine (r = 0.157, P = 0.6) and Florida to Cape Cod (r = 0.037, P = 0.95) regions, but not in the Gulf of St. Lawrence (r = 0.208, P = 0.023; Table 3). The peak of the unimodal distribution (τ) for the global mismatch analysis was 0.906 corresponding to a population expansion 26,647 to 66,618 years ago for the faster and slower mutation rate, respectively. Regional analysis of τ demonstrated population expansions for the Gulf of Maine as 17,470 to 43,676 years ago and Florida to Cape Cod region as 31,147 to 77,868 years ago. However, as the Gulf of St. Lawrence model estimated values were found significantly different than those observed, we could not estimate time since population expansion.
Discussion
Patterns of regional isolation
Phylogeographic analysis of Menidia menidia populations across the Northwest Atlantic indicates significant genetic differentiation (F ST = 0.07, P < 0.0001) among three regions: (1) Florida to Cape Cod, (2) the Gulf of Maine, and (3) the Gulf of St. Lawrence. While this structure is modest compared to some Atlantic estuarine fishes (Rocha et al. 2005; Santos et al. 2006), it is substantial compared to other studies of fishes of the western Atlantic that show limited or no evidence for regional genetic structure (e.g., sturgeon, Acipenser oxyrhynchus, Bowen and Avise 1990; Jones and Quattro 1999), including some migratory species (Cordes and Graves 2003) that have natal homing (Thorrold et al. 2001). This grouping explained more than twice the regional variation (6.38%) than the traditional Carolinian, Virginian, and Acadian biogeographic provinces (2.46%). Regional distinctions recovered in this study more closely match those of the Marine ecoregions recently proposed by Spalding et al. (2007), with the exception of the failure to recover distinct Carolinian and Virginian regions (e.g., a fourth phylogeographic region). Barrier analyses confirmed the above but suggest additional breaks in the Gulf of Maine and at Cape Hatteras, a result that may be an artifact of Barrier searching for edges between the linearly arranged sites, forcing the program to find edges between individual sites rather than groups of sites.
Previous studies using allozymes reported no detectable genetic structure in M. menidia (Johnson 1975), suggesting that the pronounced, heritable patterns of phenotypic variation across its range (Conover and Present 1990; Conover and Schultz 1995; Yamahira and Conover 2002) evolved despite high levels of gene flow that should swamp out local adaptation. However, results from this study show that there are significant limits to gene flow in M. menidia, as would be expected for adaptive phenotypic variation to evolve. That these filters to gene flow broadly correspond to distinct oceanographic regions (below) suggest that limited gene flow among these regions may facilitate local adaptation to the conditions within these regions. Limits to dispersal and gene flow M. menidia have also been recently demonstrated by Clarke et al. (in press) whose use of otolith chemistry revealed a break in migration patterns at Cape Cod suggesting evidence of natal homing.
Recovery of phylogeographic structure is remarkable given the extremely low levels of genetic diversity in M. menidia in comparison with other studies of fish using mtDNA control region (Bowen and Avise 1990). Reduced diversity may be a function of a recent expansion from a population bottleneck (below). However, lower diversity may also result from being an annual species as is observed in annual squid (Ito et al. 2006) and annual plants (Austerlitz et al. 2000), a hypothesis that should be examined further in marine taxa. However, despite this very low genetic diversity, a power analysis demonstrates the ability to detect phylogeographic structure with only mtDNA using a very limited number of samples from a limited number of sites, with reduced sampling only substantially impacting the ability to detect additional structure among sites within regions. These results attest to the power and utility of mtDNA sequence data despite recent assertions to the contrary (Bazin et al. 2006; Galtier et al. 2009).
Origins of phylogeographic structure
Patterns of regional genetic structure are largely consistent with the geological history of the Northwest Atlantic. During the last glacial maximum, the Gulf of Maine and the Gulf of St. Lawrence were entirely covered by glaciers (Shaw et al. 2002). Although glaciers slowly began to recede as early as 20 kya, it was not until 13 kya that the Gulf of Maine was entirely ice free (Shaw et al. 2002). At that time, and to this day, the Gulf of Maine formed a semi-enclosed deep basin, with Georges Bank forming a shallow-water sill to the south and Browns Bank a shallow sill to the north (Shaw et al. 2002). Given that M. menidia migrate to deeper offshore waters in the winter (Conover and Murawski 1982), the partial enclosure of the Gulf of Maine could have restricted migration and genetic exchange between the Gulf of Maine and populations south of Cape Cod, promoting differentiation among these regions.
Similarly, contemporary populations from the Gulf of St. Lawrence are restricted to the Magdalen Shelf with Nova Scotia representing a filter to return migration to the shore zone after winter, isolating northern populations from the Gulf of Maine (Kenchington et al. 2009). Furthermore, winter sea ice forms in this region of the Gulf of St. Lawrence every year, while it is largely absent from the Gulf of Maine. Sea ice would force already thermally stressed fish into even deeper waters where they face predation or freezing temperatures over the long winter months. Neither Gulf of Maine nor Florida to Cape Cod populations may be adapted to the harsh winter conditions of the Gulf of St. Lawrence, which could restrict successful recruitment and northward gene flow into this region.
While ocean currents are thought to play a major role in limiting genetic exchange around Cape Cod for benthic marine invertebrates with larval dispersal (Jennings et al. 2009), it is likely that that seasonal stability in sea surface temperature may serve a role as well. Cape Cod either serves as a physical barrier forming the northern and southern limits for many pops or may act as a filter by virtue of its position at the boundary between different oceanographic regimes limiting dispersal and gene flow (Engle and Summers 1999). Populations south of Cape Cod experience more seasonal variation in sea surface temperature but generally warmer ocean temperatures than their counterparts in the Gulf of Maine, which is cold year round (Hutchins 1947). It is possible that populations adapted to warmer temperatures south of Cape Cod may not be able to tolerate the harsh winter conditions within the Gulf of Maine, and likewise, populations from the Gulf of Maine may not be able to tolerate the large annual temperature variations experienced by southern populations. Combined, these results suggest that the environmental variables that structure marine communities in the western North Atlantic, in addition to the physical barriers, may play a role in limiting genetic exchange between populations of M. menidia, facilitating adaptation to local environmental conditions.
The presence of a clear phylogeographic break at Cape Cod but not at Cape Hatteras is difficult to explain. North of Cape Hatteras M. menidia migrate offshore in the winter to a lesser degree than populations in the Gulf of Maine and the Gulf of St. Lawrence, but more than those populations south of Cape Hatteras where there is no offshore winter migration and less potential for dispersal (Cain and Dean 1976; Shenker and Dean 1979). This difference in life history dispersal in conjunction with persistent ocean currents that collide to create one of the largest thermal gradients in the world’s ocean should, in theory, create a genetic break at Cape Hatteras similar to Cape Cod. Barrier does detect a minor genetic discontinuity in this region suggesting that there may be some limits to gene flow across this region. However, failure to see a more substantial break here suggests that limits to dispersal may not be completely a function of the physical limits to dispersal but may result from the interplay of both biotic and abiotic dispersal filters that are more likely to impact low dispersal species than high dispersal species (Díaz-Ferguson et al. 2010).
Regional fidelity and temporal stability of patterns
Further evidence of more limited genetic exchange is evidenced by the temporal stability of genetic patterns. The genetic structure between sites in the Gulf of St. Lawrence, Gulf of Maine, and Florida to Cape Cod is seen both in 2005 and in 2006. Given that M. menidia is a largely annual species with overwinter mortality that exceeds 99% (Conover and Ross 1982), there is no a priori reason to expect constancy of regional genetic patterns as bays and estuaries are recolonized each year de novo. Stability of gene frequencies over time can only be explained if offspring spawned in one region in 2005 returned to the same general region in 2006 as adults. While these results do not suggest natal homing as known in salmonids and other anadromous fishes, results do require some degree of regional fidelity of returning spawners. Regional fidelity, in conjunction with environmental filters to dispersal, may contribute to the observed regional genetic structure. Otolith analyses by Clarke et al. (in press) indicate that some adult return to their natal estuaries following their offshore winter migration, further supporting behavioral regional fidelity as a mechanism contributing to limited genetic exchange.
Even though there are consistent genetic patterns from year to year, temporal comparison of 2005 and 2006 suggests that patterns are not completely stable. For example, the New Jersey site shows two significantly different sets of haplotypes between years suggesting occasional shifts in allele levels across sites; however, this significance may just be an artifact of multiple comparison as these values were not significant after Bonferroni correction. Thus, while the data suggest broad regional fidelity, there may also be mixing with adjacent sites, a result similarly reported in weakfish (Thorrold et al. 2001) and in Clarke et al.’s otolith studies on M. menidia (Clarke et al. in press).
Evidence for post-glacial expansion
Although our data define clear phylogeographic regions, genetic diversity was remarkably low, suggesting non-equilibrium dynamics that could be attributed to demographic expansion following a bottleneck or selection. While distinguishing between these alternatives can be difficult, a number of aspects of the data suggest population expansion.
First, the relationship of haplotypes in the minimum-spanning tree is star-like, with multiple closely related haplotypes radiating from a common central haplotype. This pattern is noted to be characteristic of species that have undergone a recent range expansion or recovery from a recent population bottleneck (Brookfield 2001).
Second, results from the analyses of Fu’s F S and Fu and Li’s D* indicate that most sites have significantly negative F S values, while few significant D* values are observed. Fu’s F S values are expected to have large negative values for demographic expansion or a selective sweep (Ramirez-Soriano et al. 2008), while D* statistics are more powerful in detecting purifying selection (Fu and Li 1993). Thus, an expansion (or selective sweep) is indicated when F S is significant and D* is not as observed in our data, while the opposite pattern should be observed under selection.
Third, Menidia menidia shows a clear pattern of decreasing genetic diversity (haplotype diversity, nucleotide diversity, and theta) with increasing latitude (Fig. 4). This pattern is exactly what would be predicted if populations were extirpated from northern latitudes and were recolonized from a single southern refugia, a result consistent with other marine taxa displaced by Pleistocene glaciations (McMillen-Jackson and Bert 2004; Hare and Weinberg 2005). Taken together, these studies suggest that genetic diversity clines are a common feature of the western North Atlantic and likely represent a common result of Pleistocene glaciations (McMillan et al. 1992; Hewitt 1996; Bernatchez and Wilson 1998; Wares and Cunningham 2001; Young et al. 2002; Marko 2004).
Lastly, mismatch distributions calculated in Arlequin match the expectations under the hypothesis of population expansion, a result that is consistent with the geologic history of the Northwest Atlantic. Until 20 kya, North America experienced glacial coverage of the coastline down to Long Island and Cape Cod (Shaw et al. 2002), rendering the northern 1,000 km of M. menidia’s contemporary range uninhabitable. As M. menidia would be restricted to the southern half of its modern range, a pattern seen in many western North Atlantic marine species (Bucklin and Wiebe 1998; Wares and Cunningham 2001; Wares 2002), the transition from the LGM to the present interglacial period would necessitate a range expansion to achieve present-day distributions. The mismatch distribution analysis indicated that population increases occurred approximately 27,000–67,000 years ago (depending on the molecular clock calibration, 5%/my and 2%/my, respectively). Regional analyses showed that population expansion is the oldest in the Florida to Cape Cod region, a region that was never glaciated. In the Gulf of Maine, our analysis indicates a population increase occurred more recently 43,500–17,500 years ago, as expected, with temperatures becoming more suitable as glaciers receded northward. Although analyses could not calculate an age for the Gulf of St. Lawrence, lower diversity in this region necessitates a younger population history. The absence of detecting a population expansion in the Gulf of St. Lawrence does not mean that these populations persisted in a northern refugium as this area was either land or completely covered by glacier until the LGM. Instead, the failure to detect an expansion in this region likely results from insufficient time for these populations to evolve the necessary genetic diversity required to detect an expansion. This latitudinal gradient in time since expansion, combined with latitudinal patterns of genetic diversity, strongly suggests expansion from a single, southern refugium. This result further supports the notion of a population expansion as opposed to selective sweep or purifying selection as neither of the latter two processes would be expected to yield decreasing genetic diversity or decrease age of expansion with increasing latitude.
Conclusions
Populations of M. menidia exhibit a strong pattern of northern population expansion, following their extirpation from northern latitudes during Pleistocene glacial periods. Despite limited genetic variation, mitochondrial DNA sequence data were able to recover three phylogeographic provinces, which in part align with previously recognized marine biogeographic provinces, indicating that dispersal in this species is much less than suggested by its life history and previous genetic studies. While these results suggest the presence of filters to gene flow across recently defined marine ecoregions, temporal stability of spatial genetic heterogeneity suggests that realized dispersal in M. menidia may be even more limited than our results indicate. Temporal stability of genetic patterns and microchemistry analyses (Clarke et al. in press) support regional fidelity in M. menidia, which could further limit genetic exchange, facilitating the evolution of adaptive phenotypic clines across the strong environmental clines of the Northwest Atlantic, a hypothesis that merits further study with microsatellite-based assignment tests or additional otolith analyses.
The presence of pronounced phenotypic clines in heritable adaptive phenotypes in M. menidia following this recent population expansion attests to the power of environmental gradients in driving evolution of phenotypic gradients. This result suggests that locally adaptive phenotypes and regional genetic structure can evolve very rapidly in ecosystems with pronounced environmental heterogeneity, particularly if gene flow is limited pronounced environmental gradients as observed in M. menidia.
References
Adams SM, Lindmeier JB, Duvernell DD (2006) Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Mol Ecol 15:1109–1123
Austerlitz F, Mariette S, Machon N, Gouyon P, Godelle B (2000) Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics 154:1309–1321
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders N (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst 18:489–522
Bazin E, Gelemin S, Galtier N (2006) Population size does not influence mitochondrial genetic diversity. Science 312:570–572
Bernardi G, Sordino P, Powers DA (1993) Concordant mitochondrial and nuclear-DNA phylogenies for populations of the teleost fish Fundulus heteroclitus. Proc Natl Acad Sci USA 90:9271–9274
Bernatchez L, Wilson CC (1998) Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol 7:431–452
Billerbeck JN, Orti G, Conover DO (1997) Latitudinal variation in vertebral number has a genetic basis in the Atlantic silverside, Menidia menidia. Can J Fish Aquat Sci 54:1796–1801
Bousfield EL, Thomas MLH (1975) Postglacial changes in distribution of littoral marine invertebrates in the Canadian Atlantic Region. Proc N S Inst Sci 27:47–60
Bowen BW, Avise JC (1990) Genetic structure of Atlantic and Gulf of Mexico populations of sea bass, menhaden, and sturgeon: influence of zoogeographic factors and life-history patterns. Mar Biol 107:371–381
Bowen BW, Muss A, Rocha LA, Grant WS (2006) Shallow mtDNA coalescence in Atlantic Pygmy Angelfishes (Genus Centropyge) indicates a recent invasion from the Indian Ocean. J Hered 97:1–12
Bower AS, Rossby HT, Lillibridge JL (1985) The gulf stream—barrier or blender. J Phys Oceanogr 15:24–32
Briggs J (1974) Marine zoogeography. McGraw-Hill Inc, New York
Brody JR, Kern SE (2004) Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques 36:214–216
Brookfield JFY (2001) Population genetics: the signature of selection. Curr Biol 11:R388–R390
Bucklin A, Wiebe PH (1998) Low mitochondrial diversity and small effective population sizes of the copepods Calanus finmarchicus and Nannocalanus minor: possible impact of climatic variation during recent glaciation. J Hered 89:383–392
Cain R, Dean J (1976) Annual occurrence, abundance and diversity of fish in a South Carolina intertidal creek. Mar Biol 36:369–379
Clarke LM, Munch SB, Thorrold SR, Conover DO (in press) High connectivity among locally adapted populations of a marine fish (Menidia menidia). Ecology
Collin R (2001) The effects of mode of development on phylogeography and population structure of North Atlantic Crepidula (Gastropoda: Calyptraeidae). Mol Ecol 10:2249–2262
Conover DO (1998) Local adaptation in marine fishes: evidence and implications for stock enhancement. Bull Mar Sci 62:477–493
Conover DO, Murawski SA (1982) Offshore winter migration of the Atlantic silverside, Menidia menidia. Fish Bull 80:145–150
Conover DO, Present TMC (1990) Countergradient variation in growth-rate: compensation for length of the growing-season among Atlantic silversides from different latitudes. Oecologia 83:316–324
Conover DO, Ross MR (1982) Patterns in seasonal abundance, growth and biomass of the Atlantic silverside, Menidia menidia, in a New England estuary. Estuaries 5:275–286
Conover DO, Schultz ET (1995) Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol Evol 10:248–252
Conover DO, Arnott SA, Walsh MR, Munch SB (2005) Darwinian fishery science: lessons from the Atlantic silverside (Menidia menidia). Can J Fish Aquat Sci 62:730–737
Cordes JF, Graves JE (2003) Investigation of congeneric hybridization in and stock structure of weakfish (Cynoscion regalis) inferred from analyses of nuclear and mitochondrial DNA loci. Fish Bull 101:443–450
Crandall ED, Frey MA, Grosberg RK, Barber PH (2008) Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Mol Ecol 17:611–626
Dahlgren T, Weinberg J, Halanych K (2000) Phylogeography of the ocean quahog (Arctica islandica): Influences of paleoclimate on genetic diversity and species range. Mar Biol 137:487–495
Dawson M (2001) Phylogeography in coastal marine animals: A solution from California? J Biogeogr 28:723–736
Díaz-Ferguson E, Robinson JD, Silliman B, Wares JP (2010) Comparative phylogeography of North American Atlantic salt marsh communities. Estuaries and Coasts 33:828–839
Durand E, Jay F, Gaggiotti O, Francois O (2009) Spatial inference of admixture proportions and secondary contact zones. Mol Biol Evol 26:1963–1973
Dyke AS, Prest VK (1987) Late Wisconsonian and Holocene history of the Laurentide ice sheet. Géographie Physique et Quaternaire 41:237–263
Engle VD, Summers JK (1999) Latitudinal gradients in benthic community composition in Western Atlantic estuaries. J Biogeogr 26:1007–1023
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Galtier N, Nabholz B, Glemin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Moleclar Ecol 18:4541–4550
Gonzalez Villasenor LI, Powers DA (1990) Mitochondrial-DNA restriction-site polymorphisms in the teleost Fundulus heteroclitus support secondary intergradation. Evolution 44:27–37
Hare MP, Weinberg JR (2005) Phylogeography of surfclams, Spisula solidissima, in the western North Atlantic based on mitochondrial and nuclear DNA sequences. Mar Biol 146:707–716
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Holder K, Montgomerie R, Friesen VL (1999) A test of the glacial refugium hypothesis using patterns of mitochondrial and nuclear DNA sequence variation in rock ptarmigan (Lagopus mutus). Evolution 53:1936–1950
Hutchins LW (1947) The bases for temperature zonation in geographical distribution. Ecol Monogr 17:325–335
Ito K, Yanagimoto T, Iwata Y, Munehara H, Sakurai Y (2006) Genetic population structure of the spear squid Loligo bleekeri based on mitochondrial DNA. Nippon Suisan Gakkaishi 72:905–910
Jennings R, Shank T, Mullineaux L, Halanych K (2009) Assessment of the Cape Cod phylogeographic break using the bamboo worm Clymenella torquata reveals the role of regional water masses in dispersal. J Hered 100:86
Jensen JL, Bohonak AJ, Kelly ST (2005) Isolation by distance, web service. BMC Genetics 6, v3.04. http://ibdws.sdsu.edu
JMP 7.0 (2007) JMP. SAS Institute Inc, Cary
Johnson MS (1975) Biochemical systematics of Atherinid genus Menidia. Copeia 4:662–691
Jones ME (2005) Comparative phylogeopgraphy, population structure, and extended maternal care in three intertidal haustoriid amphipods from the western North Atlantic. M.S. Thesis, College of Charleston, SC
Jones WJ, Quattro JM (1999) Genetic structure of summer flounder (Paralichthys dentatus) populations north and south of Cape Hatteras. Mar Biol 133:129–135
Kelly DW, MacIsaac HJ, Heath DD (2006) Vicariance and dispersal effects on phylogeographic structure and speciation in a widespread estuarine invertebrate. Evolution 60:257–267
Kenchington E, Harding G, Jones M, Prodöhl P (2009) Pleistocene glaciation events shape genetic structure across the range of the American lobster, Homarus americanus. Mol Ecol 18:1654–1667
Lee WJ, Conroy J, Howell WH, Kocher TD (1995) Structure and evolution of teleost mitochondrial control regions. J Mol Evol 41:54–66
Maddison WP, Maddison DR (2002) MacClade version 4.05: analysis of phylogeny and character evolution. Sinauer, Sunderland
Manni F, Guerard E, Heyer E (2004) Geographic patters of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier’s algorithm”. Hum Biol 76:173–190
Marko PB (2004) ‘What’s larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Mol Ecol 13:597–611
McMillan WO, Raff RA, Palumbi SR (1992) Population genetic consequences of developmental evolution in sea urchins (genus Heliocidaris). Evolution 46:1299–1312
McMillen-Jackson AL, Bert TM (2004) Genetic diversity in the mtDNA control region and population structure in the pink shrimp Farfantepenaeus duorarum. J Crustac Biol 24:101–109
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Pielou EC (1991) After the ice age. University of Chicago Press, Chicago
Porter SC (1989) Some geological implications of average quaternary glacial conditions. Quat Res 32:245–261
Pratt RM, Schlee J (1969) Glaciation on the continental margin off New England. Bull Geol Soc Am 80:2335–2342
Present TMC, Conover DO (1992) Physiological basis of latitudinal growth differences in Menidia menidia—variation in consumption or efficiency. Funct Ecol 6:23–31
Ramirez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A (2008) Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179:555–567
Rice WR (1989) Analysing tables of statistical tests. Evolution 43:223–225
Rocha LA, Robertson DR, Roman J, Bowen BW (2005) Ecological speciation in tropical reef fishes. Proc R Soc Lond B Biol Sci 272:573–579
Rohling EJ, Fenton M, Jorissen FJ, Bertrand P, Ganssen G, Caulet JP (1998) Magnitudes of sea-level lowstands of the past 500,000 years. Nature 394:162–165
Rozas J, Sanchez-DelBarrio JC, Messegyer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformaticcs 19:2496–2497
Santos S, Hrbek T, Farias IP, Schneider H, Sampaio I (2006) Population genetic structuring of the king weakfish, Macrodon ancylodon (Sciaenidae), in Atlantic coastal waters of South America: deep genetic divergence without morphological change. Mol Ecol 15:4361–4373
Schmitt T (2007) Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool 4:11
Shackleton NJ, Backman J, Zimmerman H, Kent DV, Hall MA, Roberts DG, Schnitker D, Baldauf JG, Desprairies A, Homrighausen R, Huddlestun P, Keene JB, Kaltenback AJ, Krumsiek KAO, Morton AC, Murray JW, Westbergsmith J (1984) Oxygen isotope calibration of the onset of ice rafting and history of glaciation in the North Atlantic region. Nature 307:620–623
Shaw J, Gareau P, Courtney R (2002) Palaeogeography of Atlantic Canada 13–0 kyr. Quat Sci Rev 21:1861–1878
Shenker JM, Dean JM (1979) Utilization of an intertidal salt marsh creek by larval and juvenile fishes—abundance, diversity and temporal variation. Estuaries 2:154–163
Spalding M, Fox HE, Halpern BS, McManus MA, Molnar J, Allen GR, Davidson N, Jorge ZA, Lombana AL, Lourie SA, Martin KD, McManus E, Recchia CA, Robertson J (2007) Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57:573–583
Strasser CA, Barber PH (2008) Limited genetic variation and structure in softshell clams (Mya arenaria) across their native and introduced range. Conserv Genet 10:803–814
Thorrold SR, Latkoczy C, Swart PK, Jones CM (2001) Natal homing in a marine fish metapopulation. Science 291:297–299
Walsh PS, Metzger DA, Higuchi R (1991) Chelex-100 as a medium for simple extraction of DNA for PCR based typing from forensic material. Biotechniques 10:506–513
Wares JP (2002) Community genetics in the Northwestern Atlantic intertidal. Mol Ecol 11:1131–1144
Wares JP, Cunningham CW (2001) Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55:2455–2469
Watkins MW (2002) MacBonferroni [computer software]. Ed & Psych Associates, State College, PA
Yamahira K, Conover DO (2002) Intra- vs. interspecific latitudinal variation in growth: adaptation to temperature or seasonality? Ecology 83:1252–1262
Yamahira K, Lankford TE, Conover DO (2006) Intra- and interspecific latitudinal variation in vertebral number of Menidia spp. (Teleostei: Atherinopsidae). Copeia 3:431–436
Young AM, Torres C, Mack JE, Cunningham CW (2002) Morphological and genetic evidence for vicariance and refugium in Atlantic and Gulf of Mexico populations of the hermit crab Pagurus longicarpus. Mar Biol 140:1059–1066
Acknowledgments
This research is funded by NSF grants OCE-0425830 and OCE-0425728. The authors thank CJ Starger for technical assistance and the Sackler Institute for Comparative Genomics at the American Museum of Natural History for the generous use of their ABI 3730. Comments and criticisms by reviewers were extremely helpful in developing our manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Riginos.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Mach, M.E., Sbrocco, E.J., Hice, L.A. et al. Regional differentiation and post-glacial expansion of the Atlantic silverside, Menidia menidia, an annual fish with high dispersal potential. Mar Biol 158, 515–530 (2011). https://doi.org/10.1007/s00227-010-1577-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-010-1577-3