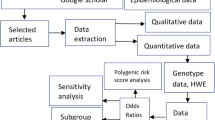
Abstract
Dozens of loci associated with fracture have been identified by genome-wide association studies (GWASs). However, most of these variants are located in the noncoding regions including introns, long terminal repeats, and intergenic regions. Although combining regulation information helps to identify the causal SNPs and interpret the involvement of these variants in the etiology of human fracture, regulation information which was truly associated with fracture was unknown. A novel functional enrichment method GARFIELD (GWAS Analysis of Regulatory of Functional Information Enrichment with LD correction) was applied to identify fracture-associated regulation information, including transcript factor binding sites, expression quantitative trait loci (eQTLs), chromatin states, enhancer, promoter, dyadic, super enhancer and Epigenome marks. Fracture SNPs were significantly enriched in exon (Bonferroni correction, p value < 7.14 × 10–3) at two GWAS p value thresholds through GARFIELD. High level of fold-enrichment was observed in super enhancer of monocyte and the enhancer of chondrocyte (Bonferroni correction, p value < 4.45 × 10–3). eQTLs of 44 tissues/cells and 10 transcription factors (TFs) were identified to be associated with human fracture. These results provide new insight into the etiology of human fracture, which might increase the identification of the causal SNPs through the fine-mapping study combined with functional annotation, as well as polygenic risk score.



Similar content being viewed by others
Data availability
All data used in this study were available online.
References
Michaëlsson K, Melhus H, Ferm H, Ahlbom A, Pedersen NL (2005) Genetic liability to fractures in the elderly. Arch Intern Med 165:1825–1830
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H (2013) The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42:D1001–D1006
Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337:1190–1195
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797
Bryois J, Garrett ME, Song L, Safi A, Giusti-Rodriguez P, Johnson GD (2018) Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun 9:3121–3121
Di Rienzo A, Kichaev G, Yang W-Y, Lindstrom S, Hormozdiari F, Eskin E (2014) Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet 10:e1004722
Ripatti S, Shi J, Park J-H, Duan J, Berndt ST, Moy W (2016) Winner’s curse correction and variable thresholding improve performance of polygenic risk modeling based on genome-wide association study summary-level data. PLoS Genet 12:e1006493
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65
Shen H, Li J, Zhang J, Xu C, Jiang Y, Wu Z (2013) Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians. PLoS ONE 8:e59494
Trynka G, Westra HJ, Slowikowski K, Hu X, Xu H, Stranger BE (2015) Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am J Hum Genet 97:139–152
Iotchkova V, Ritchie GRS, Geihs M, Morganella S, Min JL, Walter K (2019) GARFIELD classifies disease-relevant genomic features through integration of functional annotations with association signals. Nat Genet 51:343–353
Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC (2019) An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 51:258–266
GTEx Consortium, Aguet F, Brown AA, Castel SE, Davis JR, He Y (2017) Genetic effects on gene expression across human tissues. Nature 550:204
Satterlee JS, Chadwick LH, Tyson FL, McAllister K, Beaver J, Birnbaum L (2019) The NIH common fund/roadmap epigenomics program: successes of a comprehensive consortium. Sci Adv 5:eaaw6507
Hnisz D, Abraham Brian J, Lee Tong I, Lau A, Saint-André V, Sigova Alla A (2013) Super-enhancers in the control of cell identity and disease. Cell 155:934–947
Manabe N, Kawaguchi H, Chikuda H, Miyaura C, Inada M, Nagai R (2001) Connection between B lymphocyte and osteoclast differentiation pathways. J Immunol (Baltimore, Md. 1950) 167:2625–2631
Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K (2010) Development of monocytes, macrophages, and dendritic cells. Science 327:656–661
Deng FY, Lei SF, Zhang Y, Zhang YL, Zheng YP, Zhang LS (2011) Peripheral blood monocyte-expressed ANXA2 gene is involved in pathogenesis of osteoporosis in humans. Mol Cell Proteomics 10(M111):011700
UK10K Consortium, Walter K, Min JL, Huang J, Crooks L, Memari Y (2015) The UK10K project identifies rare variants in health and disease. Nature 526:82–90
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206
Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR (2015) Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47:1228–1235
Iwata T, Kawamoto T, Sasabe E, Miyazaki K, Fujimoto K, Noshiro M (2006) Effects of overexpression of basic helix-loop-helix transcription factor Dec1 on osteogenic and adipogenic differentiation of mesenchymal stem cells. Eur J Cell Biol 85:423–431
Chang HC, Kao CH, Chung SY, Chen WC, Aninda LP, Chen YH (2019) Bhlhe40 differentially regulates the function and number of peroxisomes and mitochondria in myogenic cells. Redox Biol 20:321–333
Xiao L, Naganawa T, Obugunde E, Gronowicz G, Ornitz DM, Coffin JD (2004) Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J Biol Chem 279:27743–27752
Tajima K, Takaishi H, Takito J, Tohmonda T, Yoda M, Ota N (2010) Inhibition of STAT1 accelerates bone fracture healing. J Orthopaedic Res 28:937–941
Hesslein DGT, Fretz JA, Xi Y, Nelson T, Zhou S, Lorenzo JA (2009) Ebf1-dependent control of the osteoblast and adipocyte lineages. Bone 44:537–546
Sun D, Zheng X, Chen Y, Jia C, Xu S, Lin C (2015) Enhancement of osteogenesis post-splenectomy does not attenuate bone loss in ovariectomized rats. J Orthop Res 33:1356–1363
Dhanwal DK (2011) Thyroid disorders and bone mineral metabolism. Indian J Endocrinol Metab 15:S107–S112
Meng XH, Tan LJ, Xiao HM, Tang BS, Deng HW (2019) Examining the causal role of leptin in bone mineral density: a Mendelian randomization study. Bone 125:25–29
Levy JR, Murray E, Manolagas S, Olefsky JM (1986) Demonstration of insulin receptors and modulation of alkaline phosphatase activity by insulin in rat osteoblastic cells. Endocrinology 119:1786–1792
Hickman J, McElduff A (1989) Insulin promotes growth of the cultured rat osteosarcoma cell line UMR-106-01: an osteoblast-like cell. Endocrinology 124:701–706
Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, Moorehead RA (2000) The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103:41–50
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48:481–487
Pavlides JM, Zhu Z, Gratten J, McRae AF, Wray NR, Yang J (2016) Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med 8:84
Acknowledgements
We thank all the study subjects for volunteering to participate in the study.
Funding
This study was partially supported by the Health Commission of Hunan Province (202205033867), Natural Science Foundation of Hunan Province (S2022JJMSXM3534), Natural Science Foundation of Changsha (kq2208475), and Changsha Science and Technology Bureau (kq1701016).
Author information
Authors and Affiliations
Contributions
XHM: calculation, manuscript writing. ZL, XDC: manuscript revision. AMD, ZHM: study conception and design, manuscript revision.
Corresponding authors
Ethics declarations
Disclosures
Xiang-He Meng, Zhen Liu, Xiang-Ding Chen, Ai-Min Deng, Zeng-Hui Mao have no conflict of interest to disclose.
Informed Consent
For this type of study, informed consent is not required.
Statement of Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meng, XH., Liu, Z., Chen, XD. et al. Functional Enrichment Analysis Identifying Regulatory Information Associated with Human Fracture. Calcif Tissue Int 113, 286–294 (2023). https://doi.org/10.1007/s00223-023-01108-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00223-023-01108-w