Abstract
A validated liquid chromatography–tandem mass spectrometry method was developed for the simultaneous determination of d- and l-amino acids in human serum. Under the optimum conditions, except for dl-proline, l-glutamine, and d-lysine, the enantioseparation of the other 19 enantiomeric pairs of proteinogenic amino acids and nonchiral glycine was achieved with a CROWNPAK CR-I(+) chiral column within 13 min. The lower limits of quantitation for l-amino acids (including glycine) and d-amino acids were 5–56.25 μM and 0.625–500 nM, respectively, in human serum. The intraday precision and interday precision for all the analytes were less than 15%, and the accuracy ranged from −12.84% to 12.37% at three quality control levels. The proposed method, exhibiting high rapidity, enantioresolution, and sensitivity, was successfully applied to the quantification of d- and l-amino acid levels in serum from hepatocellular carcinoma patients and healthy individuals. The serum concentrations of l-arginine, l-isoleucine, l-aspartate, l-tryptophan, l-alanine, l-methionine, l-serine, glycine, l-valine, l-leucine, l-phenylalanine, l-threonine, d-isoleucine, d-alanine, d-glutamate, d-glutamine, d-methionine, and d-threonine were significantly reduced in the hepatocellular carcinoma patients compared with the healthy individuals (P < 0.01). d-Glutamate and d-glutamine were identified as the most downregulated serum markers (fold change greater than 1.5), which deserves further attention in hepatocellular carcinoma research.

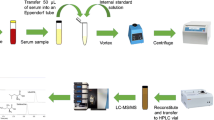
Simultaneous determination of d- and l-amino acids in human serum from hepatocellular carcinoma patients and healthy individuals. AA amino acid, HCC hepatocellular carcinoma, LC liquid chromatography, MS/MS tandem mass spectrometry, NC normal control, TIC total ion chromatogram

Similar content being viewed by others
References
Klupczynska A, Derezinski P, Dyszkiewicz W, Pawlak K, Kasprzyk M, Kokot ZJ. Evaluation of serum amino acid profiles' utility in non-small cell lung cancer detection in Polish population. Lung Cancer. 2016;100:71–6. https://doi.org/10.1016/j.lungcan.2016.04.008.
Mustafa A, Gupta S, Hudes GR, Egleston BL, Uzzo RG, Kruger WD. Serum amino acid levels as a biomarker for renal cell carcinoma. J Urol. 2011;186(4):1206–12. https://doi.org/10.1016/j.juro.2011.05.085.
Fitian AI, Nelson DR, Liu C, Xu Y, Ararat M, Cabrera R. Integrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC/MS and UPLC/MS-MS. Liver Int. 2014;34(9):1428–44. https://doi.org/10.1111/liv.12541.
Hashimoto K, Fukushima T, Shimizu E, Komatsu N, Watanabe H, Shinoda N, et al. Decreased serum levels of D-serine in patients with schizophrenia: evidence in support of the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia. Arch Gen Psychiatry. 2003;60(6):572–6. https://doi.org/10.1001/archpsyc.60.6.572.
Morikawa A, Hamase K, Zaitsu K. Determination of D-alanine in the rat central nervous system and periphery using column-switching high-performance liquid chromatography. Anal Biochem. 2003;312(1):66–72.
Rodriguez-Crespo I. d-Amino acids in the brain: pyridoxal phosphate-dependent amino acid racemases and the physiology of d-serine. FEBS J. 2008;275(14):3513. https://doi.org/10.1111/j.1742-4658.2008.06514.x.
Yamanaka M, Miyoshi Y, Ohide H, Hamase K, Konno R. d-Amino acids in the brain and mutant rodents lacking d-amino-acid oxidase activity. Amino Acids. 2012;43(5):1811–21. https://doi.org/10.1007/s00726-012-1384-x.
Billard JM. d-Amino acids in brain neurotransmission and synaptic plasticity. Amino Acids. 2012;43(5):1851–60. https://doi.org/10.1007/s00726-012-1346-3.
Kiriyama Y, Nochi H. d-Amino acids in the nervous and endocrine systems. Scientifica (Cairo). 2016;2016:6494621. https://doi.org/10.1155/2016/6494621.
Nagata Y, Sato T, Enomoto N, Ishii Y, Sasaki K, Yamada T. High concentrations of D-amino acids in human gastric juice. Amino Acids. 2007;32(1):137–40. https://doi.org/10.1007/s00726-006-0262-9.
Kimura T, Hamase K, Miyoshi Y, Yamamoto R, Yasuda K, Mita M, et al. Chiral amino acid metabolomics for novel biomarker screening in the prognosis of chronic kidney disease. Sci Rep. 2016;6:26137. https://doi.org/10.1038/srep26137.
Lorenzo MP, Dudzik D, Varas E, Gibellini M, Skotnicki M, Zorawski M, et al. Optimization and validation of a chiral GC-MS method for the determination of free D-amino acids ratio in human urine: application to a gestational diabetes mellitus study. J Pharm Biomed Anal. 2015;107:480–7. https://doi.org/10.1016/j.jpba.2015.01.015.
Pietrogrande MC. Enantioselective separation of amino acids as biomarkers indicating life in extraterrestrial environments. Anal Bioanal Chem. 2013;405(25):7931–40. https://doi.org/10.1007/s00216-013-6915-0.
Lorenzo MP, Villasenor A, Ramamoorthy A, Garcia A. Optimization and validation of a capillary electrophoresis laser-induced fluorescence method for amino acids determination in human plasma: application to bipolar disorder study. Electrophoresis. 2013;34(11):1701–9. https://doi.org/10.1002/elps.201200632.
Sanchez-Hernandez L, Serra NS, Marina ML, Crego AL. Enantiomeric separation of free L- and D-amino acids in hydrolyzed protein fertilizers by capillary electrophoresis tandem mass spectrometry. J Agric Food Chem. 2013;61(21):5022–30. https://doi.org/10.1021/jf4013345.
Karakawa S, Shimbo K, Yamada N, Mizukoshi T, Miyano H, Mita M, et al. Simultaneous analysis of D-alanine, D-aspartic acid, and D-serine using chiral high-performance liquid chromatography-tandem mass spectrometry and its application to the rat plasma and tissues. J Pharm Biomed Anal. 2015;115:123–9. https://doi.org/10.1016/j.jpba.2015.05.024.
Sakamoto T, Kuwabara R, Takahashi S, Onozato M, Ichiba H, Iizuka H, et al. Determination of D-serine in human serum by LC-MS/MS using a triazole-bonded column after pre-column derivatization with (S)-4-(3-isothiocyanatopyrrolidin-1-yl)-7- (N,N-dimethylaminosulfonyl)-2,1,3-benzoxadiazole. Anal Bioanal Chem. 2016;408(2):517–26. https://doi.org/10.1007/s00216-015-9119-y.
Song Y, Liang F, Liu YM. Quantification of D-amino acids in the central nervous system of Aplysia californica by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(1):73–7. https://doi.org/10.1002/rcm.2803.
Xie Y, Alexander GM, Schwartzman RJ, Singh N, Torjman MC, Goldberg ME, et al. Development and validation of a sensitive LC-MS/MS method for the determination of D-serine in human plasma. J Pharm Biomed Anal. 2014;89:1–5. https://doi.org/10.1016/j.jpba.2013.10.028.
Sugimoto H, Kakehi M, Jinno F. Bioanalytical method for the simultaneous determination of D- and L-serine in human plasma by LC/MS/MS. Anal Biochem. 2015;487:38–44. https://doi.org/10.1016/j.ab.2015.07.004.
Sugimoto H, Kakehi M, Jinno F. Method development for the determination of D- and L-isomers of leucine in human plasma by high-performance liquid chromatography tandem mass spectrometry and its application to animal plasma samples. Anal Bioanal Chem. 2015;407(26):7889–98. https://doi.org/10.1007/s00216-015-8999-1.
Visser WF, Verhoeven-Duif NM, Ophoff R, Bakker S, Klomp LW, Berger R, et al. A sensitive and simple ultra-high-performance-liquid chromatography-tandem mass spectrometry based method for the quantification of D-amino acids in body fluids. J Chromatogr A. 2011;1218(40):7130–6. https://doi.org/10.1016/j.chroma.2011.07.087.
Szoko E, Vincze I, Tabi T. Chiral separations for d-amino acid analysis in biological samples. J Pharm Biomed Anal. 2016;130:100–9. https://doi.org/10.1016/j.jpba.2016.06.054.
Aviles-Moreno JR, Quesada-Moreno MM, Lopez-Gonzalez JJ, Martinez-Haya B. Chiral recognition of amino acid enantiomers by a crown ether: chiroptical IR-VCD response and computational study. J Phys Chem B. 2013;117(32):9362–70. https://doi.org/10.1021/jp405027s.
Hyun MH. Development of HPLC chiral stationary phases based on (+)-(18-crown-6)-2,3,11,12-tetracarboxylic acid and their applications. Chirality. 2015;27(9):576–88. https://doi.org/10.1002/chir.22484.
Hyun MH. Liquid chromatographic enantioseparations on crown ether-based chiral stationary phases. J Chromatogr A. 2016;1467:19–32. https://doi.org/10.1016/j.chroma.2016.07.049.
Konya Y, Bamba T, Fukusaki E. Extra-facile chiral separation of amino acid enantiomers by LC-TOFMS analysis. J Biosci Bioeng. 2016;121(3):349–53. https://doi.org/10.1016/j.jbiosc.2015.06.017.
Konya Y, Taniguchi M, Fukusaki E. Novel high-throughput and widely-targeted liquid chromatography-time of flight mass spectrometry method for d-amino acids in foods. J Biosci Bioeng. 2017;123(1):126–33. https://doi.org/10.1016/j.jbiosc.2016.07.009.
Nakano Y, Konya Y, Taniguchi M, Fukusaki E. Development of a liquid chromatography-tandem mass spectrometry method for quantitative analysis of trace d-amino acids. J Biosci Bioeng. 2017;123(1):134–8. https://doi.org/10.1016/j.jbiosc.2016.07.008.
McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19(2):223–38. https://doi.org/10.1016/j.cld.2015.01.001.
Stepien M, Duarte-Salles T, Fedirko V, Floegel A, Barupal DK, Rinaldi S, et al. Alteration of amino acid and biogenic amine metabolism in hepatobiliary cancers: Findings from a prospective cohort study. Int J Cancer. 2016;138(2):348–60. https://doi.org/10.1002/ijc.29718.
Sato F, Hatano E, Kitamura K, Myomoto A, Fujiwara T, Takizawa S, et al. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6(1):e16435. https://doi.org/10.1371/journal.pone.0016435.
Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361(9361):923–9. https://doi.org/10.1016/s0140-6736(03)12775-4.
Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–91. https://doi.org/10.1200/jco.2008.20.7753.
Darpolor MM, Basu SS, Worth A, Nelson DS, Clarke-Katzenberg RH, Glickson JD, et al. The aspartate metabolism pathway is differentiable in human hepatocellular carcinoma: transcriptomics and 13C-isotope based metabolomics. NMR Biomed. 2014;27(4):381–9. https://doi.org/10.1002/nbm.3072.
Liu Y, Hong Z, Tan G, Dong X, Yang G, Zhao L, et al. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int J Cancer. 2014;135(3):658–68. https://doi.org/10.1002/ijc.28706.
Smith RJ. Nutrition and metabolism in hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2013;2(2):89–96. https://doi.org/10.3978/j.issn.2304-3881.2012.11.02.
Zhou L, Liao Y, Yin P, Zeng Z, Li J, Lu X, et al. Metabolic profiling study of early and late recurrence of hepatocellular carcinoma based on liquid chromatography-mass spectrometry. J Chromatogr B. 2014;966:163–70. https://doi.org/10.1016/j.jchromb.2014.01.057.
Ye G, Zhu B, Yao Z, Yin P, Lu X, Kong H, et al. Analysis of urinary metabolic signatures of early hepatocellular carcinoma recurrence after surgical removal using gas chromatography-mass spectrometry. J Proteome Res. 2012;11(8):4361–72. https://doi.org/10.1021/pr300502v.
Fages A, Duarte-Salles T, Stepien M, Ferrari P, Fedirko V, Pontoizeau C, et al. Metabolomic profiles of hepatocellular carcinoma in a European prospective cohort. BMC Med. 2015;13:242. https://doi.org/10.1186/s12916-015-0462-9.
Watanabe A, Higashi T, Sakata T, Nagashima H. Serum amino acid levels in patients with hepatocellular carcinoma. Cancer. 1984;54(9):1875–82.
Nojiri S, Fujiwara K, Shinkai N, Iio E, Joh T. Effects of branched-chain amino acid supplementation after radiofrequency ablation for hepatocellular carcinoma: a randomized trial. Nutrition. 2017;33:20–7. https://doi.org/10.1016/j.nut.2016.07.013.
Park JG, Tak WY, Park SY, Kweon YO, Jang SY, Lee YR, et al. Effects of branched-chain amino acids (BCAAs) on the progression of advanced liver disease: a Korean nationwide, multicenter, retrospective, observational, cohort study. Medicine (Baltimore). 2017;96(24):e6580. https://doi.org/10.1097/md.0000000000006580.
Nishizaki T, Matsumata T, Taketomi A, Yamamoto K, Sugimachi K. Levels of amino acids in human hepatocellular carcinoma and adjacent liver tissue. Nutr Cancer. 1995;23(1):85–90. https://doi.org/10.1080/01635589509514364.
Lee JC, Chen MJ, Chang CH, Tiai YF, Lin PW, Lai HS, et al. Plasma amino acid levels in patients with colorectal cancers and liver cirrhosis with hepatocellular carcinoma. Hepatogastroenterology. 2003;50(53):1269–73.
Fuchs SA, Berger R, Klomp LW, de Koning TJ. d-Amino acids in the central nervous system in health and disease. Mol Genet Metab. 2005;85(3):168–80. https://doi.org/10.1016/j.ymgme.2005.03.003.
Wolosker H, Dumin E, Balan L, Foltyn VN. d-Amino acids in the brain: d-serine in neurotransmission and neurodegeneration. FEBS J. 2008;275(14):3514-3526. https://doi.org/10.1111/j.1742-4658.2008.06515.x.
Kollipara S, Bende G, Agarwal N, Varshney B, Paliwal J. International guidelines for bioanalytical method validation: a comparison and discussion on current scenario. Chromatographia. 2011;73(3-4):201–17. https://doi.org/10.1007/s10337-010-1869-2.
Wang X, Zhang A, Han Y, Wang P, Sun H, Song G, et al. Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol Cell Proteomics. 2012;11(8):370–80. https://doi.org/10.1074/mcp.M111.016006.
He X, Zhong J, Wang S, Zhou Y, Wang L, Zhang Y, et al. Serum metabolomics differentiating pancreatic cancer from new-onset diabetes. Oncotarget. 2017;8(17):29116–24. https://doi.org/10.18632/oncotarget.16249.
Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(5):8775–84. https://doi.org/10.18632/oncotarget.14452.
Sakamoto M. Early HCC: diagnosis and molecular markers. J Gastroenterol. 2009;44(Suppl 19):108–11. https://doi.org/10.1007/s00535-008-2245-y.
Long J, Lang Z-W, Wang H-G, Wang T-L, Wang B-E, Liu S-Q. Glutamine synthetase as an early marker for hepatocellular carcinoma based on proteomic analysis of resected small hepatocellular carcinomas. Hepatobiliary Pancreat Dis Int. 2010;9(3):296–305.
Acknowledgements
We gratefully thank the National Natural Science Foundation of China (no. 81101027), the Innovation Program of Shanghai Municipal Education Commission (no. 14YZ030), and the Shanghai Young Eastern Scholar Program (QD2015009) for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was approved by the ethics committees of Eastern Hepatobiliary Surgery Hospital and Changhai Hospital and was conducted in accordance with the guidelines of the World Medical Association’s Declaration of Helsinki. Informed consent was obtained from the individual participants who provided the blood samples.
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Han, M., Xie, M., Han, J. et al. Development and validation of a rapid, selective, and sensitive LC–MS/MS method for simultaneous determination of d- and l-amino acids in human serum: application to the study of hepatocellular carcinoma. Anal Bioanal Chem 410, 2517–2531 (2018). https://doi.org/10.1007/s00216-018-0883-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-0883-3