Abstract
Currently, it is extremely important to identify and describe new alternative compounds with potential antimicrobial properties. Since various natural biological systems are capable of producing active compounds with such properties, many of them have been the subject of intensive study. The aim of this work was to heterologously overexpress, purify and preliminarily investigate the antimicrobial activity of a novel bacteriocin found in Salmonella species. Overexpressed protein shows an amino acid structure homologous to the well-known colicin M and was never expressed previously in the E. coli platform. Purified salmocin M showed an inhibition spectrum against Salmonella and E. coli strains. To determine its potential as an antimicrobial agent for use in medicine or the food industry, preliminary antimicrobial tests against pathogenic bacteria were carried out. Our research demonstrates that bacteriocin can be produced efficiently in bacterial expression systems, which are one of the cheapest and the most popular platforms for recombinant protein production. Moreover, preliminary results of microbiological tests showed its activity against most of the bacterial strains in a dose-dependent manner.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As predicted by experts, bacterial resistance to antibiotics in the coming decades will be a deepening global problem (Laxminarayan et al. 2020). Especially in food production, inappropriate antibiotic usage is linked with the development of resistant bacteria. Measures to counteract this phenomenon are currently taking place on several fronts, including legal restrictions on the use of antibiotics in animal breeding, as well as by searching for alternative antibacterial compounds (Lagadinou et al. 2020). The challenging issue of food safety is still a problem of concern to the scientific community all over the world. The European Commission Action Plan on Antimicrobial Resistance places great emphasis on developing new antimicrobials or alternatives to antibiotics, the European Medicines Agency encourages an increased level of innovation on treatment alternatives for infectious diseases, and a similar situation also takes place in the United States and other countries (Anderson et al. 2020; Mader et al. 2021). Moreover, scientists predict that the COVID-19 pandemic might even increase the rate of bacterial resistance to antibiotics and by 2050 antimicrobial resistance may be responsible for 10 million deaths per year (Strathdee et al. 2020). A large group of microbial infections consists of foodborne illnesses and among them, Salmonella is the most common cause of outbreaks in Europe as reported by European Food Safety Authority and European Centre for Disease Prevention and Control (2019). In the United States, Salmonella causes an estimated 1.2 million illnesses each year, resulting in an estimated 20,000 hospitalizations and 450 deaths reported by Centre for Disease Prevention and Control (2017). Salmonella and Escherichia coli gain resistance when exposed to the constant selective pressure of antibiotics, which are increasingly used in the treatment of livestock and then passed on with feces, water, soil, animal products, and even vegetables (Peng et al. 2018; Micallef et al. 2012). Currently, researchers are reporting that Salmonella with clinically important resistance caused 29% of outbreaks from land animals and 8% of outbreaks from plant products (Brown et al. 2017). Taking all this into account, there is an urgent need to search for novel antimicrobial agents and build up innovative strategies to combat antibiotic-resistant Salmonella causing difficult infections (Nair et al. 2018; Zhang et al. 2018; Nadi et al. 2020).
Nowadays, bacteriocins are one of the most widely described antibacterial alternatives, especially intended for the food industry (Silva et al. 2018; Juturu and Wu 2018; Sang and Blecha 2008; Cotter et al. 2013). Those ribosomal synthesized proteins are used by bacteria to compete against other closely related strains. Bacteriocins have useful features, such as the ability to kill pathogenic bacteria quickly and efficiently. They often act in pico- or nanomolar concentrations. Activity in a wide pH range and tolerance of high thermal stress are other advantages of bacteriocins (Cleveland et al. 2001). Due to their numerous beneficial qualities, they are increasingly used as innovative antimicrobial agents. Moreover, bacteriocins have a narrower spectrum of activity in comparison to antibiotics. They usually only act on a few genera or species closely related to their producer. This fact can be both an advantage and a disadvantage of their potential use. Bacteriocin activity can be very specific, which means that potential therapy would not cause a risk of microbial imbalance as in the case of antibiotic therapy. However, different bacteriocins are needed for the treatment of every group of pathogenic bacteria. Obtaining bacteriocins from strains with potential pathogenicity most often requires the use of genetic engineering tools, which involve a transfer of the bacteriocin-coding gene to a non-pathogenic strain (Arthur et al. 2014). The current results of research on bacteriocins are promising, although further analyses are needed to fully understand their huge diversity, mode of action or cytotoxicity. Therefore, research in this field will be crucial for the continued use of bacteriocins as innovative antibacterial drugs. The colicin group of bacteriocins, produced by Escherichia coli, is widely known for its antibacterial and anticancer properties (Kaur & Kaur 2015; Chumchalova et al. 2003). Colicin M is the only bacteriocin that can enzymatically degrade the peptidoglycan lipid II intermediate. Therefore, it is often described as an attractive selective antibacterial agent (Touzé et al. 2012). Colicin M-like bacteriocins were also discovered and described in Pseudomonas syringae pv. tomato strain and called syringacin M (Grinter et al. 2012), in Burkholderia such homologs were called burkhocins M1 and M2 (Ghequire and De Mot 2015), and the colicin M-like bacteriocin pectocin M2 was identified in Pectobacterium carotovorum (Grinter et al. 2014). These proteins are characterized by strong homology of their C-terminus domain to the active antibacterial domain of colicin M. They also have an antibacterial effect similar to colicin M, but extended by activity against a group of closely related bacteria: syringacin M—against Pseudomonas, burkhocin M—against strains of Burkholderia, pectocin M against Pectobacterium strains.
So far very few researchers have addressed the issue of Salmonella bacteriocins (Patankar and Joshi 1985). Previous work has only focused on Salmonella-derived bacteriocins expressed in plant-based systems and named salmocins, and their activity was also well proven in the literature (Schneider et al. 2018; Hahn-Löbmann et al. 2019). What is more, Generally Recognized As Safe (GRAS) notice for salmocins has recently been published by the FDA (Food and Drug Administration) (also for salmocin M) (GRN 824, https://www.cfsanappsexternal.fda.gov/scripts/fdcc/?set=GRASNotices&id=824&sort=GRN_No&order=DESC&startrow=1&type=basic&search=824). This paper outlines a new approach to salmocin M production, opening a faster path for manufacturing this bacteriocin.
The discovery of new effective molecules that inhibit the growth of zoonotic bacteria can help to replace at least some of the antibiotics currently in use, e.g., in agriculture. However, production systems have to be highly practical and optimized for the scaling-up process. Currently, bacteria are one of the most cost-effective, fast, and easy to scale-up recombinant protein production platforms and that fact is especially important in the case of their potential application and commercialization in agriculture and animal production (Gifre et al. 2017).
Materials and methods
Bioinformatic analysis
The search for colicin M homologs in databases was made using the BLAST program (Fig. 1). These analyses showed the presence of homologs in Salmonella genomes. The chosen protein was characterized in BLAST Global Alignment by 44% (119/273) of identities and 64% (177/273) of positives and only 2% of gaps in comparison with colicin M (cma gene). It is encoded by the AO411_2025940 gene of Salmonella enterica I (GeneBank accession number: LKLB02000036.1—Table 1). Homology analysis of colicin M and two colicin M-like proteins from Salmonella revealed a similarity in the amino acid structure (Fig. 1). The chosen protein amino acid sequence is as follows: MTDTITVVAPVPPSGSSLAGNYTASTMSSGNRISSGPTFLQFAYPYYQSPQLAVNCAKWILDFVESHDMKNANNQQIFSENVGQFCFADKNLVNYPAMKVLDAFGGDRKFIYSQDQISRLSGDVTTPITAWAHFLWGDGAARTVNLTDVGLRIQPNQISPVMDLVKGGAVGTFPVNAKFTRDTMLDGIIPASYLGNITLQTTGTLTINSLGAWSYDGVVKAYNDTYDANPSTHRGLLGEYSTSVLRHFSGTPYEIQMPGMIPVKGNGMRLVPRGSHHHHHH*.
Construction of expression vector
The DNA sequence encoding the tested protein (Table 1) was optimized with the host codon usage pattern for E. coli bias (EMBOSS Transeq Sequence Translation) to increase protein yield and then commercially synthesized (Biomatik, Cambridge, Ontario, Canada). Moreover, the addition of His-tag to the C-terminus of protein (with additional thrombin cleavage site) facilitated the purification process by Immobilized Metal Affinity Chromatography (IMAC). The salmocin-coding sequence was introduced into the pT7-MAT-2 expression vector (Sigma-Aldrich) between HindIII and KpnI restriction sites under the control of a T7 phage promoter (Fig. 2). To confirm the correct construction of the vector, a ligation mixture was used to transform chemically competent E. coli DH5-Alpha cells by heat shock (Froger and Hall 2007). Recombinant plasmids were isolated from cells using the Plasmid Mini kit (A&A Biotechnology, Poland), according to the manufacturer’s instructions. The correctness of the recombinant plasmid construction was confirmed by restriction digestion (HindIII and KpnI, data not published) and PCR with primers complementary to the AO411_2025940 cloned gene (primer F 5′-ATGAAGCTTATGACCGATACCATTACCG-3′, primer R 5′-TCAGTGATGGTGATGATGATGAGATCCC-3′).
PCR reaction
PCR was carried out in a Biometra UNOII thermocycler (Biometra, Germany) using a PCR Mix Plus Green (A&A Biotechnology, Poland), by following the instructions provided by the manufacturer. The reaction was carried out in 25 µl and the reaction mixture contained: 12.5 µl PCR Mix Plus Green, 200 nM of F and R primers (2 × 2 µl), 1 µl of template DNA and 7.5 µl of ddH2O. The cycling conditions consisted of 4 min of initial denaturation at 95 °C, followed by 30 cycles: 30 s denaturation at 95 °C, 30 s annealing at 55 °C, and a 45 s extension period at 72 °C with a final 5 min extension at 72 °C. The non-recombinant plasmid pT7-MAT-2 was used as a negative control. The amplification products were visualized after electrophoresis on a 1.5% agarose gel and an ethidium bromide staining procedure.
E. coli transformation
The pT7-MAT-2-salmocinM expression vector was used to transform chemically competent E. coli strain Rosetta (ƛDE3) cells (Novagen) by the heat shock method. Bacteria were then dispersed on Luria–Bertani (LB) agar plates supplemented with 50 mg/L ampicillin and 34 mg/L chloramphenicol and were incubated overnight at 37 °C. Single colonies were picked and cultured overnight on an LB liquid medium supplemented with 50 mg/L of ampicillin with agitation at 37 °C. The presence of a recombinant plasmid in the transgenic E. coli Rosetta cells was finally confirmed by colony PCR using a pair of AO411_2025940 gene-specific primers analogous to PCR performed on an isolated plasmid (data not shown).
Protein expression and purification
E. coli Rosetta transformed with a pT7-MAT-2-salmocinM expression vector were incubated in 5 ml liquid LB (Luria Bertani medium—10 g/L of tryptone, 5 g/L of yeast extract, and 10 g/L of NaCl) for 12 h at 37 °C, 180 rpm in the presence of 50 mg/L ampicillin and 34 mg/L chloramphenicol. 5 flasks of 100 mL TB medium (24 g/L of yeast extract, 20 g/L of tryptone, 4 ml/L of glycerol, 0.017 M KH2PO4, 0.072 M K2HPO4) were inoculated with 2 ml of the overnight culture (1:100) and grown at 37 °C, 150 rpm until reaching OD600nm = 0.6. At this point, induction was made by the addition of isopropyl β-d-1 thiogalactopyranoside (IPTG, 0.5 mM) and incubated at 37 °C, 150 rpm for 4 h. After this time, cells were harvested by centrifugation (5000 rpm for 10 min at 4 °C) and lysed by sonication in ice (Amplitude 36%, Bandelin Sonoplus) with the addition of a lysis buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl) and a 1 mM protease inhibitor—phenyl methyl sulfonyl fluoride (PMSF, Sigma). The soluble fractions of the lysed cells were collected by centrifugation (12,000 rpm for 20 min at 4 °C). The recombinant protein was purified by Immobilized Metal Affinity Chromatography on HIS-Select Nickel Affinity Gel (Sigma-Aldrich). The protein solution was loaded onto an affinity column. The column was washed with a buffer containing 20 mM Imidazole, 20 mM Tris–HCl, 200 mM NaCl. Salmocin M was eluted with 10 ml of an elution buffer containing 150 mM imidazole and finally resuspended in a sterile PBS buffer. The purified recombinant protein was filtered through columns (PD-10 columns Sephadex™ G-25, GE Healthcare) to remove possible bacterial and salt contamination and subsequently it was concentrated via column (VivaspinTM 2 MWCO 30000) up to a concentration of 500 µg/ml. The initial evaluation of the recombinant protein was conducted using SDS-PAGE and compared to the control (proteins of sonicated E. coli Rosetta cells without salmocin M expression cassette). SDS-PAGE electrophoresis assessed the effects of purification, and then the protein concentration was evaluated using a modified Bradford method (Pande and Murphy 1994).
SDS-PAGE
All samples were separated using a 12% SDS-PAGE gel. After that, the proteins were stained with colloidal Coomassie Brilliant Blue G-250 (Dyballa and Metzger 2009).
Microbiological studies
Antibacterial activity was evaluated on a set of reference Escherichia coli strains (DH5-α, NM522) and Salmonella strains (S. Enteritidis D ATTC13076, S. Paratyphi A ATCC 19150, S. Typhimurium ATCC 13311, S. Gallinarum, S. Hadar, S. Virchow) spread on a Mueller–Hinton medium agar plate. The recombinant salmocin M used for this study was dissolved in 1 × PBS. Concentrations of the salmocin M tested in a solid medium ranged from 500 to 0.24 µg/ml. The final inoculum of all the organisms studied was 1.5 × 108 CFU/mL (0.5 McFarland standard). Mueller–Hinton agar plates (150 mm) were streaked with a suspension to cover the entire surface of the plates. After the surface of the inoculated plates had dried, 10-µl drops of various concentrations (ranging from 500 to 0.24 μg/ml) of salmocin M were plated on the surface of the agar, and the plate was incubated at 37 °C for 18 h (for the control, a drop (10 µl) of 1 × PBS was used). The diameter of the growth inhibition zone was read after 18 h of incubation at 37 °C. The experiment was performed in 3 repetitions.
Results
The PCR results confirmed the presence of the insert in the constructed expression cassette. All samples of purified plasmid (and colony PCR, data not shown) that were tested confirmed the presence of the salmocin M gene by the presence of 855 bp amplicons (Fig. 3). In addition, the correctness of the construct was confirmed by restriction hydrolysis using the enzymes HindIII and KpnI, obtaining DNA fragments consistent with those expected (856 bp and 4784 bp, data not shown).
Salmocin M expression and purification
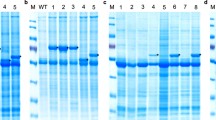
Production of recombinant Salmocin M has been observed in transformed E. coli, unlike untransformed cells. In all samples, SDS-PAGE analysis indicated the presence of an approximately 30 kDa band in the soluble protein samples (Fig. 4).
SDS-PAGE electrophoresis of proteins isolated from transgenic and non-transgenic bacterial cells. 1, 2, 3–soluble proteins of non-transgenic E. coli cells after 4 h, 3 h, 2 h, respectively from IPTG induction; 4, 5, 6—soluble proteins of transgenic E. coli cells after 4 h, 3 h, 2 h, respectively from IPTG induction 7-molecular weight marker. The arrow indicates recombinant protein salmocin M overexpressed in transgenic bacterial cells
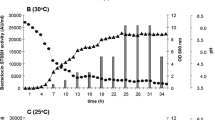
The maximal expression yield was obtained at 37 °C for 4 h after the induction (Fig. 4) without significantly affecting bacterial growth (Fig. 5). Therefore, these conditions were used for protein production. Almost all recombinant salmocin M appeared in the soluble fraction. Out of the 500 ml liquid culture of E. coli expressing pT7-MAT-2-salmocin M, 0.25 mg of salmocin M was obtained.
Our results showed that the applied recombinant protein purification strategy was effective as evidenced by the SDS-PAGE of purified protein samples (Fig. 6).
Antimicrobial activity screening
Purified recombinant salmocin M was obtained in its active form. The protein proved to have antimicrobial activity on all bacteria tested, as is illustrated in Fig. 7 Bacteriocin activity was concentration dependent although for two Escherichia coli the minimal active concentration was 250 µg/ml, while the majority of Salmonella strains were more susceptible to the protein studied. Table 2 summarizes the data on the concentration ranges that lead to the growth inhibition of bacteria on the plate. The recombinant protein was active in the concentration starting from 0.24 µg/ml for Salmonella Typhimurium, which was the most susceptible strain of all those tested, as shown in Fig. 8. Generally, Salmonella strains are more susceptible to low concentrations than the Escherichia coli strains tested, which is a promising result.
Discussion
Colicin M-like bacteriocins have gained considerable interest for their potential use as food preservatives in recent years. Given the great need to obtain new compounds with antimicrobial activity, many studies are currently being conducted to identify and use in practice such compounds from natural sources. Due to their medical and preservative properties, bacteriocins are being isolated from different organisms. However, the majority of them is derived from lactic acid bacteria (Mokoena 2017; da Costa et al. 2019; Barbosa et al. 2017; Field et al. 2018), and according to research, they can act even as growth promoters (Vieco-Saiz et al. 2019). However, different species are also considered interesting sources, for example, Bacillus (Abriouel et al. 2011), Enterococcus (Chang et al. 2013), Pediococcus (Anastasiadou et al. 2008), Gluconacetobacter (Oliveira et al. 2018), Carnobacteria (Acedo et al. 2017) and other genera (Masuda et al. 2011; Masuda et al. 2012; Marques-Bastos et al. 2020). The possibility of using advanced genetic engineering tools allows the application of various expression systems to avoid virulence factors of hosts. In our previous work, we demonstrated the possibility of colicin M biosynthesis in transgenic plants (Łojewska et al. 2020). However, due to well-developed and fast manipulation methods, E. coli can act also as an economic and efficient host for bacteriocin production with a long history of such studies (Richard et al. 2004; Tang et al. 2018; Tapia et al. 2011; Chen et al. 2012). In this study, the structural gene for salmocin M was identified from Salmonella enterica I, cloned into an expression vector for E. coli, overexpression of this recombinant protein was obtained, and then the isolated and purified protein was used for preliminary evaluation of antibacterial activity against E. coli and Salmonella. In this case, due to the speed of obtaining proteins for our preliminary antimicrobial research, as well as the high efficiency of the bacterial system, we decided to produce salmocin M in a non-pathogenic Escherichia coli Rossetta (ƛDE3) host. In this work, for the first time, salmocin M was overexpressed and purified successfully in E. coli cells. Our preliminary results confirmed the successful pT7-MAT-2 expression vector recombination resulting in the construction of an expression cassette containing the salmocin M gene under the control of the T7 promoter. Both PCR and SDS-PAGE analysis confirmed the presence of the gene in the recombinant vector and the expression of the recombinant protein in E. coli cells, respectively. Generally, it is sometimes difficult to produce antimicrobial recombinant proteins in prokaryotic expression platforms. Such proteins are potentially susceptible to proteolytic degradation and, most importantly, toxic to the host cells. However, those inconveniences can be avoided by using different techniques of optimization, starting from the expressed protein (e.g. addition of tags and/or other fusion proteins) to selecting specific conditions of culture and cell harvesting or expression induction strategy (Saida et al. 2006; Wanmakok et al. 2018). In line with the selected recommendations of many authors cited in this work, we shortened the cultivation time after induction by IPTG to obtain the optimal yield of undegraded recombinant protein. Additionally, purification of bacteriocin was achieved with Immobilized Metal Affinity Chromatography on HIS-Select Nickel Affinity Gel using a His-tag. Our preliminary data showed that protein retained its antibacterial activity despite the presence of His-tag and thrombin cleavage site allowing for His-tag removal after the purification. Our results are consistent with those of Ghachi et al., who showed that the attachment of a His-tag to colicin M did not affect its activity (El Ghachi et al. 2006).
Due to the fact that the aim of this study was the expression and only preliminary examination of the antibacterial properties of the identified protein, the protein production efficiency in this system was not optimized in detail.
The results of our preliminary antibacterial studies showed that the protein being tested was active against S. Virchow, S. Hadar, S. Typhimurium and S. Enteritidis, which are the main cause of salmonellosis, one of the most common food poisoning, possibly leading to sepsis and death. Moreover, recombinant protein was also active against S. Paratyphi, bacteria causing paratyphoid fever, a disease currently cured with antibiotics, although S. Paratyphi becomes increasingly resistant to them. Salmocin M had the highest effectiveness against Salmonella strains, among which the strongest effect was obtained for S. Typhimurium (0.24 µg/ml) and the weakest for S. Enteritidis D ATTC13076 (500 µg/ml). Among the two E. coli strains tested, recombinant salmocin M showed a similar activity at the level of 250 µg/ml. Hahn-Löbmann et al. (2019) also showed that the tested salmocins SalE1a and SalE1b revealed the strongest effect on S. Typhimurium. Contrary to our results, the salmocins studied by these authors also showed strong activity against S. Enteritidis.
Bacteriocins are natural, non-antibiotic proteins with antibacterial properties with an extremely wide potential application from medicine to the broadly understood industry. The possibility of using them to reduce or eliminate pathogenic bacteria opens up the possibility of enriching the antimicrobial arsenal alternative to commonly used and often overused antibiotics.
These results allow the assumptions about the antibacterial activity of Salmonella bacteriocins to be confirmed, but further research is necessary to investigate the mechanisms of its action, which in the future may allow for the development of a new effective alternative to combat these pathogenic bacteria.
References
Abriouel H, Franz CM, Ben Omar N, Gálvez A (2011) Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev 35:201–232
Acedo JZ, Towle KM, Lohans CT, Miskolzie M, McKay RT, Doerksen TA, Vederas JC, Martin-Visscher LA (2017) Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria. FEBS Lett 591:1349–1359
Anastasiadou S, Papagianni M, Filiousis G, Ambrosiadis I, Koidis P (2008) Pediocin SA-1, an antimicrobial peptide from Pediococcus acidilactici NRRL B5627: production conditions, purification and characterization. Bioresour Technol 99:5384–5390
Anderson M, Schulze K, Cassini A, Plauchoras D, Mossialos E (2020) Strengthening implementation of antimicrobial resistance national action plans. Eur J Public Health. 30(Supplement_5): ckaa165–1200. https://apps.who.int/iris/handle/10665/332442
Arthur TD, Cavera VL, Chikindas ML (2014) On bacteriocin delivery systems and potential applications. Future Microbiol 9(2):235–248. https://doi.org/10.2217/fmb.13.148
Barbosa AAT, Mantovani HC, Jain S (2017) Bacteriocins from lactic acid bacteria and their potential in the preservation of fruit products. Crit Rev Biotechnol 37(7):852–864
Brown AC, Grass JE, Richardson LC, Nisler AL, Bicknese AS, Gould LH (2017) Antimicrobial resistance in Salmonella that caused foodborne disease outbreaks: United States, 2003–2012. Epidemiol Infect 145(4):766–774. https://doi.org/10.1017/S0950268816002867
Centers for Disease Control and Prevention (2017) Estimated annual number of hospitalizations and deaths caused by 31 pathogens transmitted commonly by food. https://www.cdc.gov/salmonella/index.html. Accessed 20 Apr 2021
Chang SY, Chen YS, Pan SF, Lee YS, Chang CH, Chang CH, Wu HC (2013) Enterocin TW21, a novel bacteriocin from dochi-isolated Enterococcus faecium D081821. J Appl Microbiol 115:673–678
Chen H, Tian F, Li S, Xie Y, Zhang H, Chen W (2012) Cloning and heterologous expression of a bacteriocin sakacin P from Lactobacillus sakei in Escherichia coli. Appl Microbiol Biotechnol 94(2012):1061–1068
Chumchalova J, Šmarda J (2003) Human tumor cells are selectively inhibited by colicins. Folia Microbiol 48(1):111–115. https://doi.org/10.1007/BF02931286
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71(1):1–20. https://doi.org/10.1016/S0168-1605(01)00560-8
Cotter PD, Ross RP, Hill C (2013) Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11(2):95–105. https://doi.org/10.1038/nrmicro2937
da Costa RJ, Voloski FL, Mondadori RG, Duval EH, Fiorentini ÂM (2019) Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. J Food Qual 2019:1–12
Dyballa N, Metzger S (2009) Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J vis Exp 30:e1431. https://doi.org/10.3791/1431
El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem. 281(32):22761–22772 https://doi.org/10.1074/jbc.M602834200
European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) (2019) The European Union one health 2018 zoonoses report. EFSA J 17(12):e05926. https://doi.org/10.2903/j.efsa.2019.5926
Froger A, Hall JE (2007) Transformation of plasmid DNA into E. coli using the heat shock method. J vis Exp 6:253. https://doi.org/10.3791/253
Field D, Ross RP, Hill C (2018) Developing bacteriocins of lactic acid bacteria into next generation biopreservatives. Curr Opin Food Sci 20:1–6
Ghequire MG, De Mot R (2015) Distinct colicin M-like bacteriocin-immunity pairs in Burkholderia. Sci Rep 5(1):1–9. https://doi.org/10.1038/srep17368
Gifre L, Arís A, Bach À, Garcia-Fruitós E (2017) Trends in recombinant protein use in animal production. Microb Cell Fact 16(1):1–17. https://doi.org/10.1186/s12934-017-0654-4
Grinter R, Roszak AW, Cogdell RJ, Milner JJ, Walker D (2012) The crystal structure of the lipid II-degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M-like bacteriocins. J Biol Chem 287(46):38876–38888. https://doi.org/10.1074/jbc.M112.400150
Grinter R, Josts I, Zeth K, Roszak AW, McCaughey LC, Cogdell RJ, Walker D (2014) Structure of the atypical bacteriocin pectocin M 2 implies a novel mechanism of protein uptake. Mol Microbiol 93(2):234–246. https://doi.org/10.1111/mmi.12655
Hahn-Löbmann S, Stephan A, Schulz S, Schneider T, Shaverskyi A, Tusé D, Gleba Y (2019) Colicins and Salmocins–new classes of plant-made non-antibiotic food antibacterials. Front Plant Sci 10:437. https://doi.org/10.3389/fpls.2019.00437
Juturu V, Wu JC (2018) Microbial production of bacteriocins: latest research development and applications. Biotechnol Adv 36(8):2187–2200. https://doi.org/10.1016/j.biotechadv.2018.10.007
Kaur S, Kaur S (2015) Bacteriocins as potential anticancer agents. Front Pharmacol 6:272. https://doi.org/10.3389/fphar.2015.00272
Lagadinou M, Onisor MO, Rigas A, Musetescu DV, Gkentzi D, Assimakopoulos SF, Marangos M (2020) Antimicrobial properties on non-antibiotic drugs in the era of increased bacterial resistance. Antibiotics 9(3):107. https://doi.org/10.3390/antibiotics9030107
Laxminarayan R, Van Boeckel T, Frost I, Kariuki S, Khan EA, Limmathurotsakul D, Zhu YG (2020) The Lancet Infectious Diseases Commission on antimicrobial resistance: 6 years later. Lancet Infect Dis 20(4):e51–e60. https://doi.org/10.1016/S1473-3099(20)30003-7
Łojewska E, Sakowicz T, Kowalczyk A, Konieczka M, Grzegorczyk J, Sitarek P, Kowalczyk T (2020) Production of recombinant colicin M in Nicotiana tabacum plants and its antimicrobial activity. Plant Biotechnol Rep 14(1):33–43
Mader R, Damborg P, Amat JP, Bengtsson B, Bourély C, Broens EM, Madec JY (2021) Building the European Antimicrobial Resistance Surveillance network in veterinary medicine (EARS-Vet). Eurosurveillance 26(4):2001359. https://doi.org/10.2807/1560-7917.ES.2021.26.4.2001359
Marques-Bastos SLS, Coelho MLV, Ceotto-Vigoder H, Fagundes PC, Almeida GS, Brede DA, de Freire Bastos MDC (2020) Molecular characterization of aureocin 4181: a natural N-formylated aureocin A70 variant with a broad spectrum of activity. Braz J Microbiol 51(4):1527–1538
Masuda Y, Ono H, Kitagawa H, Ito H, Mu F, Sawa N, Zendo T, Sonomoto K (2011) Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl Environ Microbiol 77:8164–8170
Masuda Y, Zendo T, Sawa N, Perez RH, Nakayama J, Sonomoto K (2012) Characterization and identification of weissellicin Y and weissellicin M, novel bacteriocins produced by Weissella hellenica QU 13. J Appl Microbiol 112:99–108
Micallef SA, Goldstein RER, George A, Kleinfelter L, Boyer MS, McLaughlin CR, Sapkota AR (2012) Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ Res 114:31–39. https://doi.org/10.1016/j.envres.2012.02.005
Mokoena MP (2017) Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 22(8):1255
Nadi ZR, Salehi TZ, Tamai IA, Foroushani AR, Sillanpaa M, Dallal MMS (2020) Evaluation of antibiotic resistance and prevalence of common Salmonella enterica serovars isolated from foodborne outbreaks. Microchem J 155:104660. https://doi.org/10.1016/j.microc.2020.104660
Nair D, Venkitanarayanan K, Kollanoor JA (2018) Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 7(10):167. https://doi.org/10.3390/foods7100167
Oliveira MM, Ramos ETA, Drechsel MM, Vidal MS, Schwab S, Baldani JI (2018) Gluconacin from Gluconacetobacter diazotrophicus PAL5 is an active bacteriocin against phytopathogenic and beneficial sugarcane bacteria. J Appl Microbiol 125(6):1812–1826
Pande SV, Murthy MS (1994) A modified micro-Bradford procedure for elimination of interference from sodium dodecyl sulfate, other detergents, and lipids. Anal Biochem 220(2):424–426. https://doi.org/10.1006/abio.1994.1361
Patankar CV, Joshi LM (1985) Bacteriocin production in Salmonella. J Postgrad Med 31(1):46
Peng M, Salaheen S, Buchanan RL, Biswas D (2018) Alterations of Salmonella enterica serovar typhimurium antibiotic resistance under environmental pressure. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01173-18
Richard C, Drider D, Elmorjani K, Marion D, Prévost H (2004) Heterologous expression and purification of active divercin V41, a class IIa bacteriocin encoded by a synthetic gene in Escherichia coli. J Bacteriol 186(13):4276–4284
Saida F, Uzan M, Odaert B, Bontems F (2006) Expression of highly toxic genes in E. coli: special strategies and genetic tools. Curr Protein Pept Sci 7(1):47–56
Sang Y, Blecha F (2008) Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Anim Health Res Rev 9(2):227–235. https://doi.org/10.1017/S1466252308001497
Schneider T, Hahn-Löbmann S, Stephan A, Schulz S, Giritch A, Naumann M, Gleba Y (2018) Plant-made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci Rep 8(1):1–10. https://doi.org/10.1038/s41598-018-22465-9
Silva CC, Silva SP, Ribeiro SC (2018) Application of bacteriocins and protective cultures in dairy food preservation. Front Microbiol 9:594. https://doi.org/10.3389/fmicb.2018.00594
Strathdee SA, Davies SC, Marcelin JR (2020) Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet 396(10257):1050–1053. https://doi.org/10.1016/S0140-6736(20)32063-8
Tang X, Wu S, Wang X, Gu Q, Li P (2018) Antimicrobial activity and preliminary mode of action of PlnEF expressed in Escherichia coli against Staphylococci. Protein Expr Purif 143:28–33
Tapia E, Montes C, Rebufel P, Paradela A, Prieto H, Arenas G (2011) Expression of an optimized Argopecten purpuratus antimicrobial peptide in E. coli and evaluation of the purified recombinant protein by in vitro challenges against important plant fungi. Peptides 32(9):1909–1916
Touzé T, Barreteau H, El Ghachi M, Bouhss A, Barnéoud-Arnoulet A, Patin D, Mengin-Lecreulx D (2012) Colicin M, a peptidoglycan lipid-II-degrading enzyme: potential use for antibacterial means? Biochem Soc Trans 40(6):1522–1527. https://doi.org/10.1042/BST20120189
Vieco-Saiz N, Belguesmia Y, Raspoet R, Auclair E, Gancel F, Kempf I, Drider D (2019) Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front Microbiol 10:57
Wanmakok M, Orrapin S, Intorasoot A, Intorasoot S (2018) Expression in Escherichia coli of novel recombinant hybrid antimicrobial peptide AL32-P113 with enhanced antimicrobial activity in vitro. Gene 671:1–9
Zhang L, Fu Y, Xiong Z, Ma Y, Wei Y, Qu X, Liao M (2018) Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong. China Front Microbiol 9:2104. https://doi.org/10.3389/fmicb.2018.02104
Acknowledgements
No financial conflict of interest exists in relation to the work described. Authors certify that the above information is true and correct. All the authors contributed to the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Łojewska, E., Sakowicz, T., Korycka-Machała, M. et al. Heterologous overexpression and preliminary antimicrobial activity test of salmocin M, a novel colicin M-like bacteriocin against Salmonella sp.. Arch Microbiol 204, 154 (2022). https://doi.org/10.1007/s00203-021-02659-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-021-02659-y