Abstract
Antibiotic-resistant microorganisms causing a life-threatening infection pose a serious challenge for modern science. The rapidly growing number of incidents for which the use of standard antibiotics is ineffective forces us to develop new alternative methods of killing microorganisms. Antimicrobial proteins and peptides (AMPs) can be promising candidates to solve this problem. Colicin-M is one of the representatives of this group and is naturally produced by Escherichia coli acting on other closely related bacterial strains by disrupting their outer cell membrane. This bacteriocin has huge potential as a potent antimicrobial agent, especially, since it was recognized by the FDA as safe. In this work, we present the expression of colicin M in model transgenic Nicotiana tabacum L. plants. We demonstrate that purified colicin retains its antibacterial activity against the control Escherichia coli strains and clinical isolates of Escherichia coli and Klebsiella pneumoniae. Our results also show that plant-derived ColM is not toxic for L929 and HeLa cell line, which allows us to suppose that plant-based expression could be an alternative production method of such important proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the twentieth century, antibiotics fundamentally changed the way of dealing with bacterial infections. Soon after Alexander Fleming's discovery of penicillin in 1928, antibiotics quickly dominated the pharmaceuticals market. However, together with the beginning of the twenty-first century, voices reporting the end of the antibiotics era became more prevalent (Kåhrström 2013; Fowler et al. 2014). Currently, pathogens resistant to a number of antibiotics are known to lead to serious health complications, including death (World Health Organization 2014). Outbreaks of pan-resistant bacteria have started to be reported around the globe (Molton et al. 2013; Jones et al. 2017). Alarmingly, the gene of resistance to colistin, a last-resort antibiotic against resistant E. coli, has been found both in meat samples and in human infections, not only in China but also in India, and is reported to move freely between bacteria (Liu et al. 2016; Paveenkittiporn et al. 2017; Chen et al. 2017). The situation is difficult given the fact that a similar problem occurred in case of the fast spread of resistance to carbapenems, a previous last-resort antibiotic, which now are reported to be not active in 50% of Enterobacteriaceae infection cases (Nordmann et al. 2011).
Antibiotics have played an important role in livestock production, although their overuse was, and in many cases still is, one of the major risks for antibiotic resistance developing in bacteria. Excessive exploitation of antibiotics in farms led to the contamination of soil and water resources. Bacteria exposed to continuous selective pressure of antibiotic reservoirs in the environment are inclined to develop antibiotic resistance genes (Woolhouse et al. 2015). The threat of developing bacterial resistance to antibiotics has pushed governments to limit their usage in feed and led scientists to search for new solutions against pathogens. In the last few years alone, restrictions on antibiotic usage in agriculture have been implemented in both USA and EU law (Pugh 2002). Moreover, it is unlikely that such a reduction will eliminate already present genes of resistance, due to the fact that bacteria tend to keep plasmids with those genes, even without the presence of a drug (Ridenhour et al. 2017).
For these reasons, antibiotics should be limited where possible to keep them for effective usage for humans. During the last decade, alternatives to antibiotic treatment and the prevention of bacterial infections have become one of the most popular and urgent scientific topics. Antibiotic substances of natural origin are currently the focus of researchers’ interests. Proteins and peptides with antimicrobial activity constitute one of the alternatives (Fox 2013). Antimicrobial proteins (AMPs) are a wide group of molecules naturally occurring in almost every living organism. They are present in humans, animals, insects, plants, fungi and bacteria (Bahar and Ren 2013). What is important in terms of resistance safety is that their durability in the environment is shorter than that of antibiotics, due to the easy degradation of peptides. AMPs are usually quite small (15–50 amino acids) and they possess a positive charge, which helps with binding to the negatively charged bacterial membrane, as those proteins contribute to an organism's very first defensive mechanism. Bacterial AMPs and bacteriocins in particular have interesting properties. They act in low concentrations and are highly selective, which means that using specific bacteriocin will kill only few bacterial species (Drider et al. 2016). In contrast to antibiotics, using bacteriocins as antimicrobial drugs would not pose a danger of gut microbial imbalance due to killing commensal and beneficial bacteria (Cavera et al. 2015). Those characteristics make bacterial AMPs highly useful targets for pharmaceutical use both for humans and animals, as well as preservatives for food and feed (Ahmad et al. 2017; Cleveland et al. 2001). Obtaining bacteriocins from strains with potential pathogenicity most often requires the use of genetic engineering methods, which involve transferring the bacteriocin gene sequence to a non-pathogenic strain or another host (Arthur et al. 2014).
Plant-derived platforms are expected to be featured in such production (Ghequire and De Mot 2017). Tremendously diverse recombinant proteins have already been produced in plants (Sack et al. 2015). Reasons for the popularity of using plants for recombinant protein production include low costs of production and safety. Bearing in mind the application of antimicrobial peptides and proteins, genetically modified (GM) plants can be the platform of choice for their expression. Research shows that it is possible to successfully express a wide variety of antimicrobial proteins in plants, starting from those which can protect crops against bacteria (Lee et al. 2017), but also those which can be subsequently used as biopharmaceuticals with or without a later purification step (Paškevičius et al. 2017; Company et al. 2014). Recombinant protein production in edible plants or those used as an animal feed gives a possibility of cutting the costs of product extraction and purification. Those processes, called the downstream phase, are the most expensive part of recombinant protein production (Łojewska et al. 2016). In turn, the production of recombinant bacteriocin in plants possessing natural antimicrobial activity provides an opportunity to double the benefits of the product, as well as the production platform. However, non-food or non-feed plants are considered the safest models for the production of recombinant proteins due to reasons such as the low risk of contamination of human food plants (Ellstrand 2003). Nicotiana tabacum is a model plant for molecular farming due to its fast growth, and well understood genetic and metabolic processes. Tobacco has been considered a good choice for producing proteins with applications in different industries, as its possibilities for expressing a high yield of recombinant proteins are an uncontested advantage. However, transient expression of Nicotiana tabacum is efficient for small, laboratory amounts of protein, and stable expression of recombinant proteins is required for the mass production of pharmaceuticals and for commercialization, which is currently one of the main obstacles in recombinant protein plant platforms (Sabalza et al. 2014). However, the possibility of obtaining bacteriocins in Nicotiana benthamiana plants has been confirmed mainly by transient expression (Paškevičius et al. 2017) and in a stable way by inducible expression (Schulz et al. 2015). In this work, the possibility of producing effective antimicrobial recombinant colicin M protein by constitutive expression in Nicotiana tabacum plants is confirmed for the first time.
Materials and methods
Plant expression vector construction and Escherichia coli transformation
Construction of plant expression vector and E. coli transformation are described in Supplementary information (Figures S1, S2). The recombinant plasmid was isolated from transformed E. coli using the alkaline lysis method (Bimboim and Doly 1979).
Agrobacterium rhizogenes transformation
Recombinant pCAMBIA 1305.1-colM plasmid (500 ng) was used for the transformation of 500 μL of competent cells of the Agrobacterium tumefaciens strain LBA4404 (at OD600 = 1) using the freeze–thaw method (Jyothishwaran et al. 2007). One milliliter of liquid YEP medium was added, and the cells were incubated by shaking at 28 °C in the dark for 3 h. Then, the transformed bacteria were dispersed on YEP agar plates containing kanamycin (50 mg/L), rifampicin (100 mg/L) and streptomycin (50 mg/L). Following this, they were incubated at 28 °C in the dark for 48 h. Colonies were confirmed to be transformed using PCR and by digestion of the extracted plasmid.
Plant transformation
The ColM coding gene (cma) was introduced into Nicotiana tabacum W-38 genome via Agrobacterium-mediated transformation. Nicotiana tabacum healthy and fully expanded leaves from in vitro cultures were used for this purpose. Leaf explants of 1–2 cm2 were co-cultured on solid MS medium (Murashige and Skoog 1962) at 26 °C in the dark for 4 days with 100 µL of A. tumefaciens LBA4404 (with and without recombinant plasmid) that had been grown 24 h (to OD600 nm = 0.5). After four days, the explants were transferred to a fresh MS 0.8% agar-solidified medium supplemented with 0.1 mg/L NAA, 1 mg/L BAP, 500 mg/L of Biotaksym (cefotaxime) and 25 mg/L of hygromycin B and were maintained at 26 °C in the light (16 h daylight per day) for one month (subcultured every 2 weeks). When new shoots started to appear, they were excised and transferred to sterile glass containers with a solid MS medium and antibiotics as mentioned above. The plants were subcultured every 3 weeks on a fresh MS medium.
DNA and RNA extraction and cDNA synthesis
For each tobacco plant line, 50 mg of tissue from the upper leaf was selected via a 1 cm diameter cork borer to isolate genomic DNA. Three leaf discs from different leaves were collected from each transgenic plant line. First, the tissue was ground under liquid nitrogen and then 500 µL of CTAB isolation buffer (2% hexadecyltrimethylammonium bromide, 1.4 M NaCl, 0.2% β2-ME, 20 mM EDTA, 100 mM Tris–HCl, pH 8) was added and mixed again, then it was transferred to a 1.5-ml microcentrifuge tube. The sample was incubated at 60 °C for 15 min and after that 500 µL of chloroform/isoamyl alcohol (24:1 v/v) was added to each sample. The samples were vortexed briefly, incubated for 1 h in 4 °C and then centrifuged for 10 min at 14,000 rpm in 4 °C. The supernatant was transferred to a new 1.5-ml microcentrifuge tube, 300 μl ice-cold isopropanol was added to the tube, and the tube was inverted 5 times to precipitate the nucleic acid. The sample was centrifuged at 14,000 rpm for 10 min in a microcentrifuge, and the supernatant was discarded. The pellet was dried for 2 h and then resuspended in 100 μl of 10 mg Ribonuclease A (Sigma R642) in 10 mM Tris, pH 8.0, 1 mM EDTA (TE/RNase A buffer) (Tamari et al. 2013). Total RNA from the leaves of the transformed plants was isolated using a Plant RNA Mini Kit (Syngene, Poland) from 50 mg of the upper leaves selected via a 1 cm diameter cork borer. Three leaf discs from different leaves were collected from each transgenic plant. The procedure was conducted according to the manufacturer's recommendations. The quality and quantity of the RNA samples were measured spectrophotometrically by acquired absorbance at a wavelength of 260 nm (A260) and 280 nm (A280) using a p300 Nanophotometer (Implen, Germany). The A260/A280 ratio was higher than 1.8 for all the samples. Then, 1 ng of RNA was used to synthesize the first strand of cDNA using a TranScriba Kit (A&A Biotechnology, Poland), all in accordance with the producer's recommendations, except for scaling down the reaction mixture to 10 μl. Quantitative real-time PCR was conducted in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, USA) as follows: 10 μl of the mix contained 1 μl of cDNA, 0.313 μM of each primer and 5 μl of Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA); 10 min of initial denaturation at 95 °C, then 40 cycles of 30 s at 95 C, 60 s at primers’ annealing temperature (Tm). Each reaction was done in triplicate. Elongation factor 1α (EF-1α) was used as a reference gene, as it was previously established as a stable internal reference gene (Schmidt and Delaney 2010). Relative expression of cma was calculated using the equation 2−ΔCt, where ΔCt is the threshold cycle (Ct) values for the target gene minus Ct values obtained for EF-1α. Details of primers Tm and the sequence are presented in supplementary materials (Table 2).
Extraction of total proteins
The total soluble protein of six transgenic leaf samples (0.5 g) was extracted using the phenol extraction method (Isaacson et al. 2006). The samples were collected via a 1 cm diameter cork borer from different leaves. The resulting protein powder was suspended in 50 µL of PBS buffer. The amount of total soluble protein (TSP) extracted was determined using the modified Bradford method (Bradford 1976).
SDS-PAGE analysis
The total soluble proteins extracted from the transgenic and non-transgenic plants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 13% resolving polyacrylamide gel. Each sample contained loading buffer (250 mM Tris–HCl, pH 6.8, 8% SDS, 30% glycerol, 5% β-mercapto-ethanol and 0.04% bromophenol blue) and was heated to 95 °C for 5 min before loading onto the gel. Following electrophoresis, the gel was stained using Colloidal Coomassie G-250 Staining (Dyballa and Metzger 2009).
Western blot analysis
The total proteins extracted from the transgenic and non-transgenic plants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 13% resolving polyacrylamide gel under reducing conditions and then wet transferred to a PVDF membrane (Bio-Rad) in accordance with the manufacturer's instructions. Next, the transfer membrane was blocked by a TBS buffer with 3% BSA for 1 h. Following this, the membrane was incubated with a rabbit anti-histag antibody (GeneTex, USA) conjugated with horseradish peroxidase (HRP) in a concentration of 0.1 μg/mL for 4 h. Finally, the membrane was washed 3 times in TBS–Tween 20 and the antibody bound to the protein was detected using 3, 3′-Diaminobenzidine (DAB) (Thermo Fischer Scientific) staining, in accordance with the manufacturer's protocol.
Protein purification under native conditions
Purification of the recombinant protein was performed using the HIS-Select® Nickel Affinity Gel (Sigma-Aldrich), according to the manufacturer's protocol for native conditions column chromatography. Histidine containing colicin M was eluted from the column using 5 column volumes of Elution Buffer and concentrated on protein concentrator spin columns Vivaspin 6 (GE Healthcare) and finally dissolved in PBS buffer. Before the microbiological tests, protein solution was filtrated through 0.2 µm cellulose acetate membrane syringe filters (GE Healthcare). The protein concentration after purification was measured with the Easy Protein Assay Reagent for Bradford method (Profoldin) and a microplate reader PowerWave HT (BioTek). The maximum expression level of colicin M was 2 µg/g FW.
Biological activity
Microbial strains
Microbiological studies were conducted on a set of reference Escherichia coli strains (E. coli ATCC 25,922, NCTC 8196, DH5-α, GM, NM) and Pseudomonas aeruginosa NCTC 6749. The strains were stored in Microbank systems (Biocorp, Poland) in cryopreservation media in a freezer at − 80 °C. Each strain was revived from the Microbank system on a proliferating medium, Columbia agar (Becton Dickinson, USA), under aerobic conditions at 37 °C. After the first 18-h cultivation, the next 18-h bacterial culture was done on a new medium plate to obtain reproducibility of the method. Each experiment was performed from the same, second recultivation. Studies were also conducted on E. coli and K. pneumoniae clinical strains (n = 6) (Department of Microbiology and Laboratory Medical Immunology, Medical University of Lodz, Poland) (detailed characteristics of tested clinical straits—Supplementary material, Tables 3, 4). The species were identified on the basis of biochemical characteristics and drug susceptibility testing performed by BD Phoenix™ (PX 1902, Becton Dickinson, USA) automatic microbiological system. The findings were verified and were consistent with EUCAST system expert rules. The bacterial colonies harvested from the medium were used to obtain a suspension in McFarland standard 0.5 in sterile-buffered saline. The effect of recombinant colicin M was assessed on a bacterial suspension.
Antibacterial activity assay
The minimal inhibitory concentration (MIC) was determined using a microdilution method on 96-well microplates, according to EUCAST guidelines ISO 20,776–1. In this procedure, 100 µl of recommended Mueller–Hinton broth (BioMerieux, France) with a series of twofold dilutions of colicin M ranging from 250 to 0.49 µg/ml was inoculated with 5 µl of a microbial suspension with a final cell number concentration of approximately 5 × 105 CFU/ml in each well. The recombinant colicin M used for this assay was dissolved in 1× PBS. For all experiments, a positive control consisting of broth without colicin and a negative control consisting of broth without bacteria were prepared. The MIC values were evaluated after 18 h incubation at 37 ◦C under aerobic conditions and optical density (OD600) was measured. Subsequently, the minimal bactericidal concentration (MBC) was determined by plating out the contents of those wells that showed no visible growth of bacteria onto colicin M-free Mueller–Hinton agar. The MBC values were evaluated after 18 h incubation at 35 ˚C under aerobic conditions and they were defined as the lowest concentrations of colicin that resulted in a > 99.9% reduction in CFU. All evaluations were performed in triplicates.
Cell viability assay
The cytotoxic effect of recombinant colicin M on murine fibroblast L929 cells (ATTC® - CCL-1, mouse fibroblasts) and human tumor HeLa cells (ATTC® - CCL-2™, human epithelial cells) was detected by determining the cellular viability using MTT reduction assay. 100 μl of cells was plated in 96-well microplates at a density of 1 × 104 cells/ml and were cultivated in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml of penicillin and 100 μg/ml of streptomycin. The cell cultures were incubated at 37 °C in a humidified atmosphere with 10% CO2. After overnight incubation, the growth medium was removed and 100 μl of the medium with twofold dilutions of colicin ranging from 125 to 1.95 µg/ml was added. The recombinant protein used in this study was dissolved in PBS, which had no influence on cell viability. After 24 h incubation, the medium was removed and MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) was added to each well at a final concentration of 0.5 mg/ml and plates were incubated for the next 2 h at 37 °C, 10% CO2. Then, formazan crystals were solubilized in 150 μl DMSO and optical density was measured at 570 nm. The results of the experiments were presented as mean arithmetic values from 3 repeats in each of two independent experiments and the percentage of viability compared to the untreated control was calculated for each concentration of the protein tested. All statistics were calculated with Prism GraphPad 7 software using Two way ANOVA. A P value of < 0.05 was considered significant.
Results
Molecular characterization of the transgenic plants
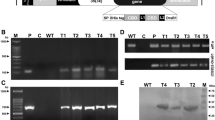
Initially, genomic DNA was isolated from transgenic (Fig. 1) and control tobacco plants to analyze the presence of a colicin M coding sequence at the genomic level by the PCR amplification using primers specific for colicin. The results indicate that the expression cassette consisting of CDR for colicin M was present in the genome of transgenic plants. An 813-bp product was amplified in transgenic lines, while in the DNA of the control plants, no amplification product was detected (Fig. 2).
aNicotiana tabacum leaf explants on MS medium (cocultivation with A. rhizogenes) b Selection of transformants on MS medium with hygromycin b. c Induced callus tissue after the transformation of Nicotiana tabacum leaf discs. d Regenerated transgenic plant. Black arrows indicate transgenic plant tissue (b) and shoot (c)
RT-PCR analysis was used to evaluate transgenic plants for the expression of colicin M gene at the transcript level. Variable levels of colicin M expression were observed in different transgenic plant lines (Fig. 3). The transgenic plant lines with the highest expression of colicin M mRNA were chosen for further SDS-PAGE and Western blot analysis.
Total protein extracts were separated on SDS-PAGE (Fig. 4). As shown in Figs. 5 and 6, colicin M expression by transgenic plants (before and after purification) was seen as a single band on the Western blot. The molecular mass of protein was approximately 29 kDa, which is consistent with the molecular mass of the colicin M protein.
Biological activity of recombinant colicin M
The purified recombinant colicin M was tested for its antimicrobial activity against a set of control E. coli strains and one P. aeruginosa strain. The in vitro antimicrobial activity of the protein at concentrations ranging from 0.49 to 250 μg/ml was screened using the microdilution method. The results showed that the majority of control E. coli strains, except for the GM strain, were sensitive to colicin M, but it was inactive against P. aeruginosa in the concentrations analyzed. The protein tested was the most active against E. coli DH5α, ATCC 25,922 and NM522 with MIC values ranging from 3.9 to 7.8 µg/ml. Considering good antibacterial activity against the control E. coli strains, we decided to determine the antimicrobial effect of colicin M against six clinical isolates of E. coli and K. pneumoniae. Both clinical K. pneumoniae strains and two from E. coli presented an ESBL + phenotype (extended-spectrum β-lactamase). The recombinant colicin M was active against all the clinical strains tested. The most sensitive were two E. coli ESBL– strains and one K. pneumoniae strain (marked as 5) with MIC values ranging from 15.6 to 31.25 µg/ml. Our results prove that colicin M not only inhibits the growth of the clinical strains tested, but in higher concentrations it has bactericidal activity. It is assumed that highly active antibacterial compounds have a bactericidal activity ratio MBC/MIC ≤ 4. In our results, the MBC/MIC ratio for all clinical strains except one E. coli (marked as 3) was 2 or 3, respectively. The in vitro results of antibacterial activity of colicin M are presented in Table 1 as a minimal inhibitory concentration (MIC) and a minimal bactericidal concentration (MBC).
While searching for a new antibacterial compound and considering its potential medical application it is necessary to evaluate its safety for mammalian cell lines. The cytotoxic activity of recombinant colicin M was assessed using L929 murine cell line (recommended by the International Standard ISO 10,993:2009 for evaluation of cytotoxic activities), as well as HeLa human tumor cell line. The percentage of viability inhibition compared to the negative control was estimated for concentrations ranging from 1.95 to 125 µg/mL of the protein. Our results demonstrated that colicin M is not toxic to either of the cell lines in the concentrations analyzed corresponding to the MIC values (IC50 > 125 µg/ml). The HeLa cells seemed to be little more sensitive to colicin than L929 cells. The in vitro results of the viability assay are presented in Fig. 7.
Effect of colicin M on viability of L929 and HeLa cells displayed as percentage of cell survival compared with untreated control. A red line corresponds to the IC30 value (70% of viability) which is considered as safe concentration for cells. No significant differences were observed (P > 0.05) No significant differences were observed (P > 0.05)
Discussion
The production of antibacterial proteins and peptides in plant expression systems is an idea undertaken by many research teams all over the world. Concept of plant made vaccines and antimicrobial proteins is well described in scientific literature (Chen et al. 2018). For this purpose, vaccine antigens have been expressed in many well-known edible plants such as: potato, tomato, lettuce, rice, carrot, soybean, corn, papaya, quinoa, banana, peas or apple (Concha et al. 2017; Sartaj Sohrab et al. 2017; Criscuolo et al. 2019). So, far, in this way it has been possible to obtain such antimicrobial proteins and peptides as human lactoferrin, cecropin A or LFchimera peptide (Mitra and Zhang 1994; Chong and Langridge 2000; Bundó et al., 2014; Chahardoli et al. 2018). Currently, there is considerable interest in the expression of bacteriocins in plant cells. Recent research shows that this group of proteins seems to have a unique potential especially as a viable alternative to antibiotics or new food processing aids (Hahn-Löbmann et al. 2019; Olishevska et al. 2019; Schneider et al. 2018; Paškevičius et al. 2017). Moreover colicin-like bacteriocins in plants could mediate resistance to bacteria in plants, however this has yet to be unveiled (Li et al. 2019).
In this study, stable expression of functionally active bacteriocin—colicin M is described for the first time in Nicotiana tabacum L. plants. We confirmed the presence of the transgene in the kanamycin-resistant plant genome of 6 plant lines (Fig. 2). RT-PCR and Western blot were performed on previously selected transgenic plants to detect cma gene transcripts (Fig. 3) and the presence of colicin M in isolated proteins (Fig. 5). Yield of recombinant colicin per fresh weight tobacco leaves was up to 2 mg/g. Yield of recombinant protein should be as high as possible, however in plant stable expression yield is usually lower than in transient expression. This is the first report describing stable expression of colicin M in a plant expression system. What is important, stable expression has its main advantage, a very little time is required for upstream processing of plants, which makes whole process more economically effective and attractive for commercialization. Currently different approaches for increasing of recombinant protein yield in stably transformed plants have been described, although each one should be tested in regard to specific protein, plant species and cultivation conditions. Therefore our results suggest that further research on stable colicin M expression methodology could have promising effects and should be proceeded. These results indicate that antimicrobial bacterial protein can be successfully expressed in a plant-based expression system. This strategy has been evaluated as an efficient source of recombinant protein production, and N. tabacum in particular is also known as a well-characterized and efficient host in terms of antibacterial molecules (Zhu et al. 2017; Hehle et al. 2015).
Colicins are a group of bacteriocins produced by Escherichia coli strains (Harkness and Braun 1989). Among them colicin M has specific properties enabling its use as an effective antibacterial protein. It is the only known colicin which blocks the synthesis of bacterial cell wall polymer peptidoglycan. The molecular size of colicin M is about 29 kDa, which makes it one of the smallest among all known colicins. Its mechanism of action relies on hydrolyzing the phosphoester bond of the peptidoglycan lipid intermediate (Touzé et al. 2012). This peptidoglycanase uses an FhuA receptor pathway to enter the susceptible cells (Kock et al. 1987). Fewer than ten molecules of colicin M can kill a single bacterial cell, which makes it a very efficient drug (Pressler et al. 1986). This peptide is composed of three functional domains, where the C-domain is the microbiologically active site of the molecule (Zeth et al. 2008). Sub-inhibitory concentration of colicin M was proven to affect the gene expression patterns involved in cell motility, osmotic stress, bacterial cell envelope, genes related to the production of exopolysaccharides, and the CreBC resistance system (Kamenšek et al. 2013). According to previous research, colicin M has high potency against different pathogenic strains of E. coli, especially STEC (O 104:H4), which caused a deadly outbreak in Europe in 2011 (Rubino et al. 2011). To date, the transient expression of colicin M has been described in tobacco, spinach and leafy beets (Schulz et al. 2015) (Stephan et al. 2017).
In this work, we describe the antibacterial properties of IMAC purified plant-derived active colicin M with stable expression, which was tested on E. coli, P. aeruginosa and K. pneumoniae strains. These results indicate that recombinant colicin M reveals the strongest antimicrobial properties against E. coli DH5α, ATCC 25,922 and NM522 with MIC values ranging from 3.9 to 7.8 µg/ml, and a weaker influence on the NCTC strain (MIC = 31.25 µg/ml). Further studies have also shown the antibacterial activity against clinical isolates of E. coli and K. pneumoniae with MIC values from 15.6 to 125 µg/ml for E. coli and from 31.25 to 62.5 µg/ml for K. pneumoniae. On the other hand, the antibacterial activity of this colicin against E. coli 2166 and P. aeruginosa NCTC 6749 was not observed (Table 1). The results of previous studies show that colicin M is active against selected Escherichia coli strains isolated from patients with urinary tract bacteraemia, inhibiting the growth of 43% of the 105 E. coli strains tested (Budič et al. 2011).
Colicin M activity was also confirmed by El Ghachi et al. 2006, who demonstrated its inhibitory effect on the growth of E. coli DH5α, which is consistent with our results. These authors also described the ability of His-tagged colM protein to degrade lipid I and lipid II peptidoglycan intermediates. What is important, colicin M is known to be highly active against 3 pathogenic strains of E. coli (O 157:H7; O 111:H8; O 26:H11) and moderately active against another 3 pathogens, all described in the “Big 7”, a list of the most harmful serotypes of E. coli based on the Center for Disease Control and Prevention analysis (Schulz et al. 2015).
In addition to antibacterial activity, our results also showed that plant-expressed colicin M has no toxicity over L929 and HeLa cell culture lines. L929 line is a good model system for pharmaceutical research and plays an important role in developing safer and more active drugs. Previous research shows that recombinant colicin M can be completely broken down in the human gastrointestinal tract by proteolytic enzymes and by detergents, which makes this molecule safe for gut microflora, as well as the environment (Schaller and Bodenmüller 1981; Schulz et al. 2015). Peptides of colicin M degrade to fragments smaller than 3 kDa, which is the threshold for triggering an allergic reaction. Moreover, its structure is not homologous with any known food proteinaceous allergens. Plant-produced colicins have recently been accepted as generally safe by FDA (GRAS status) and permitted for use with food products.
In summary, the results available in the literature and our study findings show that recombinant colicin M derived from plants could become an interesting future candidate for commercial, pharmaceutical and agricultural use.
References
Ahmad V, Khan MS, Jamal QMS, Alzohairy MA, Al Karaawi MA, Siddiqui MU (2017) Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int J Antimicrob Agents 49(1): 1–11. 10.1016/j.ijantimicag.2016.08.016
Arthur TD, Cavera VL, Chikindas ML (2014) On bacteriocin delivery systems and potential applications. Future Microbiol 9(2):235–248. https://doi.org/10.2217/fmb.13.148
Bahar AA, Ren D (2013) Antimicrobial peptides. Pharmaceuticals 6:12. https://doi.org/10.3390/ph6121543
Bimboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7(6):1513–1523. https://doi.org/10.1093/nar/7.6.1513
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Budič M, Rijavec M, Petkovšek Ž, Žgur-Bertok D (2011) Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS ONE 6(12):e28769
Bundó M, Montesinos L, Izquierdo E, Campo S, Mieulet D, Guiderdoni E et al (2014) Production of cecropin A antimicrobial peptide in rice seed endosperm. BMC Plant biol 14(1):102
Cavera VL, Arthur TD, Kashtanov D, Chikindas ML (2015) Bacteriocins and their position in the next wave of conventional antibiotics. Int J Antimicrob Agents 46(5):494–501. https://doi.org/10.1016/j.ijantimicag.2015.07.011
Chahardoli M, Fazeli A, Niazi A, Ghabooli M (2018) Recombinant expression of LFchimera antimicrobial peptide in a plant-based expression system and its antimicrobial activity against clinical and phytopathogenic bacteria. Biotechnol Biotechnol Equip 32(3):714–723. https://doi.org/10.1080/13102818.2018.1451780
Chen L, Todd R, Kiehlbauch J, Walters M, Kallen A (2017) Notes from the field: pan-resistant New Delhi metallo-beta-lactamase-producing klebsiella pneumonia—Washoe County, Nevada, 2016. MMWR Morb Mortal Wkly Rep 66(1):33. https://doi.org/10.15585/mmwr.mm6601a7
Chen Q, Dent M, Mason H (2018) Molecular pharming: plant-made vaccines. Applications, Challenges and Emerging Areas, Molecular Pharming, p 231
Chong DK, Langridge WH (2000) Expression of full-length bioactive antimicrobial human lactoferrin in potato plants. Transgenic Res 9(1):71–78
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: Safe, natural antimicrobials for food preservation. Int J Food Microbiol 71(1):1–20. https://doi.org/10.1016/S0168-1605(01)00560-8
Company N, Nadal A, La Paz JL, Martínez S, Rasche S, Schillberg S et al (2014) The production of recombinant cationic α-helical antimicrobial peptides in plant cells induces the formation of protein bodies derived from the endoplasmic reticulum. Plant Biotechnol J 12(1):81–92. https://doi.org/10.1111/pbi.12119
Concha C, Cañas R, Macuer J, Torres M, Herrada A, Jamett F, Ibáñez C (2017) Disease prevention: an opportunity to expand edible plant-based vaccines? Vaccines 5(2):14
Criscuolo E, Caputo V, Diotti RA, Sautto GA, Kirchenbaum GA, Clementi N (2019) Alternative methods of vaccine delivery: an overview of edible and intradermal vaccines. J Immunol Res, 2019.
Drider D, Bendali F, Naghmouchi K, Chikindas ML (2016) Bacteriocins: not only antibacterial agents. Probiotics Antimicrob Proteins 8(4):177–182. https://doi.org/10.1007/s12602-016-9223-0
Dyballa N, Metzger S (2009) Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J Vis Exp 30:e1431
El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem 281(32):22761–22772. https://doi.org/10.1074/jbc.M602834200
Ellstrand NC (2003) Going to “great lengths” to prevent the escape of genes that produce specialty chemicals. Plant Physiol 132(August):1770–1774. https://doi.org/10.1104/Pp.103.025908
Fowler T, Walker D, Davies SC (2014) The risk/benefit of predicting a post-antibiotic era: Is the alarm working? Ann N Y Acad Sci 1323(1):1–10. https://doi.org/10.1111/nyas.12399
Fox JL (2013) Antimicrobial peptides stage a comeback. Nature Biotechnol 31(5):379–382. https://doi.org/10.1038/nbt.2572
Ghequire MGK, De Mot R (2017) Turning over a new leaf: bacteriocins going green. Trends Microbiol xx:1–2. doi: 10.1016/j.tim.2017.11.001
Hahn-Löbmann S, Stephan A, Schulz S, Schneider T, Shaverskyi A, Tusé D, Gleba Y (2019) Colicins and salmocins–new classes of plant-made non-antibiotic food antibacterials. Front plant sci 10:437
Harkness RE, Braun V (1989) Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J Biol Chem 264(11):6177–6182
Hehle VK, Paul MJ, Roberts VA, van Dolleweerd CJ, Ma JKC (2015) Site-targeted mutagenesis for stabilization of recombinant monoclonal antibody expressed in tobacco (Nicotiana tabacum) plants. The FASEB J 30(4):1590–1598. https://doi.org/10.1096/fj.15-283226
Isaacson T, Damasceno CMB, Saravanan RS, He Y, Catalá C, Saladié M, Rose JKC (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1(2):769–774. https://doi.org/10.1038/nprot.2006.102
Jones CA, Davis JS, Looke DFM (2017) Death from an untreatable infection may signal the start of the post-antibiotic era. Med J Aust 206(7):292–293. https://doi.org/10.5694/mja17.00077
Jyothishwaran G, Kotresha D, Selvaraj T, Srideshikan SM, Rajvanshi PK, Jayabaskaran C (2007) A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens. Curr Sci 93(6):770–772. https://doi.org/10.2307/24099118
Kåhrström CT (2013) Entering a post-antibiotic era? Nat Rev Microbiol 11(3):146–146. https://doi.org/10.1038/nrmicro2983
Kamenšek S, Žgur-Bertok D (2013) Global transcriptional responses to the bacteriocin colicin M in Escherichia coli. BMC microbiol 13(1):42
Kock J, Olschlager T, Kamp RM, Braun V (1987) Primary structure of colicin M, an inhibitor of murein biosynthesis. J Bacteriol 169(7):3358–3361
Lee IH, Jung YJ, Cho YG, Nou IS, Huq MA, Nogoy FM, Kang KK (2017) SP-LL-37, human antimicrobial peptide, enhances disease resistance in transgenic rice. PLoS One 12(3). doi: 10.1371/journal.pone.0172936
Li JZ, Zhou LY, Peng YL, Fan J (2019) Pseudomonas bacteriocin syringacin M released upon desiccation suppresses the growth of sensitive bacteria in plant necrotic lesions. Microb biotechnol.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J et al (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16(2):161–168. https://doi.org/10.1016/S1473-3099(15)00424-7
Łojewska E, Kowalczyk T, Olejniczak S, Sakowicz T (2016) Extraction and purification methods in downstream processing of plant-based recombinant proteins. Protein Expr Purif 120:110–117. https://doi.org/10.1016/j.pep.2015.12.018
Mitra A, Zhang Z (1994) Expression of a human lactoferrin cDNA in tobacco cells produces antibacterial protein(s). Plant Physiol 106(3):977–981. https://doi.org/10.1104/pp.106.3.977
Molton JS, Tambyah PA, Ang BSP, Ling ML, Fisher DA (2013) The global spread of healthcare-associated multidrug-resistant bacteria: A perspective from Asia. Clin Infect Dis 56(9):1310–1318. https://doi.org/10.1093/cid/cit020
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with to- bacco tissue cultures. Physiol Plant 15:473–497
Nordmann P, Naas T, Poirel L (2011) Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis 17(10):1791–1798. https://doi.org/10.3201/eid1710.110655
Olishevska S, Nickzad A, Déziel E (2019) Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl microbiol biotechnol 103(3):1189–1215
Paškevičius Š, Starkevič U, Misiūnas A, Vitkauskienė A, Gleba Y, Ražanskienė A (2017) Plant-expressed pyocins for control of Pseudomonas aeruginosa. PLoS One 12(10). doi: 10.1371/journal.pone.0185782
Paveenkittiporn W, Kerdsin A, Chokngam S, Bunthi C, Sangkitporn S, Gregory CJ (2017) Emergence of plasmid-mediated colistin resistance and New Delhi metallo-β-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn Microbiol Infect Dis 87(2):157–159. https://doi.org/10.1016/j.diagmicrobio.2016.11.005
Petruccelli S, Otegui MS, Lareu F, Tran Dinh O, Fitchette AC, Circosta A et al (2006) A KDEL-tagged monoclonal antibody is efficiently retained in the endoplasmic reticulum in leaves, but is both partially secreted and sorted to protein storage vacuoles in seeds. Plant Biotechnol J 4(5):511–527. https://doi.org/10.1111/j.1467-7652.2006.00200.x
Pressler U, Braun V, Wittman-Liebold B, Benz R (1986) Structural and functional properties of colicin B. J Biol Chem 261(6):2654–2659
Pugh DM (2002) The EU precautionary bans of animal feed additive antibiotics. Toxicol Lett 128(1–3):35–44. https://doi.org/10.1016/S0378-4274(01)00531-8
Ridenhour BJ, Metzger GA, France M, Gliniewicz K, Millstein J, Forney LJ, Top EM (2017) Persistence of antibiotic resistance plasmids in bacterial biofilms. Evol Appl 10(6):640–647. https://doi.org/10.1111/eva.12480
Rubino S, Cappuccinelli P, Kelvin DJ (2011) Escherichia coli (STEC) serotype O104 outbreak causing haemolytic syndrome (HUS) in Germany and France. J Infect Dev Ctries 5(6):437–440. https://doi.org/10.3855/jidc.2172
Sabalza M, Christou P, Capell T (2014) Recombinant plant-derived pharmaceutical proteins: current technical and economic bottlenecks. Biotechnol Lett 36(12):2367–2379. https://doi.org/10.1007/s10529-014-1621-3
Sack M, Hofbauer A, Fischer R, Stoger E (2015) The increasing value of plant-made proteins. Curr Opin Biotechnol 32:163–170. https://doi.org/10.1016/j.copbio.2014.12.008
Sartaj Sohrab S, Suhail MA, Kamal M, Husen AI, Azhar E (2017) Recent development and future prospects of plant-based vaccines. Curr drug metab 18(9):831–841
Schaller HC, Bodenmüller H (1981) Isolation and amino acid sequence of a morphogenetic peptide from hydra. Proc Natl Acad Sci USA 78(11):7000–7004. https://doi.org/10.1073/pnas.78.11.7000
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283(3):233–241. https://doi.org/10.1007/s00438-010-0511-1
Schneider T, Hahn-Löbmann S, Stephan A, Schulz S, Giritch A, Naumann M, Gleba Y (2018) Plant-made salmonella bacteriocins salmocins for control of salmonella pathovars. Sci Rep 8(1):4078
Schulz S, Stephan A, Hahn S, Bortesi L, Jarczowski F, Bettmann U et al (2015) Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc Natl Acad Sci USA 112(40):E5454–E5460. https://doi.org/10.1073/pnas.1513311112
Sharp PM, Li WH (1987) The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15(3):1281–1295. https://doi.org/10.1093/nar/15.3.1281
Stephan A, Hahn-Löbmann S, Rosche F, Buchholz M, Giritch A, Gleba Y (2017) Simple purification of Nicotiana benthamiana-produced recombinant colicins: high-yield recovery of purified proteins with minimum alkaloid content supports the suitability of the host for manufacturing food additives. Int J Mol Sci 19(1):95. https://doi.org/10.3390/ijms19010095
Tamari F, Hinkley CS, Ramprashad N (2013) A comparison of DNA extraction methods using petunia hybrida tissues. J Biomol Tech 24(3):113. https://doi.org/10.7171/jbt.13-2403-001
Touzé T, Barreteau H, El Ghachi M, Bouhss A, Barnéoud-Arnoulet A, Patin D et al (2012) Colicin M, a peptidoglycan lipid-II-degrading enzyme: potential use for antibacterial means? Biochem Soc Trans 40(6):1522–1527. https://doi.org/10.1042/BST20120189
Woolhouse M, Ward M, van Bunnik B, Farrar J (2015) Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci 370(1670):20140083–20140083. https://doi.org/10.1098/rstb.2014.0083
World Health Organization 2014 Antimicrobial resistance: global report on surveillance 2014 WHO 1–257 9789241564748
Zeth K, Römer C, Patzer SI, Braun V (2008) Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli. J Biol Chem 283(37):25324–25331. https://doi.org/10.1074/jbc.M802591200
Zhu H, Reynolds LB, Menassa R (2017) A hyper-thermostable α-amylase from Pyrococcus furiosus accumulates in Nicotiana tabacum as functional aggregates. BMC Biotechnol 17(1):53. https://doi.org/10.1186/s12896-017-0372-3
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Łojewska, E., Sakowicz, T., Kowalczyk, A. et al. Production of recombinant colicin M in Nicotiana tabacum plants and its antimicrobial activity. Plant Biotechnol Rep 14, 33–43 (2020). https://doi.org/10.1007/s11816-019-00571-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-019-00571-y