Abstract
Strain BGMRC 2036T was isolated from rhizosphere soil of Bruguiear gymnorrhiza collected from the Beibu Gulf of China. Optimum growth occurred at 28 °C, pH 7.0, and under the conditions of 3–5% (w/v) NaCl. The phylogenetic comparisons of 16S rRNA gene sequences displayed that strain BGMRC 2036T was closely related to Martelella limonii NBRC109441T (96.6% sequence similarity), M. mediterranea CGMCC 1.12224T (96.5%), M. lutilitoris GH2-6T (96.5%), M. radicis BM5-7T (96.2%), and M. mangrove BM9-1T (95.9%), M. suaedae NBRC109440T (95.8%). The phylogenomic tree based on the up-to-date bacterial core gene set indicated that the strain BGMRC 2036T form a clade formed with members of the genera Martelella. The major polar lipids include phosphatidylmethylethanolamine, phosphatidylglycerol, phosphatidylcholine, phosphotidylinositol, two unidentified phospholipids, and three unidentified ninhydrin positive phospholipids. The major respiratory quinone is Q-10, which is similar to those of genera Martelella. The main cellular fatty acids are C18:1 ω7c, C16:0, and C12:0 aldehyde. Genome sequencing revealed a genome size of 4.99 Mbp and a G + C content of 62.3 mol%. Pairwise comparison of the genomes of the new strain BGMRC 2036T and the three reference strains M. endophytica YC 6887T, M. mediterranea CGMCC 1.12224T, and M. mangrovi USBA-857 indicated that gANI value was lower than 81% and a digital DNA–DNA hybridization value was lower than 27%. The strain BGMRC 2036T possessed genes putatively encoding riboflavin synthesis and flavodoxin A polyphasic taxonomic study suggested that strain BGMRC 2036T represented a novel species belonging to the genus Martelella, and it was named Martelella alba sp. nov. The type strain is BGMRC 2036T (=KCTC 52121T =NBRC 111908T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Martelella of the family Aurantimonadaceae was originally described by Rivas et al. (2005). To date, this genus comprises seven species with validly published names (https://lpsn.dsmz.de/genus/martelella), M. mediterranea as the type species, which were isolated from Lake Martel in Mallorca (Rivas et al. 2005). These species were isolated from different sources, including different roots of halophytes, soil from the root of a mangrove, and the water of Lake Martel in Mallorca (Lee 2019; Bibi et al. 2013; Chung et al. 2016; Zhang and Margesin 2014; Rivas et al. 2005; Kim and Lee 2019). In this study, a novel strain BGMRC 2036T was isolated from rhizosphere soil of mangrove plants B. gymnorrhiza. Mangroves are woody salt-tolerant plants that grow at tropical and subtropical coastal intertidal zones with high ecological value (Shi et al. 2020).
Materials and methods
Bacterial strain and culture condition
During our investigations of microbial biodiversity in mangrove plants, strain BGMRC 2036T was isolated from rhizosphere soil of B. gymnorrhiza, collected from the Beibu Gulf of China (21°55′ N, 108°50′ E). The rhizosphere soil was stored in a sterile plastic bottle at 4 °C as soon as it was collected and then transported to the laboratory within 12 h. Soil (2 g) was added to 20 mL sterilized seawater, shaken at 37 °C for 1 h, and then diluted by tenfold. After being incubated at 28 °C for 1 week, 100 μL of the diluent was spread on modified Yeast Malt Extract (modified ISP2; 2.0 g yeast extract, 2.0 g malt extract, 2.0 g d-(+)-glucose anhydrous, 15.0 g agar powder, and 1 L sterilized seawater). A cream colony isolation and purification on modified ISP2 medium was designed a new strain and then stored at − 80 °C with a 20% (v/v) glycerol suspension. Strains M. suaedae NBRC109440T, M. limonii NBRC109441T, and M. mediterranea CGMCC 1.12224T, obtained, respectively, from the National Biological Resource Center, NITE (NBRC) and China General Microbiological Culture Collection Center (CGMCC), were used as references.
Morphological and physiological characteristics
Morphological and physiological characteristics were observed on modified Yeast Malt Extract modified ISP2 medium unless otherwise stated. Growth and colony morphology were monitored after being induced by continuous incubation over 2 days at 28 °C. A scanning electron microscope (QUANTA 250) was used for the analysis of cell morphology. The presence of strain-flagella was checked by transmission electron microscopy (HT7700; Hitachi, Ltd, Tokyo, Japan) after 2 days of growth on modified ISP2 medium at 28 °C. Cell motility determination was realized by investigating the development process of turbidity throughout a tube using modified ISP2 semisolid medium containing 0.4% agar (Leifson 1960). Gram staining of strain BGMRC 2036T was performed as described by Smibert and Krieg (1994). Oxidase activity was examined using 1% (w/v) N, N, N′, N′-tetramethyl-p-phenylenediamine reagent, and catalase activity determination was obviously confirmed through bubble production upon the addition of 3% (w/v) hydrogen peroxide (H2O2) solution (Choi et al. 2014). Sodium chloride (NaCl) requirement and tolerance were tested at 28 °C for 7 days in modified ISP2 liquid medium with NaCl (0‐15%, w/v, with an average interval for 1.0%). The temperature range was determined by incubating cells in modified ISP2 medium broth at 4 °C, 10 °C, 15 °C, 20 °C, 25 °C, 28 °C, 37 °C, 40 °C, and 45 °C for 2 weeks. Growth at different pH values was tested in modified ISP2 liquid medium at 28 °C for 2 weeks (pH 4.0–12.0 at various intervals of 1 pH unit) with the referred buffering system of Xu et al. (2005). As to the colony color determinations, ISCC-NBS color charts were adopted (Kelly and Judd 1965). Hydrogen sulfide production and hydrolysis of substrates (cellulose, gelatin, starch, Tween 20, 40, and 80) were performed according to the description of Tindall et al. (2007). Coagulation and peptonization of milk were investigated according to the method of Gonzalez et al. (1978). Biochemical tests were performed with API ZYM, API 50CH, and API 20E strips (BioMérieux, Marcy-l’Étoile, France) according to the guidance of manufacturer. Utilization of carbon and nitrogen source was studied on Biolog GEN III MicroPlates (Biolog Hayward, CA, USA). The incubation temperature was at 28 °C and the result was monitored after 48 h.
Chemotaxonomic characterization
Cells of strain BGMRC 2036T and the reference strain were harvested after cultivation on modified ISP2 medium at 28 °C for 3 days, whose polar lipids were resulted by extraction as described by Kamekura (1993), further detection was performed through two-dimensional thin-layer chromatography plates precoated with silica gel 60 GF254 (Merck, Kenilworth, NJ, USA) (Minnikin et al. 1984). Menaquinone extraction and analysis were carried out on reversed-phase high-performance liquid chromatography (Komagata and Suzuki 1987; Nakagawa and Yamasato 1993). Cellular fatty acid composition of cell walls was extracted according to Kamekura (1993), analyzed by gas chromatography (G6890N; Agilent Technologies, Inc., Santa Clara, CA, USA), and verified through the Sherlock Microbial Identification System (version 6.0) following the instructions of manufacturer, as reported by Sasser (1990).
Genomic characterization
The DNA of strain BGMRC 2036T extraction were performed as described by Hoetzinger et al. (2017). The genome was sequenced with Illumina HiSeq 4000 system (Illumina, San Diego, CA, USA) at the Beijing Genomics Institute (Shenzhen, China). The assembly of draft genome was achieved by SOAP denovo software (version 2.04), and the short oligonucleotide of assembling results was subsequently polished by SOAP aligner software (version 2.21) (Li et al. 2008, 2015), details of which are given in Table 3. Average nucleotide identity (ANI) was analyzed with the ANI calculator tool from Ezbiocloud. The digital DNA-DNA hybridization estimate values were based on genome sequence and characterized using formula 2 at the website of Genome-to-Genome Calculator (CGGC) (http://ggdc.dsmz.de/ggdc.php) according to the study of Meier-Kolthoff et al. (2013). The obtained genome sequences were annotated by the NCBI Prokaryotic Genome Annotation Pipeline and for further comparative analyses by rapid annotation using subsystem technology version 2.0. The GenBank accession numbers of BGMRC 2036T and other genus Martelella strains are listed in Table 3.
Phylogeny analysis
The 16S rRNA gene sequence of strain BGMRC 2036T was PCR-amplified with the universal primers 27F and 1492R (Lane 1991) and sequenced using the Sanger method (Zhang et al. 2011). Bacterial DNA extraction and amplification were performed following Li et al. (2007). The 16S rRNA gene sequence similarities were determined using the EzBioCloud database (http://www.ezbiocloud.net) (Niu et al. 2018). Multiple alignments of the sequence profile were performed with Clustal X version 1.83 (Thompson et al. 1997). Phylogenetic trees were constructed through the neighbor-joining (Saitou and Nei 1987), maximum-likelihood (Felsenstein 1981), and Minimum-evolution (Rzhetsky and Nei 1992) algorithms in the MEGA software package (version 7.0) (Kumar et al. 2016). The topology of the phylogenetic tree was reasonably evaluated with bootstrap analysis based on 1000 replicates (Felsenstein 1985). The phylogenomic tree was reconstructed using the up-to-date bacterial core gene set (UBCG v.3) according to its manual (Na et al. 2018).
Results and discussion
According to API 50CH, strain BGMRC 2036T had different reactions for 198, 8, and 15 of the 49 tested substrates to M. mediterranea CGMCC 1.12224T, M. suaedae NBRC109440T, and M. limonii NBRC109441T, respectively (Table S1). There were 28, 13, and 25 different reactions of the 95 tested substrates (Biolog GEN III MicroPlate) between strains BGMRC 2036 and M. mediterranea CGMCC 1.12224T, M. suaedae NBRC109440T, and M. limonii NBRC109441T (Table S2). The differences of physiological and biochemical characteristic between strain BGMRC 2036T and its closely related type strains are listed in Table 1 and also mentioned in the species description below.
The major fatty acid of BGMRC 2036T was C18:1 ω7c (48.6%). The remaining fatty acid component (> 10%) included C16:0 (22.1%), C12:0 aldehyde (14.2%), iso-C16:1, and C14:0 3-OH (13.9%), which were similar to that of M. suaedae NBRC 109440T. However, the minor fatty acids C16:1ω7c and C16:1ω6c were discovered in BGMRC 2036T and were not present in M. suaedae NBRC 109440T. The fatty acid profile of the new isolate closely resembled those of the type strains of recognized Martelella species, although some differences in their proportions were observed. The detailed fatty acid profiles of strain BGMRC 2036T and its related reference strains are shown in Table 2. The major polar lipids consisted of phosphatidylmethylethanolamine, phosphatidylglycerol, phosphatidylcholine, phosphatidyl inositol, two unidentified phospholipids (PL1, PL3), and three unidentified ninhydrin positive phospholipids (NPL1–3) (Fig. S2). The polar lipid profile of BGMRC 2036T was similar to that of the type strains of the genus Martelella, with phosphatidylmethylethanolamine and phosphatidyl inositol as the predominant components; phosphatidylglycerol, phosphatidylcholine, one unidentified phospholipid (PL3) and three unidentified ninhydrin positive phospholipids (NPL1–3) were only detected in BGMRC 2036T. Furthermore, the absence of phosphatidylethanolamine, one unidentified phospholipid (PL2), seven unidentified ninhydrin positive lipids (AL1–7) and seven unidentified polar lipids (L1–7), along with the presence of phosphatidylglycerol, phospatidyl choline, and two unidentified ninhydrin positive phospholipids (NPL2–3) in the BGMRC 2036T lipid profile helped distinguish the strain from M. mediterranea, M. suaedae, and M. limonii (Fig. S2). Hence, from the data obtained above, strain BGMRC 2036T could clearly be differentiated from its closest phylogenetic relatives. The menaquinone was ubiquinone Q-10, which was identical to that of the Martelella genus.
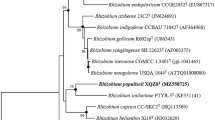
The global alignment based on 16S rRNA gene sequence in the EzBioCloud database demonstrated that strain BGMRC 2036T was closely related to M. limonii NBRC 109441T (96.6% sequence similarity), M. mediterranea CGMCC1.12224T (96.5%), M. radicis BM5-7T (96.2%), and M. suaedae NBRC109440T (95.9%). Phylogenetic analysis based on the neighbor-joining algorithm, maximum-likelihood algorithm, and minimum-evolution methods revealed that strain BGMRC 2036T formed a clade within members of the genus Martelella related to the family Aurantimonadaceae (Figs. 1, S3, S4). The phylogenomic tree based on the up-to-date bacterial core gene set also indicated that the strain BGMRC 2036T formed a robust clade within genus Martelella (Fig. 2), supporting that strain BGMRC 2036T is a novel species of the genus Martelella in agreement with the results of the 16S rRNA gene phylogenic analysis.
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences, shows the position of the BGMRC 2036T with related taxa. The sequence of Caulobacter vibrioides CB51T was used as an out group. Asterisks indicate that the corresponding branches were also recovered in trees generated with the maximum-likelihood and maximum-parsimony methods. Numbers at nodes indicated the percentage of 1000 bootstrap replicates. Only bootstrap values above 50% are shown. Bar, 0.01 substitutions per nucleotide position
Whole-genome based phylogenetic tree were constructed using UBCGs (concatenated alignment of 92 core genes) and showing the phylogenetic relationship of BGMRC 2036T with reference species in the genus Martelella. Gene support indices (GSIs) is given at branching points. Bar, 0.5 substitution per position
The genome size of strain BGMRC 2036T was 4.99 Mbp, and that of N50 was 243,156 base pairs. A total of 71 contigs were obtained (Table 3). The genome sizes of the other three reference strains M. endophytica YC6887T, M. mediterranea CGMCC 1.12224T, and M. mangrovi USBA-857 were 4.82 Mbp, 5.69 Mbp, 4.63 Mbp, respectively (Table 3). All strains had relatively high G + C contents of more than 60 mol% (Table 3). The G + C content of strain BGMRC 2036T was 62.3 mol%, which was lower than that of M. limonii NBRC 109441T and higher than that of other closely related species shown in Tables 1 and 3. The genome orthoANI value between strain BGMRC 2036T and M. endophytica YC6887T, M. mediterranea CGMCC 1.12224T, M. mangrovi USBA-857 was lower than 81% and a digital DNA-DNA hybridization value was lower than 27% (Table 3). These values were considerably lower than the recommendation of a threshold value of 96% ANI and 70% DNA-DNA relatedness as to the general species definition, indicating that the strain BGMRC 2036T does not attach to M. mediterranea and may represent a novel species.
The gene content of strain BGMRC 2036T and seven closely related species showed interesting pattern (Table 4). All strains except M. mediterranea and M. mangrovi encompassed genes putatively encoding flavodoxin and a gene cluster participating in ammonia assimilation (Table 4). Concerning the ABC-type transport systems, toxin–antitoxin replicon stabilization systems and copper transport systems of the seven strains showed different patterns. All strains except M. endophytica possessed a gene cluster participating in choline and betaine uptake and betaine biosynthesis. All strains possessed a riboflavin synthesis gene cluster that can produce 5′-phosphate decarboxylase. The strain AD-3 had been reported to possess a high phenanthrene biodegradability, which may have potential for bioremediation of PAH-contaminated hypersaline sites (Feng et al. 2012). Strain BGMRC 2036T possessed a gene cluster participating in nitrogen fixation. In addition, the related strains Martelella sp. strain 161,492 (MH001982) is a diazotroph resource in mangrove sediment, which may have relationship with the habitat of mangroves plants (Liu et al. 2020). Thus, new strain may affect mangrove ecosystems and relate to nitrogen fixation in mangrove sediment.
Description of Martelella alba sp. nov.
Martelella alba (al’ba. L. fem. adj. alba white, referring to the color of the colonies).
The Gram-negative, non-motile, and rod-shaped bacteria cells are 0.3–0.4 μm in width and 0.6–1.0 μm in length. Colonies were moist, circular, smooth, white, and 0.1–0.5 mm in diameter after being maintained on modified ISP2 agar at 28 °C for 2 days. Growth occurred at 25–37 °C (optimum, 28 °C) with pH range 6.0–11.0 (pH 7.0) and containing 0–8.0% (w/v) NaCl (3–5%). The strain was negative for nitrate reduction, hydrolysis of gelatin, cellulose, starch, Tween 20, 40, and 80, and milk coagulation and peptonization. In the API 20E, O-nitrophenyl-β-d-galactopyranoside, VP test, glucose fermentation, glucose fermentation, amygdalin, and arabinose were positive. In the API ZYM, alkaline phosphatase, esterase (C4), leucine arylamidase, yaline arylamidase, acid phosphatase, naphthol-ASBI-phosphohydrolase, β-galactosidase, α-glucanase, β-glucosidase, and N-acetyl-β-glucosaminidase activities were positive. The major fatty acids of strain BGMRC 2036T were C18:1 ω7c and C16:0, while ubiquinone Q-10 was found to be the predominant menaquinone. The main polar lipids included phosphatidylmethylethanolamine, phosphatidylglycerol, phosphatidylcholine, phosphatidyl inositol, two unidentified phospholipid (PL1 and PL3), and three unidentified ninhydrin positive phospholipid (NPL1-3). This strain type was BGMRC 2036T (=KCTC 52121T =NBRC 111908T) isolated from the rhizosphere soil of B. gymnorrhiza from the Beibu Gulf.
Abbreviations
- KCTC:
-
The Korean Collection for Type Cultures
- MCCC:
-
The Marine Culture Collection of China
- Q:
-
Ubiquinone
- PME:
-
Phosphatidylmethylethanolamine
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- PI:
-
Phosphatidyl inositol
- PC:
-
Phosphatidyl choline
- AL:
-
Unidentified aminolipid
- NPL:
-
Unidentified ninhydrin positive phospholipid
- PL:
-
Unidentified phospholipid
- L:
-
Unidentified lipid
References
Bibi F, Chung EJ, Khan A, Jeon CO, Chung YR (2013) Martelella endophytica sp. nov., an antifungal bacterium associated with a halophyte. Int J Syst Evol Microbiol 63:2914–2919
Choi JH, Seok JH, Cha JH, Cha CJ (2014) Lysobacter panacisoli sp. nov., isolated from ginseng soil. Int J Syst Evol Microbiol 64:2193–2197
Chung EJ, Hwang JM, Kim KH, Jeon CO, Chung YR (2016) Martelella suaedae sp. nov. and Martelella limonii sp. nov., isolated from the root of halophytes. Int J Syst Evol Microbiol 66:3917–3922
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: anapproach using the bootstrap. Evolution 39:783–791
Feng TC, Cui CZ, Dong F, Feng YY, Liu YD et al (2012) Phenanthrene biodegradation by halophilic Martelella sp. AD-3. J Appl Microbiol 113:779–789
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrated-metabolizing, extremely haloppilic bacterium. Can J Microbiol 24:710–715
Hoetzinger M, Schmidt J, Jezberov AJ, Koll U, Hahn MW (2017) Microdiversification of a pelagic Polynucleobacter species is mainly driven by acquisition of genomic islands from a partially interspecific gene pool. Appl Environ Microbiol 83:19
Kamekura M (1993) Lipids of extreme halophiles. In: Vreeland RH, Hochstein LI (eds) The biology of halophilic bacteria. CRC Press, Boca Raton, pp 135–161
Kelly KL, Judd DB (1965) ISCC-NBS color-name charts illustrated with centroid colors. Washington, DC: US Government Printing Office 1900–1972
Kim YJ, Lee SD (2019) Martelella lutilitoris sp. nov., isolated from a tidal mudflat. J Microbiol 57:976–981
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Chichester, John Wiley, pp 115–175
Lee SD (2019) Martelella caricis sp. nov., isolated from a rhizosphere mudflat. Int J Syst Evol Microbiol 69:266–270
Leifson E (1960) Atlas of bacterial flagellation. Q Rev Biol 242:2
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R et al (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676
Liu XY, Yang C, Yu XL, Yu H, Zhuang W et al (2020) Revealing structure and assembly for rhizophyte-endophyte diazotrophic community in mangrove ecosystem after introduced Sonneratia apetala and Laguncularia racemosa. Sci Total Environ 721:137807
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14(1):60–60
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. Appl Environ Microbiol 2:233–241
Na SI, Kim YO, Yoon SH, Ha SM, Baek I et al (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Nakagawa Y, Yamasato K (1993) Phylogenetic diversity of the genus cytophaga revealed by 16S rRNA sequencing and menaquinone analysis. J Gen Microbiol 139:1155–1161
Niu L, Xiong M, Zhang J, Xiang Y, Song L et al (2018) Bacillus camelliae sp. nov., isolated from Pu’er tea. Int J Syst Evol Microbiol 68:564–569
Rivas R, Sánchez-Márquez S, Mateos PF, Martínez-Molina E, Velázquez E (2005) Martelella mediterranea gen. nov., sp. nov., a novel alpha-proteobacterium isolated from a subterranean saline lake. Int J Syst Evol Microbiol 55:955–959
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum evolution trees. Mol Biol Evol 9:945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark, DE
Shi CC, Han K, Li LW, Inge S, Simon MYL et al (2020) Complete chloroplast genomes of 14 mangroves: phylogenetic and comparative genomic analyses. Biomed Res Int 2020:1–13
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general, molecular bacteriology. American Society for Microbiology, pp 607–654
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ, Sikorski J, Smibert RA, Krieg NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology. American Society for Microbiology, vol 365, pp 384–385
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ et al (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family Oxalobacteraceae isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Zhang DC, Margesin R (2014) Martelella radicis sp. nov. and Martelella mangrovi sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol 64:3104–3108
Zhang SM, Tian F, Huang QF, Zhao YF, Guo XK et al (2011) Bacterial diversity of subgingival plaque in 6 healthy Chinese individuals. Exp Ther Med 2(5):1023–1029
Acknowledgements
We thank Fangang Yang for carrying out the fatty acid measurements, Guanghua Wang for nomenclatural advice and Qiliang Lai for manuscript revisions advice. At the same time, we thank LetPub (http://www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This study was supported by Natural Science Foundation of China (21662006, 81903533), Natural Science Foundation of Guangxi (2018GXNSFAA281268), Development Program of High-level Talent Team under Qihuang Project of Guangxi University of Chinese Medicine (2018006), The Research Launching Fund Project from Guangxi University of Chinese Medicine Introduced the Doctoral in 2017 (2017BS039), Special Program for scientific research project under Institutes of Marine Drugs of Guangxi University of Chinese Medicine (2018ZD005), the key research and development project from Guangxi Science and Technology plan (AB16380009), Chinese postdoctoral science foundation (2016M602920XB), and the Special Fund for Bagui Scholars of Guangxi, Program for Innovative Research Team of High Education and Outstanding Scholar of Guangxi (2019-52).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical standards
This article does not describe any experimental work related to humans.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank accession number for the 16S rRNA gene sequence of strain BGMRC 2036T is MN028527. The draft geneome sequence of strain BGMRC 2036T had been submitted to and deposited in the DDBJ/ENA/GenBank with the serial number of VHLG00000000. Transmission electron micrographs of strain BGMRC 2036T, polar lipids of strain BGMRC 2036T and related type strains, Maximum-Likelihood tree and Minimum Evolution tree are available as supplementary figures in online.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Gao, C., Feng, Y. et al. Martelella alba sp. nov., isolated from mangrove rhizosphere soil within the Beibu Gulf. Arch Microbiol 203, 1779–1786 (2021). https://doi.org/10.1007/s00203-020-02178-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02178-2