Abstract
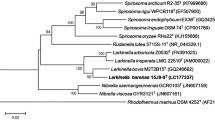
A Gram-negative, strictly aerobic, motile, and rod-shaped bacterial strain G4R7T was isolated from the roots of moso bamboo (Phyllostachys edulis) in Zhejiang, Hangzhou Province, China. After comparing 16S rRNA gene sequences, strain G4R7T exhibited the highest similarities with Massilia neuiana PTW21T (98.3%), followed by M. agri K-3-1T (98.3%), M. consociate CCUG 58010T (97.7%), M. niastensis 5516S-1T (97.7%) and M. yuzhufengensis ZD1-4T (97.6%). The phylogenetic analysis revealed that strain G4R7T belonged to the genus Massilia. The draft genome of strain G4R7T was 5.81 Mb, and the G+C content was 64.4%. The average nucleotide identity values between G4R7T and another related member of the genus Massilia ranged from 76.6 to 87.2%, and the digital DNA–DNA hybridization ranged from 20.7 to 27.9%. Strain G4R7T grew at 15–37 °C (optimum 25–30 °C) and pH 6.0–9.0 (optimum pH 7.0) in the presence of 0–3% (w/v) NaCl (optimum 0%). The respiratory quinone was Q-8, and the major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and aminophospholipid. The major cellular fatty acids were C10:0 3OH, C12:0, C12:0 2OH, and C16:0, summed feature 3 (C16:1 ω6c and/or C16:1 ω7c). As per the data from chemotaxonomic, phylogenetic, and phenotypic evidence, strain G4R7T represents a new species of genus Massilia, for which the name Massilia phyllostachyos sp. nov. is proposed. The type strain is G4R7T (=ACCC 61911T=GDMCC 1.2961T=JCM 35225T).

Similar content being viewed by others
Data Availability
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain G4R7T is MZ573411. The accession number for the draft genome sequence of strain G4R7T is GCA_021044825.1.
Code Availability
Not applicable.
Abbreviations
- ANI:
-
Average nucleotide identity;
- GGDC:
-
Genome-to-genome distance calculator
- dDDH:
-
Digital DNA-DNA hybridization
- AAI:
-
Average amino acid identity
- TYGS:
-
Type (strain) genome server
- MEGA:
-
Molecular evolutionary genetics analysis
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DPG:
-
Diphosphatidylglycerol,
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- APL:
-
Aminophospholipid
- IAA:
-
Indole acetic acid
- PGPR:
-
Plant growth-promoting rhizobacteria
References
La Scola B, Birtles RJ, Mallet MN, Raoult D (1998) Massilia timonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J Clin Microbiol 36:2847–2852. https://doi.org/10.1128/JCM.36.10.2847-2852.1998
Kämpfer P, Lodders N, MartinFalsen KE (2011) Revision of the genus Massilia La Scola et al 2000, with an emended description of the genus and inclusion of all species of the genus Naxibacter as new combinations, and proposal of Massilia consociata sp. nov. Int J Syst Evol Microbiol 61:1528–1533. https://doi.org/10.1099/ijs.0.025585-0
Singh H, Du J, Won K, Yang JE, Yin C, Kook M, Yi TH (2015) Massilia arvi sp. nov., isolated from fallow-land soil previouslycultivated with Brassica oleracea, and emended description of the genus Massilia. Int J Syst Evol Microbiol 65:3690–3696. https://doi.org/10.1099/ijsem.0.000477
Wang J, Zhang J, Pang H, Zhang Y, Li Y, Fan J (2012) Massilia flava sp. nov., isolated from soil. Int J Syst Evol Microbiol 62:580–585. https://doi.org/10.1099/ijs.0.031344-0
Raths R, Peta V, Bucking H (2020) Massilia arenosa sp. nov., isolated from the soil of a cultivated maize field. Int J Syst Evol Microbiol 70:3912–3920. https://doi.org/10.1099/ijsem.0.004266
Holochova P, Maslanova I, Sedlacek I, Svec P, Kralova S, Kovarovic V, Busse HJ, Stankova E, Bartak M, Pantucek R (2020) Description of Massilia rubra sp. nov., Massilia aquatica sp. nov., Massilia mucilaginosa sp. nov., Massilia frigida sp. nov., and one Massilia genomospecies isolated from Antarctic streams, lakes and regoliths. Syst Appl Microbiol 43:126112. https://doi.org/10.1016/j.syapm.2020.126112
Shen L, Liu Y, Wang N, Yao T, Jiao N, Liu H, Zhou Y, Xu B, Liu X (2013) Massilia yuzhufengensis sp. nov., isolated from an ice core. Int J Syst Evol Microbiol 63:1285–1290. https://doi.org/10.1099/ijs.0.042101-0
Weon HY, Kim BY, Son JA, Jang HB, Hong SK, Go SJ, Kwon SW (2008) Massilia aerilata sp. nov., isolated from an air sample. Int J Syst Evol Microbiol 58:1422–1425. https://doi.org/10.1099/ijs.0.65419-0
Sedláček I, Holochová P, Busse HJ, Koublová V, Králová S, Švec P, Sobotka R, Staňková E, Pilný J, Šedo O, Smolíková J, Sedlář K (2022) Characterisation of waterborne psychrophilic Massilia isolates with violacein production and description of Massilia antarctica sp. nov. Microorganisms 10:704. https://doi.org/10.3390/microorganisms10040704
Dahal RH, Chaudhary DK, Kim DU, Kim J (2021) Cold-shock gene cspC in the genome of Massilia polaris sp. nov. revealed cold-adaptation. Antonie Van Leeuwenhoek 114:1275–1284. https://doi.org/10.1007/s10482-021-01600-z
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family “Oxalobacteraceae” isolated from China. Int J Syst Evol Microbiol 55:1149–1153. https://doi.org/10.1099/ijs.0.63407-0
Feng GD, Yang SZ, Li HP, Zhu HH (2016) Massilia putida sp. nov., a dimethyl disulfide-producing bacterium isolated from wolfram mine tailing. Int J Syst Evol Microbiol 66:50–55. https://doi.org/10.1099/ijsem.0.000670
Lee H, Kim DU, Park S, Yoon JH, Ka JO (2017) Massilia chloroacetimidivorans sp. nov., a chloroacetamide herbicide-degrading bacterium isolated from soil. Antonie Van Leeuwenhoek 110:751–758. https://doi.org/10.1007/s10482-017-0845-3
Son J, Lee H, Kim M, Kim DU, Ka JO (2021) Massilia aromaticivorans sp. nov., a BTEX degrading bacterium isolated from Arctic Soil. Curr Microbiol 78:2143–2150. https://doi.org/10.1007/s00284-021-02379-y
Du C, Li C, Cao P, Li T, Du D, Wang X, Zhao J, Xiang W (2021) Massilia cellulosiltytica sp. nov., a novel cellulose-degrading bacterium isolated from rhizosphere soil of rice (Oryza sativa L.) and its whole genome analysis. Antonie Van Leeuwenhoek 114:1529–1540. https://doi.org/10.1007/s10482-021-01618-3
Zheng BX, Bi QF, Hao XL, Zhou GW, Yang XR (2017) Massilia phosphatilytica sp. nov., a phosphate solubilizing bacteria isolated from a long-term fertilized soil. Int J Syst Evol Microbiol 67:2514–2519. https://doi.org/10.1099/ijsem.0.001916
Rodríguez-Díaz M, Cerrone F, Sánchez-Peinado M, SantaCruz-Calvo L, Pozo C, López JG (2014) Massilia umbonata sp. nov., able to accumulate poly-β-hydroxybutyrate, isolated from a sewage sludge compost-soil microcosm. Int J Syst Evol Microbiol 64:131–137. https://doi.org/10.1099/ijs.0.049874-0
Li C, Cao P, Du C, Zhang X, Bing H, Li L, Sun P, Xiang W, Zhao J, Wang X (2021) Massilia rhizosphaerae sp. nov., a rice-associated rhizobacterium with antibacterial activity against Ralstonia solanacearum. Int J Syst Evol Microbiol 71:5009. https://doi.org/10.1099/ijsem.0.005009. (PMID: 34520338)
Janzen DH (1976) Why bamboos wait so long to flower. Annu Rev Ecol Evol S 7:347–391. https://doi.org/10.2307/2096871
Zheng X, Lin S, Fu H, Ding Y (2020) The bamboo flowering cycle sheds light on flowering diversity. Front Plant Sci 11:381. https://doi.org/10.3389/fpls.2020.00381
Pan F, Wang Y, Liu H, Wu M, Chu W, Chen D, Xiang Y (2017) Genome-wide identification and expression analysis of SBP-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). BMC Genomics 18:486. https://doi.org/10.1186/s12864-017-3882-4
Liu L, Liang LX, Zhang XX, Li LB, Sun QW (2018) Mesorhizobium ephedrae sp. nov. isolated from the roots of Ephedra przewalskii in Kumtag desert. Int J Syst Evol Microbiol 68:3615–3620. https://doi.org/10.1099/ijsem.0.003044
Liu L, Hui N, Liang L, Zhang X, Sun Q, Li L (2019) Sphingomonas deserti sp. nov., isolated from Mu Us Sandy Land soil. Int J Syst Evol Microbiol 69:441–446. https://doi.org/10.1099/ijsem.0.003168
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, pp 115–147
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Felsenstein J (1981) Evolutionary trees from DNA sequences, a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/bf01734359
Saitou N, Nei M (1987) The neighbor-joining method, a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1985) Confidence limits on phylogenies, an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646
Kelley DR, Schatz MC, Salzberg SL (2010) Quake: quality-aware detection and correction of sequencing errors. Genome Biol 11:R116. https://doi.org/10.1186/gb-2010-11-11-r116
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303:3997
Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. https://doi.org/10.1016/j.jmb.2015.11.006
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:801–807. https://doi.org/10.1093/nar/gkab902
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Lefort V, Desper R, Gascuel O (2015) FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800. https://doi.org/10.1093/molbev/msv150
Farris JS (1972) Estimating phylogenetic trees from distance matrices. Am Nat 106:645–667
Kreft L, Botzki A, Coppens F, Vandepoele K, Van Bel M (2017) PhyD3: A phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 33:2946–2947. https://doi.org/10.1093/bioinformatics/btx324
Li B, Yang X, Tan H, Ke B, He D, Ke C, Zhang Y (2017) Vibrio parahaemolyticus O4:K8 forms a potential predominant clone in southern China as detected by whole-genome sequence analysis. Int J Food Microbiol 244:90–95
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci 106:19126–19131. https://doi.org/10.1073/pnas.090641210
Jeong J, Weerawongwiwat V, Kim JH, Suh MK, Kim HS, Lee JS, Yoon JH, Sukhoom A, Kim W (2022) Arenibacterium arenosum sp. nov., isolated from sea sand. Arch Microbiol 204:147. https://doi.org/10.1007/s00203-022-02760-w
Dong XZ, Cai MY (2001) Determination of biochemical properties. In manual for the systematic identification of general bacteria. Science Press, Beijing
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207. https://doi.org/10.1016/S0580-9517(08)70410-0
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI Inc.
Glickmann E, Dessaux Y (1995) A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl Environ Microbiol 61:793–796. https://doi.org/10.1128/aem.61.2.793-796.1995
Du GX, Qu LY, Hong XG, Li CH, Ding DW, Gao P, Xu QZ (2021) Kushneria phosphatilytica sp. nov., a phosphate-solubilizing bacterium isolated from a solar saltern. Int J Syst Evol Microbiol 71:4619. https://doi.org/10.1099/ijsem.0.004619
Shen L, Liu Y, Gu Z, Xu B, Wang N, Jiao N, Liu H, Zhou Y (2015) Massilia eurypsychrophila sp. nov. a facultatively psychrophilic bacteria isolated from ice core. Int J Syst Evol Microbiol 65:2124–2129. https://doi.org/10.1099/ijs.0.000229
Acknowledgements
The authors are grateful to Prof. Xiaoxia Zhang (Key Laboratory of Microbial Resources Collection and Preservation, Ministry of Agriculture, Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences) for her help to in naming the new species, and Prof. Aharon Oren (Department of Plant and Environmental Sciences, The Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem) for his help to check the name of this new species in this paper.
Funding
This research was funded by the Fundamental Research Funds for the Central Non-profit Research Institution of Chinese Academy of Forestry (CAFYBB2020SZ006 and CAFYBB2022XE001).
Author information
Authors and Affiliations
Contributions
JY and LL: conceived the study, participated in its design and coordination, and drafted the manuscript; FY and SW: performed the measurements and analysed the data; LL and JY: conceived the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animal experiments. No specific ethical or institutional permits were required to conduct sampling and the experimental studies did not involve endangered or protected species.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yue, J., Yang, F., Wang, S. et al. Massilia phyllostachyos sp. nov., Isolated from the Roots of Moso Bamboo in China. Curr Microbiol 80, 54 (2023). https://doi.org/10.1007/s00284-022-03163-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03163-2