Abstract
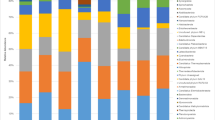
This study aimed to compare the fungal rhizosphere communities of Rhazya stricta, Enneapogon desvauxii, Citrullus colocynthis, Senna italica, and Zygophyllum simplex, and the gut mycobiota of Poekilocerus bufonius (Orthoptera, Pyrgomorphidae, “Usherhopper”). A total of 164,485 fungal reads were observed from the five plant rhizospheres and Usherhopper gut. The highest reads were in S. italica rhizosphere (29,883 reads). Species richness in the P. bufonius gut was the highest among the six samples. Ascomycota was dominant in all samples, with the highest reads in E. desvauxii (26,734 reads) rhizosphere. Sordariomycetes and Dothideomycetes were the dominant classes detected with the highest abundance in C. colocynthis and E. desvauxii rhizospheres. Aspergillus and Ceratobasidium were the most abundant genera in the R. stricta rhizosphere, Fusarium and Penicillium in the E. desvauxii rhizosphere and P. bufonius gut, Ceratobasidium and Myrothecium in the C. colocynthis rhizosphere, Aspergillus and Fusarium in the S. italica rhizosphere, and Cochliobolus in the Z. simplex rhizosphere. Aspergillus terreus was the most abundant species in the R. stricta and S. italica rhizospheres, Fusarium sp. in E. desvauxii rhizosphere, Ceratobasidium sp. in C. colocynthis rhizosphere, Cochliobolus sp. in Z. simplex rhizosphere, and Penicillium sp. in P. bufonius gut. The phylogenetic results revealed the unclassified species were related closely to Ascomycota and the species in E. desvauxii, S. italica and Z. simplex rhizospheres were closely related, where the species in the P. bufonius gut, were closely related to the species in the R. stricta, and C. colocynthis rhizospheres.










Similar content being viewed by others
References
Abarenkov K, Henrik NR, Larsson KH et al (2010) The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186(2):281–285
Al-Essa MA, Al-Mehaidib A, Al-Gain S (1998) Parental awareness of liver disease among children in Saudi Arabia. Ann Saudi Med 18(1):79–81
Allred BW (1968) Range management training handbook for Saudi Arabia. FAO, Rome
Andert J, Marten A, Brandl R, Brune A (2010) Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.). FEMS Microbiol Ecol 74(2):439–449
Assaeed AM (1996) Hammada elegans-Rhazya stricta relationships in deteriorated range site in Raudhat Al-Khafs, Saudi Arabia. J Agric Sci Mansoura Univ 21(3):957–964
Baeshen NA, Sabir JS, Zainy MM et al (2014) Biodiversity and DNA barcoding of soil fungal flora associated with Rhazya stricta in Saudi Arabia. Bothalia 44(5):301–314
Baldrian P, Kolařík M, Stursová M et al (2012) Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258
Blackwell M (2011) The Fungi: 1, 2, 3, 5.1 million species? Am J Bot 98(3):426–438
Bloemberg GV, Lugtenberg BJJ (2001) Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 4:343–350
Brucker RM, Bordenstein SR (2012) Speciation by symbiosis. Trends Ecol Evol 27(8):443–451
Buée M, Reich M, Murat C et al (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456
DeSalle R, Graham SW, Fazekas AJ et al (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3:e2802
Douglas WF, Bing M, Pawel G, Naomi S, Sandra O, Rebecca MB (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6
Elshafie AE, Al Siyabi FM, Salih FM, Ba Omar T, Al Bahry SN, Al Kindi S (2003) The mycobiota of herbal drug plants in Oman and possible decontamination by gamma radiation. Phytopathol Mediterr 42(2):149–154
Engelen B, Meinkein K, von Wintzingerode F, Heuer H, Malkomes H-P, Backhaus H (1998) Monitoring impact of a pesticide treatment on bacterial soil communities by metabolic and genetic fingerprinting in addition to conventional testing procedures. Appl Environ Microbiol 64:2814–2821
Fraune S, Bosch TC (2010) Why bacteria matter in animal development and evolution. BioEssays 32:571–580
Geml J, Gravendeel B, van der Gaag KJ et al (2014) The contribution of DNA metabarcoding to fungal conservation: diversity assessment, habitat partitioning and mapping red-listed fungi in protected coastal Salix repens communities in the Netherlands. PLoS ONE 9(6):e99852
Hamady M, Knight R (2009) Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–1152
Hirsch PR, Mauchline TH, Clark IM (2010) Culture-independent molecular techniques for soil microbial ecology. Soil Biol Biochem 42:878–887
Hollister EB, Schadt CW, Palumbo AV, Ansley RJ, Boutton TW (2010) Structural and functional diversity of soil bacterial and fungal communities following woody plant encroachment in the southern Great Plains. Soil Biol Biochem 42:1816–1824
Hongoh Y (2010) Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci Biotechnol Biochem 74:1145–1151
Hongoh Y, Deevong P, Inoue T et al (2005) Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71:6590–6599
Hongoh Y, Ekpornprasit L, Inoue T et al (2006) Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol Ecol 15:505–516
Huang XF, Bakker MG, Judd TM, Reardon KF, Vivanco JM (2013) Variations in diversity and richness of gut bacterial communities of termites (Reticulitermes flavipes) fed with grassy and woody plant substrates. Microb Ecol 65:531–536
Huse SM, Ye Y, Zhou Y, Fodor AA (2012) A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE 7:e34242
Kernaghan G (2005) Mycorrhizal diversity: cause and effect? Pedobiologia 49:511–520
Kim M, Yoon H, You Y-H et al (2013) Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. J Microbiol Biotechnol 23(10):1347–1356
Lee YK, Mazmanian SK (2010) Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330:1768–1773
Lehman RM, Lundgren JG, Petzke LM (2009) Bacterial communities associated with the digestive tract of the predatory ground beetle, Poecilus chalcites, and their modification by laboratory rearing and antibiotic treatment. Microb Ecol 57:349–358
Magoc T, Salzberg S (2011) FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Massol-Deya A, Weller R, Rios-Hernandez L, Zhou J-Z, Hickley RF, Tiedje JM (1997) Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol 63:270–276
McGuire KL, Fierer N, Bateman C, Treseder KK, Turner BL (2012) Fungal community composition in neotropical rain forests: the influence of tree diversity and precipitation. Microb Ecol 63:804–812
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190
Moussa TAA, Al-Zahrani HS, Almaghrabi OA, Abdelmoneim TS, Fuller MP (2017) Comparative metagenomics approaches to characterize the soil fungal communities of western coastal region. Saudi Arabia PLoS ONE 12(9):e0185096
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH (2008) Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinformatics Online 4:193–201
Orgiazzi A, Bianciotto V, Bonfante P et al (2013) 454 pyrosequencing analysis of fungal assemblages from geographically distant, disparate soils reveals spatial patterning and a core mycobiome. Diversity 5:73–98
Peay KG, Baraloto C, Fine PV (2013) Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J 7:1852–1861
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res 41(D1):D590–D596
Ranjard L, Poly F, Lata J-C, Mougel C, Thioulouse J, Nazaret S (2001) Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl Environ Microbiol 67(10):4479–4487
Schäfer H, Bernard L, Courties C et al (2001) Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol Ecol 34:243–253
Schmit JP, Mueller GM (2007) An estimate of the lower limit of global fungal diversity. Biodivers Conserv 16:99–111
Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E (2010) Commensal bacteria play a role in mating preference of Drosophila melanogaster. PNAS USA 107:20051–20056
Smit E, Leeflang P, Wernars K (1997) Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol Ecol 23:249–261
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449:804–810
Turner TR, Ramakrishnan K, Walshaw J et al (2013) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7:2248–2258
Verbruggen E, Kiers ET, Bakelaar PNC, Röling WFM, van der Heijden MGA (2012) Provision of contrasting ecosystem services by soil communities from different agricultural fields. Plant Soil 350:43–55
Warnecke F, Luginbuhl P, Ivanova N et al (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565
Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751
Wu YT, Wubet T, Trogisch S, Both S, Scholten T, Bruelheide H, Buscot F (2013) Forest age and plant species composition determine the soil fungal community composition in a Chinese subtropical forest. PLoS ONE 8:e66829
Yasir M, Azhar EI, Khan I et al (2015) Composition of soil microbiome along elevation gradients in southwestern highlands of Saudi Arabia. BMC Microbiol 15:65
You YH, Yoon H, Lee GS et al (2011) Diversity and plant growth-promotion of endophytic fungi isolated from the roots of plants in Dokdo islands. J Life Sci 21:992–996
You YH, Yoon H, Woo JR et al (2011) Plant growth-promoting activity of endophytic fungi isolated from the roots of native plants in Dokdo islands. J Life Sci 21:1619–1624
You YH, Yoon H, Kim H et al (2013) Plant growth-promoting activity and genetic diversity of endophytic fungi isolated from native plants in Dokdo Islands for restoration of a coastal ecosystem. J Life Sci 23:95–101
Yu L, Nicolaisen M, Larsen J, Ravnskov S (2013) Organic fertilization alters the community composition of root-associated fungi in Pisum sativum. Soil Biol Biochem 58:36–41
Zak DR, Holmes WE, White DC, Peacock AD, Tilman D (2003) Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84:2042–2050
Acknowledgment
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. (HiCi 1-363-1434H). The authors, therefore, acknowledge with thanks DSR for technical and financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Noor, S.O., Al-Zahrani, D.A., Hussein, R.M. et al. Assessment of fungal diversity in soil rhizosphere associated with Rhazya stricta and some desert plants using metagenomics. Arch Microbiol 203, 1211–1219 (2021). https://doi.org/10.1007/s00203-020-02119-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02119-z