Abstract
Purpose
As an invasive plant, Parthenium hysterophorus severely impacts the ecological environment of the Yellow River Delta and reduces biodiversity in the invaded areas. The effects of P. hysterophorus invasion on the local environment became increasingly critical, while few information was available for the effects of P. hysterophorus invasion on soil bacteria. The present study aimed to reveal the impacts of hysterophorus on the fungal communities in the Yellow River Delta.
Methods
Sixteen soil samples including four groups (ROOT group, YRR group, YNR group, and GBS group) were collected. High-throughput methods were used to explore the fungal composition of the P. hysterophorus-invaded surrounding environment and native plant-growed environment.
Results
Our results showed that the ACE (351.97) and Chao1 (351.95) values of the rhizosphere soils of P. hysterophorus (YRR group) were the highest among the four groups, whereas the non-rhizosphere soil samples of P. hysterophorus (YNR group) had the highest Shannon (7.188) and Simpson (0.984) values. The total number of operational taxonomic units (OTUs) obtained from the four groups was 1965, with 161 common OTUs among different groups. At the phylum level, both Ascomycota and Basidiomycota were the dominant fungi, with Ascomycota having the highest abundance. At the genus level, except for the endophytic fungi of P. hysterophorus roots (ROOT group), Fusarium, Mortierella, Comoclathris, and Cladosporium were the dominant fungi in three groups. The fungal communities within the roots of P. hysterophorus were distant from other groups, indicating that the composition of the fungal communities within the roots had a low degree of similarity to the other three groups. LEfSe analysis showed that Ascomycota at the phylum level and Cladosporium, Curvularia, and Alternaria at the genus level play essential roles in the ROOT group, and Comoclathris plays a vital role in the YNR group.
Conclusions
This study explored the effects of P. hysterophorus invasion on the local soil fungal communities by analyzing the fungal communities in P. hysterophorus roots, rhizosphere soil, non-rhizosphere soil, and rhizosphere soil of native plants. Generally, P. hysterophorus rhizosphere fungi specifically affect the surrounding environment.
Similar content being viewed by others
Introduction
The globalization of the economy has led to an increase in trade volume among countries, resulting in the introduction of more alien species to meet various domestic needs. As a result, the phenomenon of alien biological invasion has intensified and attracted significant attention both domestically and abroad (Dutta 2018). This invasion of alien species can cause a reduction in biodiversity and disrupt the balance of the ecosystem (Courchamp et al. 2017). The competitive ability of alien species often surpasses that of native species, leading to a loss of dominance by the latter and the eventual destruction of the local ecosystem. Given their crucial role in the biogeochemical cycle, soil microorganisms are indispensable to the wetland ecosystem (Trap et al. 2016). The microbial community in rhizosphere soil plays varying ecological roles during different stages of plant growth. Studies have shown that rhizosphere microorganisms can assist plants in absorbing soil nutrients, improving plant resistance, and protecting plants from pathogens. A favorable soil microbial composition is essential for the formation of good soil structure, ecosystem function, and material cycle (Wang et al. 2017). Invasion of exotic plants results in changes in the soil microbial community and physical and chemical properties (Shang et al. 2023), which impacts the competitive relationship between exotic and native species and further contributes to the invasion of alien plants (Shang et al. 2022a). For instance, when alien species invade new habitats, their rhizosphere probiotic communities dominate the habitats, accelerating the host’s adaptation to the environment to a certain extent (Zhang et al. 2020).
The Yellow River Delta (YRD) is recognized as one of China’s significant estuarine deltas (Zhang et al. 2016) and is renowned for its highly diverse coastal wetland ecosystem (Yu et al. 2012). However, the YRD’s ecological balance is threatened by the invasion of invasive species such as Spartina alterniflora (Shang et al. 2022b). The adaptability, reproduction, and competitive ability of invasive species negatively impact native plants, soil microbial communities, and physical and chemical properties of the local environment (Zheng and Liao 2017). Parthenium hysterophorus L., a plant species from the family Asteraceae with an erect stem and main root, is an annual plant native to the USA and northern Mexico (Batish et al. 2002). P. hysterophorus L. is known for its strong reproductive capacity via sexual or asexual reproduction, with its pollen potentially causing allergic reactions and other diseases. This invasive species can also affect the growth of other species, alter the physical and chemical properties of soil, and disrupt the ecological balance of the invaded area, subsequently reducing local biodiversity and impeding economic development. After being identified in Yunnan Province, China, P. hysterophorus rapidly spread south of the Yangtze River, and it was found in Shandong Province in 2004, inflicting damage to local crops and the environment. We investigated the bacterial relationship between the P. hysterophorus community and the native community. However, the relationship between the rhizosphere soil and the soil fungal community of P. hysterophorus is unclear. Therefore, we selected four types of micro-habitats, including the P. hysterophorus roots, P. hysterophorus rhizosphere soil, P. hysterophorus non-rhizosphere soil, and native plant-growed soil, and studied related soil physical and chemical properties (pH, EC, and total organic carbon [TOC]). Furthermore, we used high-throughput sequencing techniques conducted to analyze the fungi in these habitats. In this study, the differences in soil fungal communities and soil physical and chemical properties between invaded areas and native plant communities were compared to understand the diversity of soil fungi and the effects of P. hysterophorus invasion on soil physical and chemical properties and the fungi community. This research provides a theoretical basis for better control of the P. hysterophorus wetland ecosystem invasion.
Materials and methods
Sample site
Soil samples were collected from the vicinity of Siyuan Lake in the YRD of Binzhou (37°16′ N–38°16′ N, 118°20′ E–119°20′ E), which is situated in a warm temperate monsoon climate zone with distinct continental meteorological features and significant seasonal variations. The average annual temperature of the region is 18.3 °C. The land severely invaded by P. hysterophorus was selected as the sample site, where it formed a single community. The density of the P. hysterophorus in the plot is 58, sporadically accompanied by Setaria viridis (L.) Beauv (Shang et al. 2023).
Sample collection and determination of soil physical and chemical properties
In the sample plots, root, rhizosphere soil, and non-rhizosphere soil of P. hysterophorus and rhizosphere soil of S. viridis were collected. A rhizosphere/non-rhizosphere soil sample was mixed with five sub-samples collected. The soil samples were then divided into two parts. One part was used to determine the physical and chemical properties of the soil, whereas the other part was immediately transferred to a low-temperature refrigerator (− 80 °C) for subsequent sequencing of fungal communities. The root tissue of P. hysterophorus was also collected aseptically and a low-temperature refrigerator (− 80 °C) for subsequent sequencing of fungal communities.
The pH of the soil sample was determined using a pH meter in the supernatant of the soil–water mixture with a water/soil ratio of 5:1 (Shang et al. 2023). The EC of the sample was measured using an electrical conductivity meter in the supernatant of a soil–water mixture with a water/soil ratio of 5:1 (Shang et al. 2023). The TOC in the soil samples was determined by the volumetric potassium dichromate external heating method, as described in a previous study (Shang et al. 2023).
High-throughput sequencing of soil microbes
The soil samples stored at − 80 °C were dispensed into 1.5-ml centrifuge tubes and placed on an ultra-clean workbench. Each treatment was repeated four times. The endophytic fungi of P. hysterophorus roots were labeled as ROOT (ROOT1, ROOT2, ROOT3, and ROOT4), the fungi in the rhizosphere soil of P. hysterophorus were labeled as YRR (YRR1, YRR2, YRR3, and YRR4), the non-rhizosphere soil samples of P. hysterophorus were labeled as YNR (YNR1, YNR2, YNR3, and YNR4), and the rhizosphere soil of native plants was labeled as GBS (GBS1, GBS2, GBS3, and GBS4).
The DNA of the soil was extracted by using the soil DNA kit. PCR amplification of ITS1F and ITS2R (ITS1F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′; ITS2R: 5′-GCTGCGTTCTTCATCGATGC-3′) was performed using universal primers for PCR amplification of the fungal ITS region (Cui et al. 2019). The PCR products from the same sample were mixed and used AxyPrep DNA Gel Extraction Kit recycle product purification. Furthermore, the NEXTFLEX Rapid DNA-Seq Kit was used to build the library. The library was sequenced using Illumina's NovaSeq 6000.
Data analysis
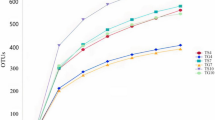
The alpha diversity index was calculated using the Qiime software (version 2.0) and SPSS 23.0 Student’s t test (IBM SPSS Inc., USA) (Bolyen 2019), and dilution curves and PCoA graphs were generated using R package ggplot (version 3.4.4) (McMurdie and Holmes 2013). The sequencing data obtained in this study are available at NCBI (accession number: PRJNA956238).
Results
Alpha diversity analysis
Alpha diversity is used to measure the species abundance and diversity of individual samples, and there are various measuring indices, including Chao1, ACE, Shannon, and Simpson (Fig. 1). Chao1 and ACE indices were used to estimate species abundance, whereas Shannon’s and Simpson’s indices were used to measure diversity of species abundance and community evenness. The YRR group had the highest ACE (351.97) and Chao1 (351.95) indices among the four groups, indicating that the abundance of the rhizosphere soil fungal communities of P. hysterophorus was higher than that of the other three groups. The YNR group had the second-highest ACE (339.33) and Chao1 (335.75) indices.
Alpha diversity indices of fungi among different groups. a ACE index. b Chao1 index. c Shannon index. d Simpson index. GBS, rhizosphere soil of native plants (S. viridi); Root, endophytic fungi of P. hysterophorus roots; YNR, non-rhizosphere soil samples of P. hysterophorus; YRR, fungi in the rhizosphere soil of P. hysterophorus. The number on the line between the columns is the P value of the T test (if the P value > 0.05, the P value is not displayed by default)
The YNR group showed the highest Shannon index (7.188) and Simpson index (0.984) among the four groups, indicating a higher diversity of fungal communities in the non-rhizosphere soil of P. hysterophorus. The Chao1 (277.25), ACE (278.30), Shannon (5.947), and Simpson (0.944) indices of the ROOT group were the lowest among the four groups, indicating the lowest abundance and diversity of fungal communities in the roots of P. hysterophorus.
OTU abundance analysis
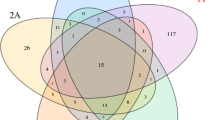
In our study, we identified 967 OTUs in the YRR group, 943 OTUs in the YNR group, 787 OTUs in the GBS group, and 835 OTUs in the ROOT group (Fig. 2). The total number of OTUs obtained from different groups was 1965, with 161 common OTUs found between the four groups. Specifically, 448 OTUs were shared between the YNR and YRR groups, 425 between the YRR and GBS groups, and 368 between the YNR and GBS groups. The number of OTUs in the rhizosphere and non-rhizosphere soils of P. hysterophorus was higher than that in P. hysterophorus roots.
Venn diagram showing the number of shared and unique OTUs among different groups. Different colors mean different groups. GBS, rhizosphere soil of native plants (S. viridi); Root, endophytic fungi of P. hysterophorus roots; YNR, non-rhizosphere soil samples of P. hysterophorus; YRR, fungi in the rhizosphere soil of P. hysterophorus
Soil microbial community structure analysis
Based on species annotation results, the top 10 species ranked by richness at the phylum level (excluding “others”) were used to generate a column accumulation map, as shown in Fig. 3. At the phylum level, the dominant fungi in the rhizosphere and non-rhizosphere of P. hysterophorus were Ascomycota, Basidiomycota, and Mortierellomycota, with a total relative abundance of 83.8% and 85.8%, respectively. The dominant fungi within the P. hysterophorus roots were Ascomycota and Basidiomycota, with a total relative abundance of 88.1%. In the rhizosphere soil of the native plants, the dominant fungi were Ascomycota, Basidiomycota, and Glomeromycota, with a total relative abundance of 85.1%. Ascomycota and Basidiomycota were also dominant in all four groups, with Ascomycota having the highest abundance at 54.7% (YRR), 64.9% (YNR), 78.1% (GBS), and 56.5% (ROOT).
The top 10 genera ranked in abundance, except for unclassified species, were selected to generate a column accumulation map, as shown in Fig. 4. At the genus level, the dominant fungi were Fusarium, Mortierella, Comoclathris, and Cladosporium in three groups. In the P. hysterophorus root, the dominant fungi were Fusarium, Comoclathris, Cladosporium, and Alternaria, with a total relative abundance of 41.7%.
PCoA analysis
PCoA analysis can be used to visualize the differences or similarities between different groups and compare the variability of fungal communities among samples. Our results showed that the contributions of PC1 and PC2 were 10.12% and 8.82%, respectively (Fig. 5). The fungal communities of P. hysterophorus were similar in composition to those of the rhizosphere soil of native plants, as they were close in distance. Conversely, the fungal communities within the roots of P. hysterophorus were distant from the other groups, indicating that the composition of the fungal communities within the roots had a low degree of similarity to the other three groups.
The PCoA based on the Bray–Curtis distance shows the variation in bacterial community structure.GBS, rhizosphere soil of native plants (S. viridi); Root, endophytic fungi of P. hysterophorus roots; YNR, non-rhizosphere soil samples of P. hysterophorus; YRR, fungi in the rhizosphere soil of P. hysterophorus
LEfSe analysis
LEfSe analysis can identify species with significant differences between different groups. In the ROOT group, Ascomycota at the phylum level and Cladosporium, Curvularia, and Alternaria at the genus level play important roles. In contrast, Comoclathris plays a crucial role in the YNR group (Fig. 6).
LEfSe analysis at different fungal taxonomic levels among different groups. Different colored dots mean the taxa with significant differences among different samples. GBS, rhizosphere soil of native plants (S. viridi); Root, endophytic fungi of P. hysterophorus roots; YNR, non-rhizosphere soil samples of P. hysterophorus; YRR, fungi in the rhizosphere soil of P. hysterophorus
Correlation between soil fungal species and soil physicochemical properties
The clustered heat map displays the effects of different environmental factors on soil microorganisms. Our results indicated that Olpidiomycota and Mucoromycota are correlated with pH, whereas Ascomycota is associated with EC. Additionally, Mucoromycota and Mortierellomycota are related to TOC levels (Fig. 7). The pH, EC, and TOC data were submitted as a supplement (Table S1).
Anosim (analysis of similarities), also known as similarity analysis, is a statistical method primarily used to analyze the similarity between groups of multidimensional data. The Binary–Jaccard algorithm is used to calculate the inter-sample distance, and both the R value and P value are calculated. A higher R value indicates that the difference between groups is more significant than the difference within groups. Conversely, a smaller R value suggests no significant difference between or within groups. In this study, the R value was 0.292 (P value = 0.002), indicating that the difference between groups is significant and substantial (Fig. 8).
Discussion
Previous studies have shown that plant invasion can significantly increase the pH of invaded soils (Dzurendova et al. 2020). However, in the present study, we found that the pH of the rhizosphere soil of P. hysterophorus did not differ significantly from that of the non-rhizosphere soil, and the TOC content was significantly lower than that of the non-rhizosphere soil. The dominant flora is a crucial factor in maintaining the stability of microbial communities and can significantly influence the composition and structure of such communities. In this study, the dominant phyla found in the soil samples were Ascomycota and Basidiomycota, which was consistent with the previously reported dominant fungal taxa in soil (Deveau et al. 2018). Interestingly, during the invasion of P. hysterophorus, the phylum Glomeromycota, which was the dominant fungal group in the rhizosphere soil of native plants, was gradually replaced by the saprophytic fungus Mortierellomycota. Mortierellomycota can decompose lignin and other difficult-to-decompose substances, thereby accelerating soil nutrient circulation and improving soil quality (Landinez-Torres et al. 2020). We speculate that the rhizosphere soil microorganisms of P. hysterophorus may be responsible for accelerating the decomposition of soil organics. At the genus level, the content of Mortierella in the rhizosphere soil of P. hysterophorus was higher than that of the native plants. Mortierella is known to produce polyunsaturated fatty acids and has been reported to act as a beneficial flora, inhibiting the growth of pathogenic bacteria in some crops (Sun et al. 2013). In our study, we investigated the endophytic fungi in the roots of P. hysterophorus. Endophytic fungi have been reported as significant contributors to the competitiveness of exotic plants, and their high diversity indicates that they possess complex and versatile functional characteristics. Our previous study found that only 377 OTUs were identified in the Root group (Shang et al. 2023), while 835 OTUs of the ROOT group were identified in the present study. We speculated that compared with endophytic bacteria, the endophytic fungi might have more important functions in the root of P. hysterophorus. Ultimately, we found that Alternaria was the dominant group of root endophytes and a potentially pathogenic fungus at the genus level. Further analysis using LEfSe showed that Alternaria plays an essential role within the roots of P. hysterophorus. Thus, Alternaria in the roots of P. hysterophorus may indirectly serve as a weapon to invade in the proper direction. Our previous study found that the Acidobacteriota might be necessary for P. hysterophorus invasion (Shang et al. 2023). In general, the invasion strategy of P. hysterophorus was not only reflected at the bacterial level, but also at the fungal level.
There are some disparities in microbial diversity and richness between the rhizosphere soil and non-rhizosphere soil of P. hysterophorus. In the present study, the diversity of rhizosphere soil fungi is lower than that of non-rhizosphere soil. Whereas for the bacterial diversity in our previous study, the richness is higher than that of non-rhizosphere soil (Shang et al. 2023). Exotic plants can alter the soil microbial diversity and community structure at the invasion site during the invasion process. This enhances their ability to adapt to the environment and facilitates their invasion and growth (Sun et al. 2013). Soil fungi play a crucial role in the ecosystem as decomposers, contributing to the degradation of organic matter and the conversion of soil nutrients (Frac et al. 2018). The results showed that the invasion of P. hysterophorus caused changes in soil microbial composition and structure, which, in turn, affected the physical and chemical properties of the soil. This favored the growth of P. hysterophorus, allowing it to form a dominant community and impacting the changes in plant community diversity at the invaded site. Finally, High-throughput sequencing technology helps us better understand the impacts of plant invasions on the surrounding environment. Previous studies found that the sequencing alone could not adequately assess population size and dynamics (Beule et al. 2021). Thus, we need to increase the number of samples and detection frequency in subsequent experiments.
Conclusions
This study explored the effects of P. hysterophorus invasion on the local soil fungal communities by analyzing the fungal communities in P. hysterophorus roots, rhizosphere soil, non-rhizosphere soil, and rhizosphere soil of native plants. Ascomycota at the phylum level and Cladosporium, Curvularia, and Alternaria at the genus level play essential roles in the ROOT group, and Comoclathris plays a vital role in the YNR group. Generally, P. hysterophorus rhizosphere fungi specifically affect the surrounding environment.
Availability of data and materials
The obtained sequencing data was available in NCBI (accession numbers PRJNA956238).
References
Batish DR, Kohli RK, Singh HP (2002) Invasive potential of ragweed parthenium (Parthenium hysterophorus L.) with reference to its biology and ecology
Beule L, Arndt M, Karlovsky P (2021) Relative abundances of species or sequence variants can be misleading: soil fungal communities as an example. Microorganisms 9:589
Bolyen E (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 (vol 37, pg 852, 2019). Nat Biotechnol 37:1091–1091
Courchamp F, Fournier A, Bellard C, Bertelsmeier C, Bonnaud E, Jeschke JM, Russell JC (2017) Invasion biology: specific problems and possible solutions. Trends Ecol Evol 32:13–22
Cui YX, Bing HJ, Fang LC, Wu YH, Yu JL, Shen GT, et al (2019) Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338:118-27. https://doi.org/10.1016/j.geoderma.2018.11.047
Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, Hacquard S, Herve V, Labbe J, Lastovetsky OA, Mieszkin S, Millet LJ, Vajna B, Junier P, Bonfante P, Krom BP, Olsson S, van Elsas JD, Wick LY (2018) Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev 42:335–352
Dutta H (2018) Insights into the phenomenon of alien plant invasion and its synergistic interlinkage with three current ecological issues. J Asia-Pacific Biodivers 11:188–198
Dzurendova S, Zimmermann B, Tafintseva V, Kohler A, Ekeberg D, Shapaval V (2020) The influence of phosphorus source and the nature of nitrogen substrate on the biomass production and lipid accumulation in oleaginous Mucoromycota fungi. Appl Microbiol Biotechnol 104(18):8065-76. https://doi.org/10.1007/s00253-020-10821-7
Frac M, Hannula SE, Belka M, Jedryczka M (2018) Fungal biodiversity and their role in soil health. Front Microbiol 9:707
Landinez-Torres AY, Abril JLB, Tosi S, Nicola L (2020) Soil microfungi of the colombian natural regions. Int J Environ Res Public Health 17: 8311
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. Plos One 8:e61217
Shang S, Hu SX, Liu XX, Zang Y, Chen J, Gao N, Li LY, Wang J, Liu LX, Xu JK, Zhang YM, Wu T, Tang XX (2022a) Effects of Spartina alterniflora invasion on the community structure and diversity of wetland soil bacteria in the Yellow River Delta. Ecology Evol 12:e8905
Shang S, Li LY, Zhang ZW, Zang Y, Chen J, Wang J, Wu T, Xia JB, Tang XX (2022b) The effects of secondary growth of Spartina alterniflora after treatment on sediment microorganisms in the Yellow River Delta. Microorganisms 10:1722
Shang S, Zhang Z, Zhao L, Liu L, Shi D, Xu H, Zhang H, Xie W, Zhao F, Zhou Z, Xu J, Wang J (2023) Effect of Parthenium hysterophorus L. Invasion on soil microbial communities in the Yellow River Delta, China. 11;18
Sun X, Gao C, Guo LD (2013) Changes in soil microbial community and enzyme activity along an exotic plant Eupatorium adenophorum invasion in a Chinese secondary forest. Chin Sci Bull 58:4101–4108
Trap J, Bonkowski M, Plassard C, Villenave C, Blanchart E (2016) Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 398:1–24
Wang R, Zhang HC, Sun LG, Qi GF, Chen S, Zhao XY (2017) Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci Rep 7:343
Yu Y, Wang H, Liu J, Wang Q, Shen TL, Guo WH, Wang RQ (2012) Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River Estuary. Eur J Soil Biol 49:12–21
Zhang H, Chen XB, Luo YM (2016) An overview of ecohydrology of the Yellow River delta wetland. Ecohydrol Hydrobiol 16:39–44
Zhang ZJ, Liu YJ, Brunel C, van Kleunen M (2020) Soil-microorganism-mediated invasional meltdown in plants. Nat Ecol Evol 4:1612–1621
Zheng YL, Liao ZY (2017) High-density native-range species affects the invasive plant Chromolaena odorata more strongly than species from its invasive range. Sci Rep 7:16075.
Informed consent
Not applicable.
Funding
This work was supported by the Natural Science Foundation of Shandong Province (ZR2021QD082).
Author information
Authors and Affiliations
Contributions
Methodology, L.G. and X.X.; Software, W.S.; Writing—original draft preparation, L.G., X.X. and S.S.; writing—review and editing, Z.Z. and J.Z. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the agreed to publish.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Soil physical and chemical property data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gong, L., Xin, X., Song, W. et al. Impact of Parthenium hysterophorus L. invasion on soil fungal communities in the Yellow River Delta. Ann Microbiol 73, 33 (2023). https://doi.org/10.1186/s13213-023-01735-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13213-023-01735-6