Abstract
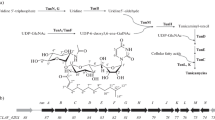
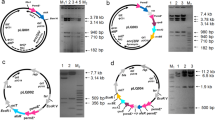
Streptomyces albus J1074 is one of the most popular and convenient hosts for heterologous expression of gene clusters directing the biosynthesis of various natural metabolic products, such as antibiotics. This fuels interest in elucidation of genetic mechanisms that may limit secondary metabolism in J1074. Here, we report the generation and initial study of J1074 mutant, deficient in gene bldA for tRNALeu UAA, the only tRNA capable of decoding rare leucyl TTA codon in Streptomyces. The bldA deletion in J1074 resulted in a highly conditional Bld phenotype, with depleted formation of aerial hyphae on certain solid media. In addition, bldA mutant of J1074 was unable to produce endogenous antibacterial compounds and two heterologous antibiotics, moenomycin and aranciamycin, whose biosynthesis is directed by TTA-containing genes. We have employed a new TTA codon-specific β-galactosidase reporter system to provide genetic evidence that J1074 bldA mutant is impaired in translation of TTA. In addition, we have discussed the possible reasons for differences in the phenotypes of bldA mutants described here and in previous studies, providing knowledge to study bldA-based regulation of antibiotic biosynthesis.




Similar content being viewed by others
References
Anagnostopoulos C, Spizizen J (1961) Requirements for transformation of Bacillus subtilis. J Bacteriol 81(5):741–746
Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37(8):759–772. doi:10.1007/s10295-010-0730-9
Bilyk B, Luzhetskyy A (2014) Unusual site-specific DNA integration into the highly active pseudo-attB of the Streptomyces albus J1074 genome. Appl Microbiol Biotechnol 98(11):5095–5104. doi:10.1007/s00253-014-5605-y
Braña AF, Rodríguez M, Pahari P, Rohr J, García LA, Blanco G (2014) Activation and silencing of secondary metabolites in Streptomyces albus and Streptomyces lividans after transformation with cosmids containing the thienamycin gene cluster from Streptomyces cattleya. Arch Microbiol 196:345–355. doi:10.1007/s00203-014-0977-z
Chater K, Chandra G (2008) The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J Microbiol 46:1–11
Chater KF, Wilde LC (1980) Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol 116(2):323–334
Datsenko K, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Fedoryshyn M, Petzke L, Welle E, Bechthold A, Luzhetskyy A (2008) Marker removal from actinomycetes genome using Flp recombinase. Gene 419(1–2):43–47. doi:10.1016/j.gene.2008.04.011
González A, Rodríguez M, Braña AF, Méndez C, Salas JA, Olano C (2016) New insights into paulomycin biosynthesis pathway in Streptomyces albus J1074 and generation of novel derivatives by combinatorial biosynthesis. Microb Cell Fact 21:56. doi:10.1186/s12934-016-0452-4
Gramajo H, Takano E, Bibb M (1993) Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolorA3(2) is transcriptionally regulated. Mol Microbiol 7(6):837–845
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546
Guthrie EP, Chater KF (1990) The level of a transcript required for production of a Streptomyces coelicolor antibiotic is conditionally dependent on a tRNA gene. J Bacteriol 172:6189–6193. doi:10.1128/jb.172.11.6189-6193.1990
Herrmann S, Siegl T, Luzhetska M, Petzke L, Jilg C, Welle E, Erb A, Leadlay PF, Bechthold A, Luzhetskyy A (2012) Site-specific recombination strategies for engineering actinomycete genomes. Appl Environ Microbiol 78(6):1804–1812. doi:10.1128/AEM.06054-11
Horbal L, Fedorenko V, Luzhetskyy A (2014) Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria. Appl Microbiol Biotechnol 98(20):8641–8655. doi:10.1007/s00253-014-5918-x
Iqbal HA, Low-Beinart L, Obiajulu JU, Brady SF (2016) Natural product discovery through improved functional metagenomics in Streptomyces. J Am Chem Soc 138(30):9341–9344. doi:10.1021/jacs.6b02921
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
King AA, Chater KF (1986) The expression of the Escherichia coli lacZ gene in Streptomyces. J Gen Microbiol 132(6):1739–1752. doi:10.1099/00221287-132-6-1739
Kuzniar A, van Ham RC, Pongor S, Leunissen JA (2008) The quest for orthologs: finding the corresponding gene across genomes. Trends Genet 24:539–551. doi:10.1016/j.tig.2008.08.009
Lamichhane TN, Blewett NH, Crawford AK, Cherkasova VA, Iben JR, Begley TJ, Farabaugh PJ, Maraia RJ (2013) Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 33:2918–2929. doi:10.1128/MCB.00278-13
Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP (2013) Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154:416–429. doi:10.1016/j.cell.2013.06.043
Leskiw BK, Lawlor EJ, Fernandez-Abalos JM, Chater KF (1991) TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc Natl Acad Sci USA 88:2461–2465
Leskiw BK, Mah R, Lawlor EJ, Chater KF (1993) Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J Bacteriol 175:1995–2005. doi:10.1128/jb.175.7.1995-2005.1993
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77(1):112–143. doi:10.1128/MMBR.00054-12
Lopatniuk M, Ostash B, Luzhetskyy A, Walker S, Fedorenko V (2014) Generation and study of the strains of streptomycetes—heterologous hosts for production of moenomycin. Russ J Genet 50(4):360–365
Luzhetskyy A, Mayer A, Hoffmann J, Pelzer S, Holzenkämper M, Schmitt B, Wohlert SE, Vente A, Bechthold A (2007) Cloning and heterologous expression of the aranciamycin biosynthetic gene cluster revealed a new flexible glycosyltransferase. ChemBioChem 8(6):599–602
Majer J, Chater KF (1987) Streptomyces albus G produces an antibiotic complex identical to paulomycins A and B. J Gen Microbiol 133:2503–2507. doi:10.1099/00221287-133-9-2503
Makitrynskyy R, Rebets Y, Ostash B, Zaburannyi N, Rabyk M, Walker S, Fedorenko V (2010) Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37:559–566
Makitrynskyy R, Ostash B, Tsypik O, Rebets Y, Doud E, Meredith T, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V (2013) Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol 3:130121. doi:10.1098/rsob.130121
Moukadiri I, Garzón MJ, Björk GR, Armengod ME (2014) The output of the tRNA modification pathways controlled by the Escherichia coli MnmEG and MnmC enzymes depends on the growth conditions and the tRNA species. Nucleic Acids Res 42:2602–2623. doi:10.1093/nar/gkt1228
Myronovskyi M, Rosenkränzer B, Luzhetskyy A (2014) Iterative marker excision system. Appl Microbiol Biotechnol 98(10):4557–4570. doi:10.1007/s00253-014-5523-z
Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ (2003) Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J Bacteriol 185:7291–7296. doi:10.1128/JB.185.24.7291-7296.2003
Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA (2014) Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol 7:242–256. doi:10.1111/1751-7915.12116
Ostash B, Campbell J, Luzhetskyy A, Walker S (2013) MoeH5: a natural glycorandomizer from the moenomycin biosynthetic pathway. Mol Microbiol 90:1324–1338. doi:10.1111/mmi.12437
Pettersson BM, Kirsebom LA (2011) tRNA accumulation and suppression of the bldA phenotype during development in Streptomyces coelicolor. Mol Microbiol 79:1602–1614. doi:10.1111/j.1365-2958.2011.07543.x
Pope MK, Green BD, Westpheling J (1996) The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell–cell signalling. Mol Microbiol 19:747–756. doi:10.1046/j.1365-2958.1996.414933.x
Rebets YV, Ostash BO, Fukuhara M, Nakamura T, Fedorenko VO (2006) Expression of the regulatory protein LndI for landomycin E production in Streptomyces globisporus 1912 is controlled by the availability of tRNA for the rare UUA codon. FEMS Microbiol Lett 256(1):30–37
Rodriguez L, Oelkers C, Aguirrezabalaga I, Braña AF, Rohr J, Méndez C, Salas JA (2000) Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J Mol Microbiol Biotechnol 2(3):271–276
Rodríguez A, Olano C, Vilches C, Méndez C, Salas JA (1993) Streptomyces antibioticus contains at least three oleandomycin-resistance determinants, one of which shows similarity with proteins of the ABC-transporter superfamily. Mol Microbiol 8(3):571–582
Rokytskyy I, Koshla O, Fedorenko V, Ostash B (2016) Decoding options and accuracy of translation of developmentally regulated UUA codon in Streptomyces: bioinformatics analysis. Springerplus 5:982. doi:10.1186/s40064-016-2683-6
Sakai Y, Miyauchi K, Kimura S, Suzuki T (2016) Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res 44:509–523. doi:10.1093/nar/gkv1470
Sambrook J, Russell DW (2001) Molecular cloning, a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New-York
Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF (2003) A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486
Trepanier N, Jensen S, Alexander D, Leskiw B (2002) The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology 148:643–656
Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Björk GR (2003) Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9:760–768. doi:10.1261/rna.5210803
Wang J, Schully KL, Pettis GS (2009) Growth-regulated expression of a bacteriocin, produced by the sweet potato pathogen Streptomyces ipomoeae, that exhibits interstrain inhibition. Appl Environ Microbiol 75:1236–1242. doi:10.1128/AEM.01598-08
Xu D, Kwon HJ, Suh JW (2008) S-Adenosylmethionine induces BldH and activates secondary metabolism by involving the TTA-codon control of bldH expression in Streptomyces lividans. Arch Microbiol 189:419–426. doi:10.1007/s00203-007-0336-4
Zaburannyi N, Rabyk M, Ostash B, Fedorenko V, Luzhetskyy A (2014) Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom 15:97. doi:10.1186/1471-2164-15-97
Zaburannyy N, Ostash B, Fedorenko V (2009) TTA Lynx: a web-based service for analysis of actinomycete genes containing rare TTA codon. Bioinformatics 25:2432–2443. doi:10.1093/bioinformatics/btp402
Acknowledgements
This work was supported by grant BG-41Nr from the Ministry of Education and Science of Ukraine (to B.O.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koshla, O., Lopatniuk, M., Rokytskyy, I. et al. Properties of Streptomyces albus J1074 mutant deficient in tRNALeu UAA gene bldA . Arch Microbiol 199, 1175–1183 (2017). https://doi.org/10.1007/s00203-017-1389-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-017-1389-7