Abstract
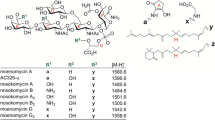
Moenomycin, a natural phosphoglycolipid product that has a long history of use in animal nutrition, is currently considered an attractive starting point for the development of novel antibiotics. We recently reconstituted the biosynthesis of this natural product in a heterologous host, Streptomyces lividans TK24, but production levels were too low to be useful. We have examined several other streptomycetes strains as hosts and have also explored the overexpression of two pleiotropic regulatory genes, afsS and relA, on moenomycin production. A moenomycin-resistant derivative of S. albus J1074 was found to give the highest titers of moenomycin, and production was improved by overexpressing relA. Partial duplication of the moe cluster 1 in S. ghanaensis also increased average moenomycin production. The results reported here suggest that rational manipulation of global regulators combined with increased moe gene dosage could be a useful technique for improvement of moenomycin biosynthesis.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baizman ER, Branstrom AA, Longley CB, Allanson N, Sofia MJ, Gange D, Goldman RC (2000) Antibacterial activity of synthetic analogues based on the disaccharide structure of moenomycin, an inhibitor of bacterial transglycosylase. Microbiology 146:3129–3140
Baltz RH (1998) Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol 6:76–83
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Champness W (2000) Actinomycete development, antibiotic production and phylogeny: questions and challenges. In: Brun YV, Skimkets LJ (eds) Prokaryotic development. Am Soc Microbiol, Washington, DC, pp 11–31
Chater KF, Wilde LC (1980) Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol 116:323–334
Chen L, Lu Y, Chen J, Zhang W, Shu D, Qin Z, Yang S, Jiang W (2008) Characterization of a negative regulator AveI for avermectin biosynthesis in Streptomyces avermitilis NRRL8165. Appl Microbiol Biotechnol 80:277–286
Datsenko K, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Fedorenko V, Golets L, Yu Demydchuk, Kriugel H (1998) Analysis of genome rearrangements in Streptomyces kanamyceticus mutants. Antibiot Khimitoter (Rus) 43:14–19
Floriano B, Bibb M (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol Microbiol 21:385–396
Goldman RC, Gange D (2000) Inhibition of transglycosylation involved in bacterial peptidoglycan synthesis. Curr Med Chem 7:801–820
Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN (2005) Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol 58:1276–1287
Kieser T, Bibb M, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Lee J, Hwang Y, Kim S, Kim E, Choi C (2000) Effect of a global regulatory gene, afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng 89:606–608
Lian W, Jayapal KP, Charaniya S, Mehra S, Glod F, Kyung YS, Sherman DH, Hu WS (2008) Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2). BMC Genomics 9:56
Lovering AL, de Castro LH, Lim D, Strynadka NC (2007) Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315:1402–1405
Luzhetskyy AM, Ostash BO, Fedorenko VO (2001) Intergeneric conjugation Escherichia coli–Streptomyces globisporus 1912 with using of integrative plasmid pSET152 and its derivative. Rus J Genet 37:1340–1347
McKenzie NL, Nodwell JR (2007) Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J Bacteriol 189:5284–5292
Ostash B, Walker S (2005) Bacterial transglycosylase inhibitors. Curr Opin Chem Biol 9:459–466
Ostash B, Saghatelian A, Walker S (2007) A streamlined metabolic pathway for the biosynthesis of moenomycin A. Chem Biol 14:257–267
Ostash B, Makitrinskyy R, Walker S, Fedorenko V (2009) Identification and characterization of Streptomyces ghanaensis ATCC14672 integration sites for three actinophage-based plasmids. Plasmid 61:171–175
Ostash B, Doud EH, Lin C, Ostash I, Perlstein DL, Fuse S, Wolpert M, Kahne D, Walker S (2009) Complete characterization of seventeen-step moenomycin biosynthetic pathway. Biochemistry 48:8830–8841
Ostash I, Ostash B, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V (2008) Coordination of export and glycosylation of landomycins in Streptomyces cyanogenus S136. FEMS Microbiol Lett 285:195–202
Sambrook J, Russel DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sánchez C, Butovich IA, Braña AF, Rohr J, Méndez C, Salas JA (2002) The biosynthetic gene cluster for the antitumor rebeccamycin: characterization and generation of indolocarbazole derivatives. Chem Biol 9:519–531
Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K (1996) Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J Bacteriol 178:7276–7284
Stinchi S, Azimonti S, Donadio S, Sosio M (2003) A gene transfer system for glycopeptide producer Nonomuraea sp. ATCC39727. FEMS Microbiol Lett 225:53–57
Sun J, Hesketh A, Bibb M (2001) Functional analysis of relA and rshA, two relA/spoT homologues of Streptomyces coelicolor A3(2). J Bacteriol 183:3488–3498
Vogtli M, Chang PC, Cohen SN (1994) afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol Microbiol 14:643–653
Taylor J, Li X, Oberthür M, Zhu W, Kahne D (2006) The total synthesis of moenomycin A. J Am Chem Soc 128:15084–15085
Yanai K, Murakami T, Bibb M (2006) Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc Natl Acad Sci USA 103:9661–9666
Yuan Y, Fuse S, Ostash B, Sliz P, Kahne D, Walker S (2008) Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design. ACS Chem Biol 3:429–436
Acknowledgments
This work was supported by grant Bg-01F from the Ministry of Education and Science of Ukraine (to V. F.) and NIH grant AI50855 (to S. W.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Makitrynskyy, R., Rebets, Y., Ostash, B. et al. Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37, 559–566 (2010). https://doi.org/10.1007/s10295-010-0701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0701-1