Abstract
Key message
A new functional Pm21 haplotype, Pm21(8#), was cloned from the new wheat-H. villosa translocation line T6VS(8#)·6DL, which confers the same strong resistance to powdery mildew through a different resistance mechanism.
Abstract
Broad-spectrum disease resistance genes are desirable in crop breeding for conferring stable, durable resistance in field production. Pm21(4#) is a gene introduced from wild Haynaldia villosa into wheat that confers broad-spectrum resistance to wheat powdery mildew and has been widely used in wheat production for approximately 30 years. The discovery and transfer of new functional haplotypes of Pm21 into wheat will expand its genetic diversity in production and avoid the breakdown of resistance conferred by a single gene on a large scale. Pm21(4#) previously found from T6VS(4#)·6AL has been cloned. In this study, a new wheat-H. villosa translocation, T6VS(8#)·6DL, was identified. A new functional Pm21 haplotype, designated Pm21(8#), was cloned and characterized. The genomic structures and the splicing patterns of Pm21(4#) and Pm21(8#) were different, and widespread sequence diversity was observed in the gene coding region and the promoter region. In the field, Pm21(8#) conferred resistance to Blumeria graminis f. sp. tritici (Bgt), similar to Pm21(4#), indicating that Pm21(8#) was also a resistance gene. However, Bgt development during the infection stage was obviously different between Pm21(4#)- and Pm21(8#)-containing materials under the microscopic observation. Pm21(4#) inhibited the formation of haustoria and the development of hyphae in the initial infection stage, while Pm21(8#) limited the growth of hyphae and inhibited the formation of conidiophores in the late infection stage. Therefore, Pm21(8#) is a new functional Pm21 haplotype that provides a new gene resource for wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat powdery mildew is a worldwide fungal disease caused by the obligate biotrophic plant pathogen Blumeria graminis f. sp. tritici (Bgt), which belongs to the Ascomycotas group Erysiphales in the fungal kingdom (Takamatsu 2004). The disease can occur in leaves, stems and spikes, leading to a severe effect on wheat growth and a great decease in grain yield (Griffey et al. 1993; Randhawa et al. 2019). Therefore, the breeding and cultivation of powdery mildew resistant varieties is a highly efficient and eco-friendly way to ensure food security in major wheat-producing countries (Randhawa et al. 2019; Dreiseitl 2021). The identification of powdery mildew (Pm) genes and the development of germplasms containing Pm genes are the basis for breeding resistant varieties. To date, more than 100 Pm genes have been explored, among which 25 came from the wild relatives of wheat (Zhu et al. 2022), but only a few of these genes have been used in disease resistance breeding. Most of the Pm genes confer race-specific resistance, which is short-lived due to the rapid changes in virulence of the Bgt population (Zeng et al. 2014; Jin et al. 2021; Wu et al. 2021). In addition, Pm genes from the wild species are usually accompanied by unfavorable traits; therefore, a few Pm genes from the wild species, such as Pm8 from Secale cereale and Pm21 from Haynadia villosa, have been widely used in wheat production (Wu et al. 2021). Therefore, the identification and utilization of Pm genes with broad-spectrum, strong effect are the most urgent and challenging tasks.
In China, wheat powdery mildew is one of the most devastating fungal diseases in all of the ecological areas of wheat production. Resistance genes have played an important role in preventing powdery mildew in wheat. Pm21 confers high-level, broad-spectrum resistance to powdery mildew, which was transferred from H. villosa to wheat through the cytogenetical development of the wheat-H. villosa translocation T6VS·6AL (Chen et al. 1995). The T6VS·6AL line has been widely used and has made significant contributions to the disease control in the main wheat-producing regions of China. In the southwestern wheat planting area, the proportions of elite lines carrying Pm21 are high, and in the middle and lower reaches of the Yangtze River, approximately 28.3% of commercial varieties carry the Pm21 gene (Wu et al. 2021). According to the statistical data from 2003 to 2012, the planting area of Pm21-carrying varieties was more than 4 million hectares (Xing et al. 2018). In recent years, due to the increase in the number of released varieties carrying Pm21, the planting area has also significantly expanded. There have been many cases in which resistance genes, including Pm1, Pm3a, Pm3b, Pm3c, Pm3f, Pm5, Pm7 and Pm8, have been overcome after large-scale application due to the emergence of new virulent isolates (Wang et al. 2005). For example, Pm8, a resistance gene transferred to wheat through the development of the wheat-S. cereale translocation T1RS·1BL, has played a major role in the prevention of powdery mildew worldwide (Hsam et al. 1997). In China, the percentage of Pm8-carrying varieties was as high as 70% in the 1980s (Zhou et al. 2004). However, Pm8 lost its resistance in most areas due to large-scale application in the end of 1980s, leading to the extreme lack of powdery mildew resistant varieties all over the country (Sheng et al. 1995). Currently, the large-scale, frequent use of a single Pm21 gene in some ecological areas of China is imposing strong selection pressure on Bgt, which will probably lead to the resistance breakdown. Therefore, it is particularly important to explore and reasonable layout new genes or new functional haplotypes of known genes with broad-spectrum resistance to powdery mildew.
Haynaldia villosa (2n = 14, VV), an annual cross-pollinated diploid species indigenous to the Mediterranean and Caucasus regions, provides a valuable tertiary gene pool for wheat improvement. More than 300 H. villosa accessions have been collected from their native habitats, and these accessions show high genetic diversity and displaying extremely high value for the improvement of tolerance to biotic and abiotic stresses and agronomic traits (De Pace et al. 2011; Chen et al. 2002; Grądzielewska et al. 2006). However, only a few accessions have been used to develop wheat-H. villosa alien chromosome lines, and even fewer H. villosa accessions have played roles in successful wheat breeding through the developed germplasms. According to De Pace (2011), seven accessions have been successively used to synthesize hexaploid amphidiploids, including Hv(1#) (Sears 1953), Hv(2#) (Lukaszewski 1988), Hv(3#) (Urbano 1988), Hv(4#) (Liu et al. 1988; Chen et al. 1995), Hv(5#) (Chen et al. 1996; Li et al. 2005), Hv(6#) (Ma et al. 1997) and Hv(7#) (De Pace 2001).
Previously, the total number of Pm genes was reported to be more than 100 because of multiple haplotypes at some loci, including Pm1, Pm2, Pm3, Pm4, Pm5 and Pm24, etc. Genetic diversity provides an opportunity to explore new functional resistance haplotypes for breeding. For example, more than 17 Pm3 haplotypes have been identified, and some haplotypes still show high breeding value, even though some haplotypes have completely lost their ability to confer resistance (Brunner et al. 2012; Koller et al. 2018). Therefore, the mining of new functional haplotypes from previously identified loci is a promising method for addressing resistance gene deficiency.
Xing et al. (2018) cloned Pm21(4#) from Hv(4#) via combinational approaches, and Li et al. (2020) cloned Pm21(5#) from Hv(5#) through a homology-based approach. H. villosa displays high genetic diversity, and more importantly, the Pm21 haplotypes show high sequence polymorphism. Therefore, it is possible to identify new Pm21 haplotypes conferring resistance mediated by alternative molecular mechanisms.
In this study, to identify a novel functional Pm21 haplotype, a new wheat-H. villosa translocation line, T6VS(8#)·6DL, was developed and characterized. The high and broad-spectrum resistance of T6VS(8#)·6DL is mediated by Pm21(8#). However, Pm21(8#) shows dramatic sequence polymorphism, different transcriptional patterns, and a special resistance mechanism. Therefore, the identified Pm21(8#) haplotype, with high genetic diversity and a distinct resistance mechanism, provides a new resistance haplotype and new resistant germplasm for breeding.
Materials and methods
Plant materials
The wheat variety Nannong9918, named as T6VS(4#)·6AL, was developed by the Cytogenetic Institute of Nanjing Agricultural University (CINAU). The wheat variety Yangmai 22, named as T6VS(5#)·6DL, was introduced from the Lixiahe Agricultural Research Institute of Jiangsu Academy of Agricultural Sciences. T6VS (8#)·6DL was developed by CINAU through chromosome engineering. The 6VS fragments of T6VS(4#)·6AL, T6VS(5#)·6DL and T6VS(8#)·6DL were derived from different H. villosa accessions, and Pm21 haplotypes display high diversity in their nucleotide and protein sequences. Powdery mildew-susceptible cv. Nannong0686 was developed by CINAU and used as the recurrent parent to develop the Nannong0686-Pm21(8#) and Nannong0686-Pm21(5#) lines.
Inoculation of Bgt and evaluation of powdery mildew resistance
The powdery mildew resistance of T6VS(8#)·6DL was evaluated at the adult stage in the field and at the seedling stage in the greenhouse. In the field, T6VS(8#)·6DL was infected by the local mixed races of Bgt at the Baima Experimental Station of Nanjing Agricultural University (Nanjing, China). The Bgt isolate E26 was obtained from Dr. Yilin Zhou of the Chinese Academy of Agricultural Sciences and maintained on seedlings of the susceptible variety Nannong0686 in the greenhouse under a 14/10-h light/dark photoperiod (24/18 °C temperature, 70% relative humidity). In the greenhouse, Bgt inoculation was performed by spraying fresh conidiospores of E26 from the susceptible host Nannong0686 onto the seedling leaves. T6VS(4#)·6AL, T6VS(5#)·6DL and T6VS (8#)·6DL plants at the two-leaf stage were inoculated with E26 and harvested at different time points to observe Bgt development, detect H2O2 accumulation and analyze gene expression. The infection types (ITs) were scored on a 0–4 scale, with 0 indicating no visible symptoms; 1 indicating minute colonies with few conidia; 2 indicating colonies with moderately developed hyphae, but few conidia; 3 indicating colonies with well-developed hyphae and abundant conidia, but colonies not joined together; and 4 indicating colonies with well-developed hyphae and abundant colonies that are mostly joined together (Liu et al. 1999).
Silencing of Pm21(8#) by barley stripe mosaic virus induced gene silencing (BSMV-VIGS)
BSMV-VIGS was performed as described by Hu et al. (2018) and Xing et al. (2018). Pm21(8#) was silenced by BSMV-VIGS in T6VS(8#)·6DL to evaluate the powdery mildew resistance function. Previously, the 225 bp fragment specific to Pm21(4#) was amplified and inserted into the γ-strain of BSMV to produce the recombinant vector for silencing Pm21(4#) (Xing et al. 2018), and this recombinant vector was also used for silencing Pm21(8#) in this study. The second fully expanded leaves were infected with the in vitro-transcribed virus BSMV:Pm21(8#), and BSMV:TaPDS- and BSMV:γ-infected leaves were used as controls. Then, the fourth fully expanded leaves with clear virus infection symptoms were used to evaluate powdery mildew resistance and to analyze the target gene silencing efficiency. The leaves were placed on the 20 mg/l 6-BA agar medium and inoculated with fresh Bgt E26 in a greenhouse with a 14-h light (25 °C)/10-h dark (18 °C). Then the resistance level was recorded at 7 days after Bgt inoculation. Leaves were bleached with ethanol:acetic acid (3:1) and stained with Coomassie blue (6 mg/ml) to observe fungal development under an Olympus BX-60 microscope (Olympus, Tokyo, Japan). The gene silencing efficiency was analyzed by qRT‒PCR on an LC 480II instrument (Roche, Colorado Springs, CO, USA).
Cloning and sequence analysis of Pm21 haplotypes
The genomic sequences and transcripts of Pm21(8#) were obtained according to the sequences of Pm21(4#) through homology-based cloning. The nucleotide and codon protein sequences of Pm21(4#), Pm21(5#) and Pm21(8#), were aligned by using DNAMAN software (https://www.lynnon.com/). The cis-element prediction in promoters from the three genes was performed on PlantCare (https://www.plantcaretools.com/). The putative domains of the cloned genes were analyzed using SMART (http://smart.embl-heidelberg.de/) and visualized with TBtools software (https://github.com/CJ-Chen/TBtools). The phylogenetic tree was constructed by using MEGA7 software base on neighbor-joining algorithms, and bootstrapping was performed 1000 times to obtain support values for each branch (Kumar et al. 2016). The tree was visualized using iTOL (Letunic et al. 2021). The conserved motifs of the NLR protein were annotated using MAST (version 4.9.1) (Bailey et al. 2009) and the ‘motif 1’ to ‘motif 20’ were defined by Jupe (2012).
Genomic in situ hybridization (GISH) and florescence in situ hybridization (FISH)
To identify the introgressed 6VS chromosomal fragment in the translocation lines, genomic in situ hybridization (GISH) and fluorescence in situ hybridization (FISH) were performed following Du et al. (2017), using root-tip cells at mitotic metaphase with AMP soaking and nitrous oxide treatment as described by Komuro et al. (2013). Total genomic DNA of H. villosa (91C43) labeled with 5’ FAM was used as the GISH probe, which produced green fluorescence signals. Oligo-pAs1-1 and oligo-pAs1-4 labeled with 5’ TAMRA in a mixture were used as the probes for FISH analysis, which produced red fluorescence signals. Moreover, the wheat chromosomes were counterstained with DAPI and visualized based on blue fluorescence. Oligo-painting as described by Wang et al. (2017), using pAs1-1, pAs1-3, pAs1-4, pAs1-6, AFA-3, AFA-4, pSc119.2–1 and (GAA)10 as the mixed probes, was performed to identify the polymorphism of these repeat sequences among the three 6VS chromosome arms. The hybridized slides were observed under an Olympus BX60 fluorescence microscope with a CCD camera DP72 (Olympus, Tokyo, Japan) for image acquisition, and individual chromosomes with hybridization signals were cropped using the Adobe Photoshop software CS 7.0 (Adobe Systems, San Jose, CA, USA).
Development of molecular markers and analysis of the genetic population
Molecular markers specific to Pm21(8#), including Pm21_588, Pm21_1178 and Pm21_2683, were developed according to the sequences comparison of Pm21(8#) with Pm21(5#) and Pm21(4#), and the primers are listed in Table S1. For the allelic testing of Pm21(5#) and Pm21(8#), an F2 population was derived by crossing T6VS(5#)·6DL and T6VS(8#)·6DL. The Pm21 haplotypes of the two translocation parents, which were recombinant plants homozygous for Pm21(5#) and Pm21(8#) (designated F2-Pm21(5#) and F2-Pm21(8#), respectively) were analyzed based on specific markers. PCR amplification was performed using a T100TM Thermal Cycler (Bio-Rad Laboratories, Hercules, California, USA) in a 10 μL reaction solution containing 80–100 ng of genomic DNA, 2.5 mmol L−1 of each dNTP, 2 μmol L−1 of each primer, 2.5 mmol L−1 MgCl2 and 0.2 U Taq DNA polymerase. The PCR cycling conditions were as follows: 94 °C for 4 min, followed by 32 cycles of 94 °C for 15 s, 58 to 60 °C for 45 s, and 72 °C for 60 s, with a final extension at 72 °C for 10 min. The PCR products were separated on 8% nondenaturing PAGE gels (Acr:Bis = 39:1) and visualized by silver staining.
Observation of Bgt development and detection of H2O2 accumulation
To compare Bgt development and H2O2 accumulation mediated by different Pm21 haplotypes, the third leaves of T6VS(4#)·6AL, T6VS(5#)·6DL and T6VS(8#)·6DL and plants of the selected individuals from the F2 population with homozygous Pm21(5#) and Pm21(8#) were inoculated with Bgt isolate E26 at 24, 48, 72 and 120 h after inoculation. Then, the detached leaves were bleached in ethanol:acetic acid (3:1, v/v) at the 37 °C incubator overnight and finally stained with Coomassie Brilliant Blue R250 for 5 min and examined under an Olympus BX40 microscope (Olympus, Tokyo, Japan). The leaves were also stained with DAB solution (1 mg/ml, pH 3.8) for 8 h, decolorized in ethanol:acetic acid (3:1), and observed under the same microscope. The numbers of whole cells in which H2O2 was distributed were counted in approximately 200 to 300 Bgt-interacted epidermal cells of each leaf in both the resistant plants and the susceptible plants, and three leaves of each plant were sampled as an experimental repetition. Three independent replicate experiments were performed, and their results were statistically analyzed using SPSS 16.0 software.
Analysis of the haustorium index via a single-cell GUS expression assay
The single-cell transient overexpression assay was performed according to Hu et al. (2018) described. The vector pWMB002 carrying the β-glucuronidase (GUS) gene driven by the ubiquitin promoter was used as the reporter of the transformed epidermal cells. Seedlings of the Nannong0686 and Nannong9918, Nannong0686-Pm21(8#) and Nannong0686-Pm21(5#) lines and the F2-Pm21(5#) and F2-Pm21(8#) lines were grown under the growth conditions with a 16-h light (18 °C)/8-h dark (16 °C) cycle and constant relative humidity of 70%. In the two-leaf stage, primary leaves were cut and placed on 20 mg/l 6-BA agar plates. The plasmid pWMB002 was extracted with a High Purity Plasmid Extraction Kit (TIANGEN, Beijing, China), coated with the particles and delivered into wheat epidermal cells through bombardment (PDS-1000/He Bio-Rad, California, USA). The bombarded leaves were incubated in the dark at 18 °C for 4 to 6 h before high-density inoculation with Bgt conidiospores. The leaves were stained for GUS at 24 h after Bgt inoculation, and after 18–24 h of GUS staining, both the GUS-expressing cells and the haustoria of the Bgt-infected cells were visible based on blue coloration under an Olympus BX40 microscope (Olympus, Tokyo, Japan). The haustorium index was calculated as described by Hu et al. (2018). Three independent replicated experiments were carried out, and for each replicate included the examination of 100–300 successfully GUS-expressing cells interaction with Bgt. The data were statistically analyzed using SPSS 16.0 software.
Results
Development of the new wheat-H. villosa translocation T6VS(8#)·6DL
Phenotypic evaluation of different accessions of H. villosa (2n = 14, VV) revealed the high morphological polymorphism, indicating high genetic diversity. H. villosa 01I140, Hv(8#), which originated from Italy, was selected as the male parent, and the tetraploid wheat cv. Zhongyin1286 was used as the female parent to synthesize T. durum-H. villosa amphidiploid (2n = 14, AABBVV) (Zhang et al. 2018). Then, the amphidiploid was used as the V genome for backcrossing with Nannong0686 to induce wheat-H. villosa translocation involved different chromosome arms of V genome. In the backcross-derived offspring, the wheat-H. villosa translocation line T6VS·6DL was identified, which showed high resistance to powdery mildew. The results of cytogenetic analysis showed that seven pairs of chromosomes displayed a red signal, indicating that they were D genome since pAs1-1 and pAs1-4 were the D-genome specific probes. However, one pair of chromosomes showed a strong green signal on the short arm indicating that they were translocated chromosome between wheat and H. villosa (Fig. 1a–d). Because the long arm of the translocated chromosome was 6DL, so we proposed that the short arm was 6VS, produced by the recombination between 6D and 6V. Previously, we developed a molecular marker, CINAU15, to distinguish the 6AS, 6BS, 6DS and 6VS (Cao et al. 2006). The marker analysis showed that the bands specific to 6AS, 6BS, and 6DS were amplified from Nannong0686, bands specific to 6DS, 6BS, and 6VS were amplified from the wheat-H.villosa translocation line T6VS(4#)·6AL, and bands specific to 6AS, 6BS, and 6VS were amplified from the wheat-H.villosa translocation line T6VS(5#)·6DL and T6VS(8#)·6DL (Fig. 1e). These results indicated that in T6VS(8#)·6DL, the 6DS was substituted by 6VS(8#). Interestingly, the size of the bands produced from 6VS of T6VS(8#)·6DL was different from those of T6VS (5#)·6DL and T6VS (4#)·6AL, indicating the existence of genetic diversity between different V genomes. Therefore, the newly developed germplasm T6VS(8#)·6DL is a novel wheat-H. villosa translocation line.
Cytogenetic identification and molecular marker analysis of the new wheat-H. villosa translocation T6VS(8#)·6DL, a–b Genomic in situ hybridization (GISH) and fluorescence in situ hybridization (FISH) analysis of T6VS(5#)·6DL and T6VS(8#)·6DL on root tip metaphase chromosomes. Green fluorescence represents GISH analysis signal obtained using the total genomic DNA of H. villosa as a probe, and the red fluorescence represents the FISH analysis signal obtained using oligo-pAs as a probe. Chromosomes were counterstained with DAPI (blue). The translocation chromosomes are indicated with white arrows. c–d GISH-FISH patterns of the D-genome chromosomes of the two translocation lines. The chromosomes were extracted from a and b respectively, and the two translocation chromosomes, 6VS(5#)·6DL and 6VS(8#)·6DL, were identified and are indicated. e PCR analysis of the codominant molecular marker CINAU15. The chromosome specific bands are labeled on the right, and the band of 6VS(8#) was different from that of 6VS(4#) or 6VS(5#) (color figure online)
Cytogenetic comparison of the T6VS(8#)·6DL with other translocations
Previously, two wheat-H. villosa translocation lines involving 6VS, T6VS(4#)·6AL developed by CINAU and T6VS(5#)·6DL developed by CAAS, have been reported to display complete immunity to powdery mildew. Both germplasms have been used in breeding pipelines to develop many commercial cultivars. To study whether the 6VS(4#), 6VS(5#) and 6VS(8#) display diversity at the level of cytogenetic analysis, oligo-painting was performed to identify the polymorphism of the repeat sequences among the three 6VS chromosome arms (Fig. S1a–c). The results indicated that the green signals, produced by oligo-pSc119.2–1 and oligo-(GAA)10, displayed similar distribution patterns in 6VS(4#), 6VS(5#) and 6VS(8#). However, the red signals, produced by oligo-pAs1-1, oligo-pAs1-3, oligo-pAs1-4, oligo-pAs1-6, oligo-AFA-3 and oligo-AFA-4, showed different distribution patterns in 6VS(4#) from those in 6VS(5#) and 6VS(8#) (Fig. S1d–e). Therefore, the results from both molecular marker and cytogenetic marker analyses identified diversity among different V genomes.
Identification of a new Pm21 haplotype in T6VS(8#)·6DL
Evaluation of the powdery mildew resistance of T6VS(8#)·6DL
The resistance of T6VS(8#)·6DL to powdery mildew was evaluated using the naturally mixed Bgt isolates in the field (Fig. 2b), and T6VS(8#)·6DL showed complete immunity to powdery mildew both in the seedling stage and the adult plant stage. Then, the resistance level of T6VS(8#)·6DL to powdery mildew was compared to that of T6VS (4#)·6AL and T6VS(5#)·6DL in the greenhouse (Fig. 2a). The three translocation lines showed a high level of resistance to the Bgt isolate E26, without visible colonies on their leaf surfaces (Fig. 2). Therefore, T6VS(8#)·6DL showed the same high level of resistance to powdery mildew as T6VS(4#)·6AL and T6VS(5#)·6DL.
Evaluation of the powdery mildew resistance in the wheat-H. villosa translocation line T6VS(8#)·6DL, a Powdery mildew resistance evaluation in T6VS(8#)·6DL at the seedling stage in the greenhouse, using T6VS(4#)·6AL and T6VS(5#)·6DL as the resistant controls and Nannong0686 as the susceptible control. b Powdery mildew resistance evaluation in T6VS(8#)·6DL at the adult plant stage in the field, using T6VS(5#)·6DL as the resistant control
Identification of a new Pm21 allele, Pm21(8#), in T6VS(8#)·6DL
Since both T6VS(4#)·6AL and T6VS(5#)·6DL contain functional Pm21 alleles, designated Pm21(4#) and Pm21(5#), respectively, it was hypothesized that there was a functional Pm21(8#) allele was present in T6VS(8#)·6DL. T6VS(8#)·6DL was crossed with T6VS(5#)·6DL to produce an F2 population for the allelism test of Pm21(5#) in T6VS(5#)·6DL with Pm21(8#) in the T6VS (8#)·6DL. All the F2 individuals showed a high level of resistance similar to those of both parents, with no visible colonies, and no susceptible individual was produced due to chromosome recombination (Fig. S2). Therefore, this indicates that a new Pm21 allele, Pm21(8#), is present in T6VS(8#)·6DL, as inferred from the genetic analysis.
To further verify that Pm21(8#) is a functional allele of Pm21, a probe that was previously used to silence Pm21(4#) in T6VS(4#)·6AL was used to silence Pm21(8#) in T6VS(8#)·6DL by virus gene induced gene silencing (VIGS) mediated by BSMV. The expression level of Pm21(8#) in T6VS(8#)·6DL was significantly decreased based on qRT‒PCR analysis (Fig. 3b). The results indicated that silencing of Pm21(8#) could completely abolish the resistance of T6VS(8#)·6DL, and a small number of colonies could be observed on the leaf surface (Fig. 3a). Microscopic observation also showed that Bgt produced many branches of hyphae and even conidiophores (Fig. 3c–h). This indicates that the resistance of T6VS(8#)·6DL is mediated by Pm21(8#) and that Pm21(8#) is a functional Pm21 allele.
Functional analysis of Pm21(8#) using BSMV-VIGS, a Evaluation of Bgt resistance conferred by Pm21(8#) in T6VS(8#)·6DL by silencing of Pm21(8#). Seven days after Bgt inoculation, no Bgt colonies were visible on the BSMV:γ infected leaves and on the BSMV:PDS infected leaves, while a small number of Bgt colonies were visible on the BSMV:Pm21(8#)-infected leaves, and a large number of Bgt colonies were visible in the susceptible control Nannong0686. b qRT‒PCR analysis of Pm21(8#) expression levels in both BSMV:γ and BSMV: Pm21(8#)-infected individuals. The expression level of Pm21(8#) was decreased significantly in the BSMV:Pm21(8#) infected leaves. c–h Observation of Bgt development under the microscope. Bgt developed more slowly and produced fewer hyphal branches in the BSMV:γ and BSMV:PDS infected leaves than in the BSMV:Pm21(8#)-infected leaves (c and d). Moreover, a large number of conidial chains could be produced in the BSMV:Pm21(8#)-infected leaves but not in the BSMV:γ and BSMV:PDS-infected leaves (e and g). Figures f and h show enlarged views of the red rectangles in e and g, respectively. The white line represents the scale bar 200 μm
Sequence analysis of Pm21(8#) with its haplotypes
Nucleotide sequence and gene structure of Pm21(8#)
We cloned the alleles of Pm21 from 110 accessions of H. villosa using the primers designed according to the Pm21(4#) sequence that we previously reported (Xing et al. 2018), and 99 alleles corresponding to the coding region and 76 alleles corresponding to the promoter were successfully cloned. Sequence analysis indicated that there were high polymorphisms in both the gene coding region and in the promoter region. Using primers specific to Pm21(4#), NLR1-F (5′-ATTGAGATGTCTGCACCGGTCG-3′) and NLR1-R (5′-CTCTCTTCGTTACATAATGTAGTG-3′), the genomic sequences of Pm21(5#) from T6VS(5#)·6DL were cloned, but the amplification of Pm21(8#) from T6VS(8#)·6DL failed, implying the existence of polymorphism between Pm21(8#) and Pm21(4#). Then, another pair of primers, NLR1-F1 (5′-CTACAACCGCATCCTCAATCATACT-3′) and NLR1-R1 (5′-TTAAAGTAAAACTGGGACCACATT-3′), were designed based on the 5′-UTR and 3′-UTR region of Pm21(4#), and the genomic sequence of Pm21(8#) was amplified and cloned. Sequence diversity was widespread in the coding region of genes (Fig. S3). At the same time, significant differences were also identified in the promoter sequences and cis-elements arrangements (Fig. S4a, S4b). The genomic sequence comparison showed that Pm21(8#) displayed 89.2% homology with Pm21(4#) and 90.0% homology with Pm21(5#).
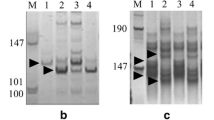
The CDS sequence of Pm21(5#) was amplified from T6VS(5#)·6DL using NLR1-F (5′-ATTGAGATGTCTGCACCGGTCG-3′) and NLR1-R (5′-CTCTCTTCGTTACATAATGTAGTG-3′), and Pm21(8#) was amplified from T6VS(8#)·6DL using NLR1-F1 (5′-CTACAACCGCATCCTCAATCATACT-3′) and NLR1-R1 (5′-TTAAAGTAAAACTGGGACCACATT-3′). Only one type of transcript corresponding to Pm21(5#) was identified, while two types of transcripts corresponding to Pm21(8#) were identified. In our previous study, we identified only one type of transcript corresponding to Pm21(4#) (Xing et al. 2018). The alignment of the genomic sequence with the transcripts indicated that the splicing pattern of Pm21(8#) was different from those of Pm21(4#) and Pm21(5#). Pm21(4#) and Pm21(5#) showed only one type of splicing pattern and contained three exons and two introns, while Pm21(8#) presented two types of splicing patterns. The splicing pattern of Pm21(8#)-1 was the same as those of Pm21(4#) and Pm21(5#), while Pm21(8#)-2 differed in the presence of one more intron in exon 2 (Fig. 4a, Fig. S6a). The RT‒PCR primers were designed such that one primer was located in exon 2 and the other located in exon 3 using samples of T6VS(8#)·6DL inoculated with Bgt for different times, and the results showed that two bands corresponding to Pm(8#)-1 and Pm(8#)-2 were produced, indicating that the alternative splicing did in fact occur in Pm(8#) (Fig. S6b). Then, qRT‒PCR was performed to analyze the expression pattern of Pm(8#)-1 and Pm(8#)-2, and it was found that both transcripts could be induced by Bgt infection (Fig. S6c).
Comparison of the gene structures and protein compositions of the Pm21(4#), Pm21(5#) and Pm21(8#), a Gene structures of Pm21(4#), Pm21(5#) and Pm21(8#) by alignment of the genomic sequences with the corresponding transcripts. An alternative splicing pattern was identified in Pm21(8#) but not in Pm21(4#) and Pm21(5#). Exons are represented by black rectangles, and introns are represented by lines. Their sizes are marked in bp. b Protein compositions of Pm21(4#), Pm21(5#), and Pm21(8#) based on searching the conserved motifs. Pm21(4#), Pm21(5#) and Pm21(8#)-1 were complete NLR proteins, while Pm21(8#)-2 was a truncated protein with a premature stop codon at 200 aa. The Rx_N (CC), NB-ARC and LRR domains were identified respectively, and each conserved motif distributed in the three domains is displayed in a different color (color figure online)
Amino acid sequence comparison of Pm21(8#) with Pm21(4#) and Pm21(5#)
The putative amino acid sequences of Pm21(4#), Pm21(5#) and Pm21(8#) differ in length; Pm21(4#), Pm21(5#), and Pm21(8#)-1 contain 909, 907, and 916 aa, respectively, while Pm21(8#)-2 contains 200 aa due to frameshift causing premature termination by introducing a stop codon (Fig. 4b). Pm21(4#), Pm21(5#) and Pm21(8#)-1 were also scanned to identify the conserved motifs found in NLR proteins using Pfamscan and Interproscan. In the N-terminus, three proteins shared a high level of conservation. The regions corresponding to the amino acids 1–413 of Pm21(4#), 1–413 of Pm21(5#) and 1–423 of Pm21(8#) contained a coiled-coil and NB-ARC domains, with identical motif compositions and arrangements between these proteins. An extra motif_16 was present in the linker region of Pm21(4#) but not in the regions of Pm21(5#) and Pm21(8#). In the C-terminus, the conservation of the motif composition was relatively low; Pm21(4#) consists of Motif_9, Motif_19 and two Motif_11, Pm21(5#) consists of Motif_11, Motif_19 and two Motif_9, while Pm21(8#)-1 consists of three Motif_9 and two Motif_19 (Fig. 4b). Furthermore, the sequence identity of Pm21(8#) with Pm21(4#) is 90.1% and that with Pm21(5#) is 92.7% in the CC-NB-ARC region, while the sequence identity of Pm21(8#) with Pm21(4#) is only 79.0%, and that with Pm21(5#) is 81.7% in the LRR region.
Motif comparisons among all Pm21 alleles
Using the primers specific to Pm21(4#), 101 sequences from 100 H. villosa accessions were cloned. The CDSs of all the alleles were predicted based on the splicing pattern of Pm21(4#), and the corresponding protein sequences were obtained. In total, 70 full-length proteins were predicted with both NBS-ARC and LRR domains, 30 intact proteins were predicted to show only NBS-ARC but no LRRs, and one abnormal protein was predicted to show only 13 amino acids. Then, the redundant proteins were removed and 33 unique proteins were used to construct the phylogenetic tree (Fig. S7). From the diagram of protein compositions, it was found that the NB-ARC domains were highly conserved, while the LRR domains were highly diverse. The tree showed that all Pm21 alleles were divided into three branches, among which Pm21(4#) belonged to the type I, while Pm21(5#) and Pm21(8#) belonged to type III (Fig. S7). It was also found that the protein compositions of Pm21(8#) were unique among all the alleles.
Microscopic observation of the pathogen development in different Pm21 haplotype backgrounds
Although T6VS(8#)·6DL was as resistant to Bgt as T6VS(4#)·6AL and T6VS(5#)·6DL, it was interesting to study whether there was a difference in the resistance process in T6VS(8#)·6DL because of the distinct transcriptional and motif diversity of Pm21(8#). The Bgt development on the leaves of the three lines was observed under a microscope, and an obvious difference in hyphal growth was detected in T6VS(8#)·6DL in the initial Bgt infection stage (Fig. 5). Then, the percentage of conidia producing secondary hyphae on the leaves of the three lines were statistical analyzed at 48 hpi. Pm21(4#) in T6VS(4#)·6AL and Pm21(5#) in T6VS(5#)·6DL showed stronger ability to inhibit the formation of the haustoria and the development of the hyphae than that of Pm21(8#) in T6VS (8#)·6DL (Fig. S9). Although Pm21(8#) showed weaker ability to inhibit the formation of haustoria and the development of hyphae, it reduced the growth of hyphae and inhibited the formation of conidiophores (Fig. 5) based on the comparison of Bgt development in T6VS(8#)·6DL and Nannong0686. This indicated that the resistance mechanism mediated by Pm21(8#) was different from that of Pm21(4#) and Pm21(5#).
Observation of the Bgt development on the leaves of the three translocation lines at different time points after Bgt inoculation, An obvious difference was detected in T6VS(8#)·6DL, in which more conidia could produce hyphae. On the left side of the figure, 24 hpi, 48 hpi, 72 hpi and 120 hpi are the hours post Bgt inoculation, respectively. The white line represents the scale bar 100 μm
Because genetic backgrounds of T6VS(5#)·6DL and T6VS(8#)·6DL are different, it needs to study whether the difference between Pm21(5#) and Pm21(8#) was induced by an element of the genetic backgrounds other than the Pm21 haplotypes themselves. The polymorphic molecular markers, Pm21_588, Pm21_1178, and Pm21_2683, were developed (Fig. S5) for identifying the Pm21(5#) homozygous lines and Pm21(8#) homozygous lines from the F2 population crossing between T6VS(5#)·6DL and T6VS (8#)·6DL (Fig. S8). Then, the randomly selected lines from the Pm21(5#) homozygous lines, and Pm21(8#) homozygous lines were used to compare the Bgt development process. The development of Bgt in the Pm21(5#) homozygous lines was similar as that in T6VS(5#)·6DL, and the development of Bgt in the Pm21(8#) homozygous lines was similar to that in T6VS(8#)·6DL (Fig. S10). Therefore, it was verified that Pm21(8#) inhibited pathogen development in a distinct pattern, different from that of Pm21(4#) and Pm21(5#).
Analysis of the haustorium index in a single-cell GUS-expression assay
To further examine whether the ability to inhibit haustorium formation by Pm21 haplotypes was different, single-cell transient expression of GUS gene was performed to statistically analyze the percentage of haustorium formation in the Pm21(5#) and the Pm21(8#) genetic backgrounds based on counting the visible haustoria in the GUS-expressed cells. The haustorium index in the susceptible Nannong0686 line was 53.6%, the haustorium indexes in Nannong9918 with Pm21(4#), and in Nannong0686-Pm21(5#) and F2-Pm21(5#) were similar, while the haustorium indexes in Nannong0686-Pm21(8#) and F2-Pm21(8#) were significantly higher than those of materials containing Pm21(4#) and Pm21(5#) (Fig S11). Therefore, the observation of Bgt development and the statistical analysis of the haustorium index indicated that the ability of Pm21(8#) to prevent haustorium formation was different from that of Pm21(4#) and Pm21(5#).
ROS-inducing activity of different Pm21 haplotypes
To test the ROS-inducing activity of different Pm21 haplotypes, ROS accumulation at the Bgt interaction sites was observed at 24 hpi and 48 hpi. It was found that ROS accumulation cells after Bgt inoculation were less in materials containing Pm21(8#) than in materials containing Pm21(4#) and Pm21(5#) (Fig. 6). Then, the ROS-induced activity was statistically analyzed in Pm21(5#)- and Pm21(8#)-containing materials, Nannong0686-Pm21(5#) (T6VS(5#).6DL-Pm21(5#)) and Nannong0686-Pm21(8#) (T6VS(8#).6DL-Pm21(8#)), at 24 hpi and 48 hpi. Two types of Bgt-interacting cells were observed; one type with ROS accumulation around the penetration pegs (Type a) and the other type with ROS accumulation in the whole Bgt-interacting cell (Type b) (Fig. S12 a, b). The numbers of Bgt corresponding to ‘Type a’ and ‘Type b’ cells were counted, and the ratio of ROS-producing Bgt spores to the total number of germinated Bgt spores was statistically compared. The ROS-inducing activity of Nannong0686-Pm21(8#) and F2-Pm21(8#) was significantly lower than that of Nannong0686-Pm21(5#) and F2-Pm21(5#). (Fig. S12 c, d). Therefore, it was indicated that the ROS-inducing activity observed after the Bgt infection of Pm21(8#) was lower than that observed after the Bgt infection of Pm21(5#), and this difference was not produced by the genetic background.
Detection of ROS accumulation by DAB staining of Bgt-infected leaves, An obvious difference was detected in materials containing Pm21(8#), including T6VS(8#)·6DL and F2-Pm21(8#), in which fewer Bgt-interacting cells showed ROS accumulation. In the up portion of the figure, 24 hpi and 48 hpi are the hours post Bgt inoculation. The white line represents the scale bar 2 mm
Discussion
Different Pm21 haplotypes regulate powdery mildew resistance by inhibiting Bgt growth at different developmental stages
The Bgt infection process in leaves includes primary tube germination, attached tube germination, appressorium formation, penetration peg development in the tip of appressorium, penetration peg entry into epidermal cells, haustorium formation under the penetration peg, primary hyphae development, secondary penetration peg and secondary haustorium formation under the primary hyphae, secondary hyphae development into large colonies with many branches of hyphae (Glawe 2008). If a conidial chain forms and new conidia are released, there will be a second (or more) round of infection. In the leaves of susceptible Nannong0686, a large number of haustoria formed in the early infection stage, and white colonies produced due to the high density of hyphae and conidia chains in the later infection stage. Pm21(4#) and Pm21(5#) inhibited the formation of haustoria strongly, leading to very few hyphal production. Most powdery mildew resistance genes play a role in disease resistance by inhibiting the formation of haustoria (Zou et al. 2018; Hurni et al. 2013; Xie et al. 2020) However, the haustorium inhibition activity of Pm21(8#) was significantly lower than those of Pm21(4#) and Pm21(5#). So, how does Pm21(8#) exert disease resistance? Hyphal development was inhibited by Pm21(8#) leading to a reduced number of branches, and no conidial chain was formed; thus, no visible white colonies could be observed, and no further rounds of infection could be induced. For inhibiting Bgt development, Pm21(8#) works mainly at the late secondary hyphae producing stage, but Pm21(4#) and Pm21(5#) works mainly at the early haustoria formation stage. Therefore, it was suggested that the resistance mechanism of Pm21(8#) was different from that of Pm21(4#) and Pm21(5#).
The LRR domain of Pm21 haplotypes affects the resistance patterns
Because of the rapid variation in pathogens, maintaining the diversity of NLRs is necessary for plants to withstand the evolutionary battle (Baggs et al. 2017). A number of studies have revealed that natural variations in protein structures and copy number of the NLR genes could change the resistance spectrum to plant pathogens, especially when the mutations and expansions occur in LRR domains. The LRR domains in NLR genes have long been speculated to be involved in determining the plant’s ability to recognize specific pathogen effectors (DeYoung et al. 2006). In maize (Zea mays L.), variations in the LRR domains of the Rp1 locus could alter race-specific resistance to rust (Collins et al. 1999; Sun et al. 2001). Similarly, variations in the LRR domains of the Pm3 and Pm60 loci in wheat and the Mla locus in barley (Hordeum vulgare L.) were tightly related to the resistance spectrum to powdery mildew (Bhullar et al. 2009; Seeholzer et al. 2010; Zou et al. 2022). In the last 20 years, the structure of the R protein has been analyzed to identify important protein domains and key amino acid sites, which has facilitated the molecular design of new variations to obtain new resistance genes. It has been reported that the artificial mutation of NBS or the LRR region of NLR resistance proteins, such as RX and R3a, could change the recognition of effectors and expand the resistance spectrum (Harris et al. 2013; Chapman et al. 2014; Segretin et al. 2014; Stirnweis et al. 2014). Natural diversity of the LRR domains in different Pm21 haplotypes was found in this study, which was conjectured to affect the resistance pattern. Therefore, the resistance mechanism of the Pm21 haplotypes should be identified.
ROS accumulation influenced by Pm21 may be tightly related to the resistance signal
In recent years, with the rapid development of high-throughput sequencing technology, molecular biology and bioinformatics, an increasing number of Pm genes have been cloned. Among the successfully cloned Pm genes, most of them encode NLR-type proteins, except that Pm38 encodes an abscisic acid transporter, Pm46 encodes a hexose transporter, Pm24 encodes a tandem kinase and Pm4a encodes an MCTP-kinase (Krattinger et al. 2009; Moore et al. 2015; Lu et al. 2020; Sánchez-Martín et al. 2021). The diversity of PM resistance proteins indicates that the struggle between wheat and powdery mildew pathogens involves a complex mechanism. ROS production not only is a phenomenon accompanying the interaction between plants and pathogens but also plays an important role in the plant defense response. ROS accumulation at infection sites and in whole cells was shown to be significantly different in Pm21 haplotypes after Bgt inoculation, which indicates that the resistance effects and the underlying resistance mechanisms of Pm21 haplotypes should be different. The powdery mildew fungus is an obligate biotrophic pathogen that extracts nutrients from plant host cells via its haustoria; therefore, the formation of haustoria is a crucial step of Bgt development in the host. In this study, the inhibition of haustoria formation by Pm21(5#) was stronger than by the inhibition mediated by Pm21(8#), while the percentage of ROS accumulation was higher in Pm21(5#) than in Pm21(8#). Therefore, we inferred that ROS accumulation induced by different Pm21 haplotypes after Bgt infection might correlate with its ability to inhibit haustoria formation. However, the inhibition of hyphal growth and the suppression of further conidial chains formation were observed during the late Bgt infection period in Pm21(#8)-containing materials. It was speculated that different intercellular resistance regulation pathways were involved in Pm21(#8) in the post-haustorium formation stage, which needs to be investigated.
Genetically diverse Pm21 haplotypes and 6VS chromosome arms provide new resources for wheat resistance breeding
The large-scale and long-term use of a single or a few disease resistance genes makes it easy to lose resistance. Therefore, the exploration and utilization of genetically diverse disease resistance genes to control crop disease has attracted much attention for a long time. Multiple strategies have been adopted in wheat breeding to increase the durability of resistance genes, such as pyramiding diverse resistance genes in one variety, developing multiline varieties with different resistance genes in the same genetic backgrounds, the rational distribution and rotation of the valuable resistance genes, and the utilization of varieties with different resistance patterns (Brunner et al. 2012; Koller et al. 2018; Hafeez et al. 2021). These strategies can make full use of the diversity of disease resistance genes and have been used in disease resistance breeding. The wild relatives of wheat contain precious gene resources for increasing genetic diversity and belong to the tertiary gene pool for wheat genetic improvement (Qi et al. 2007; Li et al. 2016). The broad-spectrum resistance mediated by Pm21(4#) gene from H. villosa has powerful resistance effect and is easily selected in the field; therefore, overreliance on Pm21(4#) by breeders is responsible for an increasing risk of the resistance breakdown. In addition, all the varieties harboring Pm21(4#) contained the same chromosome arm 6VS(4#), thus reducing the genetic diversity of this chromosome. One of the most effective and economical approaches to combat the high genetic variability of pathogens is to efficiently mine and appropriately deploy broad-spectrum resistance gene haplotypes. In this study, a new chromosome arm 6VS(8#) with a novel Pm21(8#) haplotype was developed and transferred to wheat, which provides both diverse broad-spectrum resistance gene resource Pm21(8#) and genetic germplasm resources including 6VS(8#) for the improvement of disease resistance in the future.
Data availability
The nucleotide sequences of complete CDS with two transcript splicing patterns and the genomic sequence for Pm21(8#) were submitted to NCBI GenBank with accession number: ON505440, ON505441, ON505442. The plant materials reported in the manuscript are freely available to all the readers on reasonable request.
Change history
23 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00122-023-04323-z
References
Baggs E, Dagdas G, Krasileva KV (2017) NLR diversity, helpers and integrated domains: making sense of the NLR identity. Curr Opin Plant Biol 38:59–67. https://doi.org/10.1016/j.pbi.2017.04.012
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci U S A 106:9519–9524. https://doi.org/10.1073/pnas.0904152106
Brunner S, Stirnweis D, Quijano CD, Buesing G, Herren G, Parlange F, Barret P, Tassy C, Sautter C, Winzeler M, Keller B (2012) Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J 10:398–409. https://doi.org/10.1111/j.1467-7652.2011.00670.x
Cao AZ, Wang XE, Chen YP, Zou XW, Chen PD (2006) A sequence-specific PCR marker linked with Pm21 distinguishes chromosomes 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa. Plant Breed 125:201–205. https://doi.org/10.1111/j.1439-0523.2006.01222.x
Chapman S, Stevens LJ, Boevink PC, Engelhardt S, Alexander CJ, Harrower B, Champouret N, McGeachy K, Van Weymers PSM, Chen X, Birch PRJ, Hein I (2014) Detection of the virulent form of AVR3a from phytophthora infestans following artificial evolution of potato resistance gene R3a. PLoS One 9:e110158. https://doi.org/10.1371/journal.pone.0110158
Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ (1995) Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet 91:1125–1128. https://doi.org/10.1007/bf00223930
Chen Q, Conner RL, Li H, Laroche A, Graf RJ, Kuzyk AD (2002) Expression of resistance to stripe rust, powdery mildew and the wheat curl mite in Triticum aestivum x Haynaldia villosa lines. Can J Plant Sci 82:451–456. https://doi.org/10.1094/pdis.2002.86.4.423
Chen X, Xu HJ, Du LP, Shang LM, Han B, Shi A, Xiao S (1996) Transfer of gene resistant to powdery mildew from H.villosa to common wheat by tissue culture. Sci Agric Sin 29:1–8
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376. https://doi.org/10.1105/tpc.11.7.1365
De Pace C, Snidaro D, Ciaffi M, Vittori D, Ciofo A, Cenci A, Tanzarella OA, Qualset CO, Mugnozza GTS (2001) Introgression of Dasypyrum villosum chromatin into common wheat improves grain protein quality. Euphytica 117:67–75. https://doi.org/10.1023/A:1004095705460
De Pace C, Vaccino P, Cionini PG, Pasquini M, Bizzarri M, Qualset CO (2011) Dasypyrum. Springer, Berlin, Heidelberg
DeYoung BJ, Innes RW (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol 7:1243–1249. https://doi.org/10.1038/ni1410
Dreiseitl A (2021) Powdery mildew resistance phenotypes of wheat gene bank accessions. Biol (Basel) 10:846. https://doi.org/10.3390/biology10090846
Du P, Zhuang LF, Wang YZ, Yuan L, Wang Q, Wang DR, Dawadondup TLJ, Shen J, Xu HB, Zhao H, Chu CG, Qi ZJ (2017) Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60:93–103. https://doi.org/10.1139/gen-2016-0095
Glawe DA (2008) The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol 46:27–51. https://doi.org/10.1146/annurev.phyto.46.081407.104740
Gradzielewska A (2006) The genus Dasypyrum-Part 2. Dasypyrum villosum—a wild species used in wheat improvement. Euphytica 152:441–454. https://doi.org/10.1007/s10681-006-9245-x
Griffey CA, Das MK, Stromberg EL (1993) Effectiveness of adult-plant resistance in reducing grain-yield loss to powdery mildew in winter-wheat. Plant Dis 77:618–622. https://doi.org/10.1094/PD-77-0618
Hafeez AN, Arora S, Ghosh S, Gilbert D, Bowden RL, Wulff BBH (2021) Creation and judicious application of a wheat resistance gene atlas. Mol Plant 14:1053–1070. https://doi.org/10.1016/j.molp.2021.05.014
Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci U S A 110:21189–21194. https://doi.org/10.1073/pnas.1311134110
Hsam SLK, Zeller FJ (1997) Evidence of allelism between genes Pm8 and Pm17 and chromosomal location of powdery mildew and leaf rust resistance genes in the common wheat cultivar “Amigo.” Plant Breed 116:119–122. https://doi.org/10.1111/j.1439-0523.1997.tb02164.x
Hu P, Liu JQ, Xu JF, Zhou CY, Cao SQ, Zhou WH, Huang ZP, Yuan SF, Wang XE, Xiao J, Zhang RQ, Wang HY, Zhang SZ, Xing LP, Cao AZ (2018) A malectin-like/leucine-rich repeat receptor protein kinase gene, RLK-V, regulates powdery mildew resistance in wheat. Mol Plant Pathol 19:2561–2574. https://doi.org/10.1111/mpp.12729
Hurni S, Brunner S, Buchmann G, Herren G, Jordan T, Krukowski P, Wicker T, Yahiaoui N, Mago R, Keller B (2013) Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J 76:957–969. https://doi.org/10.1111/tpj.12345
Jin YL, Shi FY, Liu WH, Fu XY, Gu TT, Han GH, Shi ZP, Sheng Y, Xu HX, Li LH, An DG (2021) Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2,978 wheat accessions. Plant Dis 105:3918–3926. https://doi.org/10.1094/pdis-03-21-0532-re
Jupe F, Pritchard L, Etherington GJ, MacKenzie K, Cock PJA, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JDG, Hein I (2012) Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom 13:75. https://doi.org/10.1186/1471-2164-13-75
Koller T, Brunner S, Herren G, Hurni S, Keller B (2018) Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor Appl Genet 131:861–871. https://doi.org/10.1007/s00122-017-3043-9
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137. https://doi.org/10.1139/gen-2013-0003
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363. https://doi.org/10.1126/science.1166453
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Li H, Chen X, Xin ZY, Ma YZ, Xu HJ, Chen XY, Jia X (2005) Development and identification of wheat-Haynaldia villosa T6DL.6VS chromosome translocation lines conferring resistance to powdery mildew. Plant Breed 124:203–205. https://doi.org/10.1111/j.1439-0523.2004.01062.x
Li S, Jia Z, Wang K, Du L, Li H, Lin Z, Ye X (2020) Screening and functional characterization of candidate resistance genes to powdery mildew from Dasypyrum villosum#4 in a wheat line Pm97033. Theor Appl Genet 133:3067–3083. https://doi.org/10.1007/s00122-020-03655-4
Li YH, Song N, Zhao CZ, Li F, Geng MM, Wang YH, Liu WH, Xie CJ, Sun QX (2016) Application of glycerol for induced powdery mildew resistance in Triticum aestivum L. Front Physiol 7:413. https://doi.org/10.3389/fphys.2016.00413
Liu DJ, Chen PD, Pei GZ, Wang YN, Qiu BX, Wang SL (1988) Transfer of Haynaldia villosa chromosomes into Triticum aestivum. In: Miller TE (ed) Proceedings of the 7th international wheat genetics symposium. Institute of Plant Sciences Research, Cambridge, England, pp 355–361
Liu ZY, Sun QX, Li HJ, Ni ZF, Yang ZM, Tang BR, Yang AD, Jia X (1999) Molecular identification and marker-assisted selection of Pm21 gene conferring resistance to powdery mildew in wheat. Acta Genet Sin 26:673–682
Lukaszewski AJ (1988) A comparison of several approaches in the development of disomic alien addition lines of wheat. In: Miller TE (ed) Proceedings of the 7th international wheat genetics symposium, vol 1. Inst Plant Sci Res, Cambridge England, pp 363–367
Lu P, Guo L, Wang ZZ, Li BB, Li J, Li YH, Qiu D, Shi WQ, Yang LJ, Wang N, Guo GH, Xie JZ, Wu QH, Chen YX, Li MM, Zhang HZ, Dong LL, Zhang PP, Zhu KY, Yu DZ, Zhang Y, Deal KR, Huo NX, Liu CM, Luo MC, Dvorak J, Gu YQ, Li HJ, Liu ZY (2020) A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun 11:680. https://doi.org/10.1038/s41467-020-14294-0
Ma JA, Zhou RH, Jia JZ (1997) Identification of wheat-Haynaldia villosa substitution lines conferring resistance to powdery mildew using genomic in situ hybridization (GISH) and RFLP markers. Acta Geol Sin 24:447–452
Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S, Kong X, Spielmeyer W, Talbot M, Bariana H, Patrick JW, Dodds P, Singh R, Lagudah E (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet 47:1494–1498. https://doi.org/10.1038/ng.3439
Qi LL, Friebe B, Zhang P, Gill BS (2007) Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res 15:3–19. https://doi.org/10.1007/s10577-006-1108-8
Randhawa MS, Bhavani S, Singh PK, Huerta-Espino J, Singh RP (2019) Disease resistance in wheat: present status and future prospects. In: Wani S (ed) Disease resistance in crop plants. Springer, Cham, pp 61–81
Sanchez-Martin J, Widrig V, Herren G, Wicker T, Zbinden H, Gronnier J, Sporri L, Praz CR, Heuberger M, Kolodziej MC, Isaksson J, Steuernagel B, Karafiatova M, Dolezel J, Zipfel C, Keller B (2021) Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat Plants 7:327–341. https://doi.org/10.1038/s41477-021-00869-2
Sears ER (1953) Addition of the genome of Haynaldia villosa to Triticum aestivum. Am J Bot 40:168–174. https://doi.org/10.1002/j.1537-2197.1953.tb06464.x
Seeholzer S, Tsuchimatsu T, Jordan T, Bieri S, Pajonk S, Yang W, Jahoor A, Shimizu KK, Keller B, Schulze-Lefert P (2010) Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant Microbe Interact 23:497–509. https://doi.org/10.1094/mpmi-23-4-0497
Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JI, Banfield MJ, Kamoun S (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol Plant Microbe Interact 27:624–637. https://doi.org/10.1094/mpmi-02-14-0040-r
Sheng BQ, Duan XY, Zhou YL (1995) Present situation and suggestion of wheat powdery mildew resistance resources (in Chinese). Crops 11(6):20
Stirnweis D, Milani SD, Jordan T, Keller B, Brunner S (2014) Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the Pm3f resistance spectrum in wheat. Mol Plant Microbe Interact 27:265–276. https://doi.org/10.1094/mpmi-10-13-0297-fi
Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH (2001) Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158:423–438. https://doi.org/10.1093/genetics/158.1.423
Takamatsu S (2004) Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience 45:147–157. https://doi.org/10.1007/S10267-003-0159-3
Urbano M, Resta P, Benedettelli S, Blanco A. (1988). A Dasypyrum villosum (L.) Candargy chromosome related to homoeologous group 3 of wheat. In: Miller TE (eds) Proceedings of the 7th international wheat genetics symposium. Inst Plant Sci Res, Cambridge England, pp 169–173
Wang D, Pei D, Pei ZY, Zhuang LF, Qi ZJ (2017) Development and application of high resolution karyotypes of wheat “Chinese Spring” aneuploids. Crop J 43:1575. https://doi.org/10.3724/SP.J.1006.2017.01575
Wang ZL, Li LH, He ZH, Duan XY, Zhou YL, Chen XM, Lillemo M, Singh RP, Wang H, Xia XC (2005) Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis 89:457–463. https://doi.org/10.1094/pd-89-0457
Wu XX, Bian Q, Gao Y, Ni XY, Sun YQ, Xuan YH, Cao YY, Li TY (2021) Evaluation of resistance to powdery mildew and identification of resistance genes in wheat cultivars. Peer J 9:e10425. https://doi.org/10.7717/peerj.10425
Xie JZ, Guo GH, Wang Y, Hu TZ, Wang LL, Li JT, Qiu D, Li YH, Wu QH, Lu P, Chen YX, Dong LL, Li MM, Zhang HZ, Zhang PP, Zhu KU, Li BB, Deal KR, Huo NX, Zhang Y, Luo MC, Liu SZ, Gu YQ, Li HJ, Liu ZY (2020) A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol 228:1011–1026. https://doi.org/10.1111/nph.16762
Xing LP, Hu P, Liu JQ, Witek K, Zhou S, Xu JF, Zhou W, Gao L, Huang ZP, Zhang RQ, Wang XE, Chen PD, Wang HY, Jones JDG, Karafiatova M, Vrana J, Bartos J, Dolezel J, Tian YC, Wu YF, Cao A (2018) Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol Plant 11:874–878. https://doi.org/10.1016/j.molp.2018.02.013
Zeng FS, Yang LJ, Gong SJ, Zhang XJ, Wang H, Xiang LB, Xue MF, Yu DZ (2014) Virulence and diversity of Blumeria graminis f. sp. tritici populations in China. J Integr Agric 13:2424–2437. https://doi.org/10.1016/S2095-3119(13)60669-3
Zhang RQ, Fan YL, Kong LN, Wang ZJ, Wu JZ, Xing LP, Cao AZ, Feng YG (2018) Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor Appl Genet 131:2613–2620. https://doi.org/10.1007/s00122-018-3176-5
Zhou Y, He ZH, Zhang GH, Xia LQ, Chen XM, Gao YC, Jing ZB, Yu GJ (2004) Utilization of 1BL/1RS Translocation in Wheat Breeding in China. Acta Agron Sin 30:531–535
Zhu KY, Li MM, Wu HB, Zhang DY, Dong LL, Wu QH, Chen YX, Xie JZ, Lu P, Guo GH, Zhang HZ, Zhang PP, Li BB, Li WL, Dong L, Wang QF, Zhu JH, Hu WL, Guo LQ, Wang RG, Yuan CG, Li HJ, Liu ZY, Hua W (2022) Fine mapping of powdery mildew resistance gene MlWE74 derived from wild emmer wheat (Triticum turgidum ssp. dicoccoides) in an NBS-LRR gene cluster. Theor Appl Genet 135:1235–1245. https://doi.org/10.1007/s00122-021-04027-2
Zou SH, Wang H, Li YW, Kong ZS, Tang DZ (2018) The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol 218:298–309. https://doi.org/10.1111/nph.14964
Zou SH, Shi WQ, Ji JH, Wang HM, Tang YS, Yu DZ, Tang DZ (2022) Diversity and similarity of wheat powdery mildew resistance among three allelic functional genes at the Pm60 locus. Plant J 110:1781–1790. https://doi.org/10.1111/tpj.15771
Acknowledgements
This work was supported by the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS [2021] 013, JBGS [2021] 007), Natural Science Foundation of China (Grant No. 32171973, 31971933), Special Fund for Independent Innovation of Agricultural Science and Technology in Jiangsu (No. CX[22] 1006) and the Joint Research of Wheat Variety Improvement of Anhui (2021-2025). The bioinformatics analysis was supported by the high-performance computing platform of Bioinformatics Center, Nanjing Agricultural University.
Author information
Authors and Affiliations
Contributions
LX and AC participated in the design of the experimental plan. ZH and JL performed the gene cloning and functional analysis, XL performed the bioinformatics analysis, YG and YL performed disease resistance evaluation and the specific marker analysis. RZ performed the cytogenetic stocks development. ZH, JL, LX and AC wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Beat Keller.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, Z., Liu, J., Lu, X. et al. Identification and transfer of a new Pm21 haplotype with high genetic diversity and a special molecular resistance mechanism. Theor Appl Genet 136, 10 (2023). https://doi.org/10.1007/s00122-023-04251-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04251-y