Abstract
Background
Date palm, oasis pivot, plays a vital socio-economic part in the southern area of Morocco. However, with climate change and drought intensity and frequency increasing, the Moroccan palm grove is threatened with significant genetic degradation. Genetic characterization of this resource is key element for the development of effective conservation and management strategies in the current circumstances of climate change and various biotic and abiotic stresses. To evaluate the genetic diversity of date palm populations collected from different Moroccan oases, we used simple sequence repeats (SSR) and directed amplification of mini-satellite DNA (DAMD) markers. Our results showed that used markers could efficiently assess genetic diversity in Phoenix dactylifera L.
Results
A total of 249 and 471 bands were respectively scored for SSR and DAMD, of which 100% and 92.9% were polymorphic. The polymorphic information content (PIC = 0.95), generated by the SSR primer was nearly identical to that generated by the DAMD primer (PIC = 0.98). The resolving power (Rp) was higher in DAMD than SSR (29.46 and 19.51, respectively). Analysis of the molecular variance (AMOVA) based on the combined data sets for both markers revealed a higher variance within populations (75%) than among populations (25%). Principal coordinate analysis (PCoA) and the ascendant hierarchical classification showed that the population of Zagora and Goulmima regions were the closest populations. The STRUCTURE analysis clustering of the 283 tested samples into seven clusters based on their genetic composition.
Conclusion
The results drawn from this study will orient genotypes selection strategies for a successful future breeding and conservation program, particularly under climate change context.
Similar content being viewed by others
Background
The date palm (Phoenix dactylifera L.) is a diploid (2n = 36), monocotyledonous plant [1]. It is a dioecious species and a member of the Arecaceae family [2]. It is one of the world's oldest plants, and has been farmed for over 6000 years, mainly for its fruit [3]. Dates are well-known for containing a complex mixture of nutritive and non-nutritive bioactive molecules. These include antioxidants [4], bioactive phytochemicals, water- and fat-soluble vitamins, non-starch polysaccharides, and minerals [5]. It is a key element in food security, and known as a “tree of life” [6].
Phoenix dactylifera L. significantly contributes towards the sustenance of the desert ecosystem. It combats desertification and modulates macro- and micro-climate temperatures. It also protects the soil from different types of degradation, promotes biodiversity, and reduces pollutants [7].
In Morocco, date palm represents about 4.5% of the world’s date population [8, 9]. Additionally, Morocco stands seventh in the world in area devoted to date-cultivation, eleventh in number of date palms, and ranks twelfth in the production of dates [8].
In 2018, the global production of dates reached 8.5 million tons (Mt) from a total cultivated area of 1.09 million hectares. For Morocco, the production is considered to be 1 million tons over an area of 59.127 ha [10].
Despite the availability of several cultivars, local date palm groves are seriously threatened by abiotic and biotic stresses such as, salinity, prolonged drought, and diseases [8] which could induce genetic degradation of this resource. In addition, the lack of data relating to the genetic diversity of the Moroccan date palm. It is therefore imperative to assess the genetic diversity and population structure of Moroccan date palms in order to establish strategies for the conservation of this resource and the selection of tolerant and adapted genotypes.
Phenotypic and biochemical markers have been used to assess genetic diversity of date palm, [11–19]. However, those markers are limited, and often affected by environmental conditions or date palm developmental stage. For this reason, molecular markers have been applied to unmask genetic differentiation. In date palm germplasm, different molecular markers such as amplified fragment length polymorphism(AFLP) [20–25], random restriction fragment length polymorphisms (RFLP) [26, 27], random amplified polymorphic DNA (RAPD) [28–35], fingerprints inter simple sequence repeat (ISSR) [36–40], start codon target (Scot) [41, 42], simple sequence repeats (SSR) [43–55], and single nucleotide polymorphism (SNP) [56] were largely used for diversity assessment. Recently, Directed Amplification of Minisatellite-region DNA (DAMD) markers have been employed, for the first time, to characterize the germplasm of the date palm [2].
Indeed, a fitting molecular marker ought to be significantly heritable, and applicable to any part of the genome and should be sufficiently polymorphic to allow the discrimination of narrowly related genotypes [57]. SSRs and DAMDs meet these criteria. They appear, easy to set up and are also remarkably reproducible. In order to acquire a more thorough knowledge of the genetic organization of Moroccan date palm therefore, in this work, we have used both markers. The use of SSR markers is assumed to be one of the best effective strategies in rating date palm genetic diversity. DAMD has been proven as a powerful tool for studding the genetic diversity. In fact, the combination of DAMD and SSR markers would bestow additional and complementary information to measure date palm genetic diversity and population structure.
Description and genetic diversity assessment are important requirements for genetic improvement approaches, and resource management strategies. Hitherto, little is known about genetic diversity and population structure of Moroccan date palm. This research is conducted to assess genetic diversity and genetic structure present in Moroccan date palm population collected from five important Oases in southern Morocco.
The current study is the first report to my knowledge on the genetic diversity assessment and population structure of 276 date palm collected from 5 different zones of Morocco. Additionally, here we also provide a correlation between Moroccan date palm genotypes and some Iraqi cultivars using DAMD and SSR markers.
Methods
Plant material
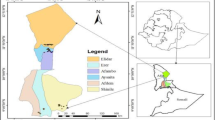
A set of 276 date palm leaves were sampled from five important regions of Phoenix dactylifera L. population in Morocco: Zagora, Errachidia, Goulmima, Tata, Tinghir. Multiple collection sites were selected from each region (Fig. 1 map). Each location was recorded using a Global Positioning System (GPS) receiver (Additional file 1). The plants were collected to cover the greatest possible genetic diversity in the five traditional regions. The young leaves were selected and picked from healthy adult plants. Then the fresh leaf samples were preserved in a portable refrigerator at – 20 °C and transported to the laboratory for further processing. In addition, we used in this research 10 Moroccan cultivars (five female cultivars contain Majhoul, Boufgouss, Najda, Bouskri, Aziza, Gharass, Sedrat, and three male cultivars that include Nebch-Bouskri NP3, Nebch-Boufeggous NP4, and GS), and one Tunisian accession; Deglet Nour, besides seven Iraqi cultivars (which are female including Maktoum, Khestaoui, Hamrawi, Zahdi, Tebarzal, Bahri, and Habhab).
DNA extraction
About 60 mg of lyophilized leaves were mechanically ground using a ball mill (Mill: SPEX Sample Prep). Thereafter, DNA was extracted from the ground tissues by using the CTAB (Cetyl Trimethyl Ammonium Bromide) method as described by Saghai-Maroof [58] with minor modifications.
The crushed plant tissue was combined with 1 ml pre-warmed of 2% CTAB buffer (1 M Tris–HCl (pH 8.0), 5 M NaCl, 0.5 M EDTA ethylene diamine tetra acetic acid, 2% CTAB, and 0.2% of β-mercaptoethanol, 0.2% polyvinylpyrrolidone (PVP)), and then the mixture was maintained at 65 °C in a water bath for 60 min; next, the blend was cooled in an ice bucket, and after 700 μl of an equal volume of chloroform isoamyl alcohol (24:1) were added. The tubes which contain the mixture were inverted several times for 15 min to ensure that reagents were mixed well. The mixture was then centrifuged at 13,000 rpm for 10 min.
Next, the supernatant was moved to a new tube and the RNA was eliminated by adding 1 μl of RNA-ase to the mixture, which was incubated at 37 °C for 30 min. Thereafter, 700 μL of ice-cold isopropanol was added to the samples, the mixture was shaken gently and stored overnight to ensure precipitation of nucleic acids.
DNA recovery was completed out by centrifuging at 13,000 rpm for 10 min at 4 °C. and washing twice with ice-cold ethanol (70%). The resulting DNA was dissolved in 200 μL of pure sterile water.
The DNA concentration and quality were assessed by using a NanoDrop spectrophotometer. DNA was deemed pure when the A260/A280 ratio ranged between 1.80 and 2.0 ([53], p. 20). The quality of DNA was also evaluated by electrophoresis, on 1% (w/v) agarose gel. Thereafter all the DNA extracts were stored at – 20 °C.
SSR analysis
At first, a set of 30 SSR primers were screened on 10 DNA random templates of Phoenix dactylifera L. samples. Based on the best amplification results, only 10 SSR primers that gave clear and reproducible bands were eventually chosen for PCR amplification of the entire range of 283 Phoenix dactylifera L. genotypes.
The ten polymorphic microsatellites selected were antecedently developed in date palm by [59], the forward and reverse primers of theses markers are presented in Table 1.
The SSR loci’s were performed in a reaction volume of 10 μl consisting of 1 × buffer, 0.5 U of Taq DNA polymerase (BIOLINE, London, UK), 0.5 μM (10 pmol) of each forward and reverse primers and approximately 25 ng of genomic DNA.
The amplification reactions were performed out in the Eppendorf Master cycler with initial denaturation for 1 min at 94 °C. This was followed by 35 cycles of each cycle with 30 s denaturation at 94 °C, 1 min annealing at 52 °C, 2 min extension at 72 °C. and the final extension was carried out at 72 °C for 8 min. The reactions were then held at 4 °C.
DAMD analysis
Nine minisatellite DAMDs were chosen and used in this study (Table 2) based on their polymorphic information content in published plant diversity analysis researches [2].
The PCR composition was the same as SSR analysis. Polymerase chain reactions (PCR) were performed out according to the program as follows: initial denaturation for 3 min at 94, then 35 cycles of 45 s denaturation at 92 °C, 2 min at annealing temperature (depending on the primer’s Tm), 2 min extension at 72 °C and the Final extension was carried out at 72 °C for 5 min followed by cooling at 4 °C for an unlimited period.
Gel electrophoresis
In order to detect polymorphism among accessions, the PCR product was transferred to 6% polyacrylamide gel (for higher resolution), using 1 × TBE as running buffer at 120 V. The amplified bands were detected by staining in an ethidium bromide solution for 5 min(10 mg/ml) and visualized under ultraviolet light in Molecular Imager_ Gel DocTM XR System. The gel profiles were photographed and recorded as digital images in a Gel Documentation System for later scoring.
Statistical analyses
Each visual, clear and distinct band amplified was considered as a single allele and scored manually as present (1) or absent (0) from each SSR and DAMD profiles. Then, a binary data matrix was developed that is used for all analyses.
The effectiveness of each primer used (SSR and DAMD) was assessed by calculating the polymorphic information content (PIC) using the equation \(\mathrm{PICvalue}=1-\sum_{\mathrm{n}=1}^{\mathrm{n}}{\mathrm{pi}}^{2}\) where pi is the frequency of the ith allele [60]. Also, (RP) was calculated as a parameter used to detect the ability of each primer to distinguish between individuals, according to Prevost and al [61, 62]. Additionally, the effective marker ratio (EMR) was calculated with the formula EMR = np(np/n), where “np” represents the number of polymorphic loci and "n" represents the total number of loci [63]. Finally, the marker index (MI) was calculated using the formula MI = EMR × PIC, which characterizes the capacity of each primer to detect polymorphic loci among the genotypes.
GenAlex ver. 6.5 software [64] was used to carry out genetic diversity analysis of Phoenix dactylifera L. by calculating Nei’s genetic diversity index (H), percentage of polymorphic loci (PPL), number of effective alleles (Ne), expected heterozygoty (He), and Shannon’s information index (I). Molecular variance (AMOVA) was performed, to measure the partitioning of genetic variability between and within populations by the same software. The resulting distance matrices of squared Euclidean distances [65] between all pairwise genotypes, principal coordinate analysis (PCoA) was executed. Based on 999 permutations, the significance of these genetic differentiations was tested. A dendrogram was generated based on Jaccard’s dissimilarity coefficient and UPGMA (Unweighted Pair Group Method of Arithmetic averages) clustering method using XLSTAT 5.14 software.
Finally, to characterize the overall genetic structure and assign individuals to populations, molecular data were analyzed using the STRUCTURE software v.2.3.4 [66] based on Bayesian clustering method. The predefined numbers of populations (K) was set from 1 to 10, and each simulation was performed in 100,000 Monte-Carlo Markov Chain (MCMC) iterations after a burn-in period length of 50,000. The analyzed data resulting from STRUCTURE were then exported to the STRUCTURE HARVESTER online tool [67]. The Evanno approach was applied to determine the most possible structure with the best K value.
Results
Characterization and efficiency of SSR markers
Thirty SSR primers were initially screened for amplification. From these, 10 primers produced sharp, clear, and reproducible banding profile (Fig. 2a) in 283 genotypes of Phoenix dactylifera L. The variation of the SSRs amplification frequency based on the availability of motifs in genomic DNA of different genotypes with an average of 24.9 distinct scorable bands were produced by these primers. The primer Cir 50 produced a maximum of 41 scorable bands while the primer Cir 93 generated only 15 fragments. A total of 249 DNA fragments were obtained across all 283 genotypes using the 10 SSR primers (Table 3).
The highest number of SSR bands occurred in the Zagora and the Goulmima populations with 232 and 227 respectively, and the lowest number of SSR bands observed in Moroccan varieties and Iraqi varieties. In the latter case, on one private band was observed.
The effectiveness of each primer in tracing polymorphism was evaluated by calculating their PIC and Rp values. PIC value ranged from 0.83 to 0.99, and Rp value ranged from 12.26 to 20.74. The maximum PIC (0.99) and Rp (20.74) values were observed for the Cir 25 and Cir 48 primers, respectively; minimum values for PIC (0.83) and Rp (12.26) were recorded for primers Cir 35 and Cir 25, respectively. In general, the 10 SSR primers utilized in this research recorded an average PIC of 0.954 and an average Rp of 19.51 (Table 4).
Characterization and efficiency of DAMD markers
Wholly eight DAMD markers were reproducible and displayed good banding patterns (Fig. 2b). These markers yielded a varied number of bands. The eight DAMDs in this study produced a total of 471 bands with a mean of 58 (Table 3). The maximum number of DAMD bands were registered for the Zagora population with 330 alleles and Goulmima with 299 bands.
The primer URP25F produced a maximum of 94 bands; URP2R primer produced the minimum of 32 bands. PIC values were calculated for all fragments of DAMD markers which ranged from 0.95 (URP2R) to 0.99 (HBV5 and HBV3) with an average PIC of 0.98. Rp values were evaluated for the eight primers, the lowest Rp value was observed in URP2R (16.87) and the highest one (36.96) in HBV5 with mean of 29.46.
The presence of private alleles is a further indication of population differentiation. Private alleles were detected in Zagora, Goulmima, Errachidia, Tinghir, and Tata (39, 29, 18, 16, and 8 respectively) with DAMD primers, while none were detected in Moroccan and Iraqi varieties (Table 4).
Genetic diversity
SSR and DAMD markers disclosed various degrees of genetic variability within and between Phoenix dactylifera L. populations.
For the SSR markers, the highest values of Genetic parameters viz- observed number of alleles (Na) number of effective alleles (Ne), Shannon’s information index (I), expected heterozygosity (He), and unbiased expected heterozygosity (uHe) were obtained in the Tata population, with values of 1.984, 1.408, 0.402, 0.255, and 0.258, respectively. While the lowest values were recorded in Moroccan varieties, with values 1.394, 1.285, 0.288, 0.181, and 0.190, respectively (Table 4).
For DAMD markers, the data showed that the upper diversity indexes values (Na, Ne, I, He, uHe, and %P) were observed in Zagora population with values 1.349, 1.254, 0.252, 0.160, 0.160, and 67.15% (Table 4). And the lowest values were recorded in Moroccan varieties, with values 0.96, 1.148, 0.139, 0.09, and 0.094, respectively.
Whereas the combination of data from both markers indicated the high genetic diversity indices were recorded in the Zagora population and low values in Moroccan varieties (Table 4).
AMOVA (analysis of molecular variance) analysis of Phoenix dactylifera L.
AMOVA analysis showed significant variation within population, with 88% for SSR and 65% for DAMD marker (Table 5). While among populations less variance was observed, only 12% and 35% respectively for the both markers. Combined SSR and DAMD data showed 75% of variation within populations and 25% of variations among population (Fig. 3). PhiPT for 283 date palm accessions was 0.253 (p < 0.001). Pairwise population PhiPT values for seven clusters ranged from 0.163 (Zagora-Goulmima) to 0.391 (Tinghir-varieties) (Table 6).
Genetic distance
Genetic distance and identity coefficient were evaluated by the method Nei [68].
Based on the combined SSR and DAMD data, the values of the genetic identity were high with the mean genetic similarities among populations ranged from 0.898 to 0.966. In view of the genetic distance, the values vary from 0.035 to 0.108. The Genetic identity and genetic distance between the six regions and varieties found that Zagora and Goulmima populations had the highest genetic identity (0.966), and the lowest genetic distance (0.035; P = 0.163). Whereas Errachidia population and Moroccan varieties had the lowest similarity (0.898) and the maximum genetic distance (0.108; P = 0.382) (Table 7).
Cluster analysis
Combined matrix of SSR and DAMD data was employed to build an UPGMA tree. The cluster analysis classified the investigated genotypes into two distinct clusters. The first cluster (I) comprised Moroccan varieties and Iraqi varieties. The second cluster (II) was segregated into two sub clusters. The first one (IIa) comprised of Tinghir and Errachidia populations, and the second sub-cluster (IIb) was split also into two groups, the first group contained Tata population and the second Group encompassed Zagora and Goulmima populations. The UPGMA tree showed in the majority of the individuals were separated according to their populations (Fig. 4).
Principal coordinate analysis (PCoA)
The PCoA allowed the study of the correlation between different Phoenix dactylifera L. genotypes studied. Depending on combined data from SSR and DAMD markers, PCoA illustrated 17.43% of the total variations, with the first three axes (1, 2, and 3) accounted for 7.3%, 5.75%, and 4.38%, respectively.
The screen plot generated by PCoA highlighted, the grouping of Tata population with Iraqi varieties. The rally of Zagora population with Goulmima population and Moroccan varieties and one genotype from Iraq and the closeness of Errachidia population with Tinghir population (Fig. 5).
Population genetic structure
To characterize the overall genetic structure, we applied a Bayesian clustering approach implemented in STRUCTURE software. The output was extracted with Structure Harvester, following the method of (Evanno et al. 2005) for the estimation of the most likely number of genetic clusters (K), we investigated the range from K = 1 to K = 10. The maximum value of Δk was detected at K = 7. This suggested that the most likely structure of tested samples is their distribution into seven principal clusters (Fig. 6).
Individuals are separated by vertical columns and identified by colors. The same color in different individuals shows that they are part of the same genetic cluster. Different colors in the same individual indicate the likelihood to pertain to different genetic groups.
Our result indicated that the genetic structure of Phoenix Dactylifera L. could be grouped into seven populations. The first cluster mainly included of Tinghir and Errachidia population. The second cluster consisted of the Goulmima population. The major group of the Zagora population was split into three clusters. One of the seven clusters was containing the population of Tata and the other genetic cluster comprised Moroccan varieties, while the populations of Iraq varieties were found into both clusters.
Discussion
For all species evolution and adaptation, genetic variability plays an important role. So, genetic diversity assessment is a key to an effective management and breeding program. Therefore, in this study, we combined SSR and DAMD molecular markers to assess genetic variation among and within Phoenix dactylifera L. collection from five Moroccan regions populations.
The results of this investigation revealed that both molecular markers SSR and DAMD generated high polymorphism level with 100% of PP for DAMD primers, and 92.9% for SSR primers. This polymorphism level was higher than that reported by Mirbahar et al. [69] and Srivashtav et al. [70] with 84% and 39.77% polymorphism level using ISSR and RAPD markers respectively. The PIC averages 0.954 and 0.98 obtained using SSR and DAMD markers respectively were informative according to Botstein and al [71]. The SSR primers recorded 19.51 average Rp, while Rp mean value for the DAMD primers was 29.46, showing that DAMD were more discriminant than SSR primers. This data indicates the right choice of primers in the current research for the rating of genetic diversity.
The genotyping data in this study have shown that date palm is rich in allelic variation as confirmed by previous studies [72, 73]. In this research, 249 bands were obtained from 10 SSR primers with a mean of 25 (range from 15 to 41) alleles per locus. This is relatively comparable to an earlier study [74] of date palm from Sudan with female cultivars from Morocco, where the paper reported a total of 343 alleles at the 16 primers with a mean of 21.4 (range from 14 to 44) alleles per locus. A high number of alleles was also detected by al Najm et al. [75] in the assessment of date palm cultivars from Australia and the Middle East by SSR markers which they found 313 bands.
Amplified band number by DAMD were higher than those amplified by SSR markers, indeed 471 bands were amplified with an average of 59. However, the average numbers of each marker in the current study were higher than those reported earlier [53]. The Zagora population had a higher number of private alleles with 39 bands. This result is likely due to Zagora being the largest population and most genetically diverse compared with the other populations.
The genetic parameters detected from the two markers in our findings (SSR: Na = 1,786(0,015) Ne = 1,368(0.008), I = 0.365(0.005) and He = 0.231(0,004) uHe = 0.237(0.004) %P = 88.87%; DAMD: Na = 1,016(0.017) Ne = 1,231(0.006), I 0,215(0,004) and He = 0.140(0.003) uHe = 0.143(0,003) %P = 49.49%; Combined Na = 1,277(0.013) Ne = 1,277(0.005), I = 0.266(0.004) and He = 0.171(0.003) uHe = 0.175(0.003) %P = 62.81%) are a little moderate and higher in SSR. Based on these parameters, the studied populations ranked from the most diverse to the least diverse as follows: Zagora, Tata, Errachidia, Goulmima, Tinghir, Iraqi, and Moroccan varieties. Hence, our findings suggest that the Zagora and Tata populations have the highest genetic variability level. These observations suggest that Zagora and Tata populations could be involved in an effective date palm improvement and conservation strategy.
When assessing genetic relationships of the date palm cultivars using the dominant marker system iPBS Al-Najm et al. [76] noted a Shannon’s Information Index of 0.330, expected heterozygosity of 0.218 and unbiased expected heterozygosity of 0.229). This is in line with the SSR results obtained in the present study (Table 4). Our parameters are higher than the ones obtained from earlier studies in Iranian of date palm cultivars using SCoT molecular markers [42] (Na = 0.671 Ne = 1.179, I = 0.156 and He = 0.104 uHe = 0.114%P = 29.67%). While the Al-Najm et al. [75] reported a much higher Shannon’s Information Index (2.067), expected heterozygosity (0.841) and unbiased expected heterozygosity (0.883) in the Australian germplasm when using SSR markers.
Results of AMOVA test have shown that the genetic diversity of Moroccan date palms is highly represented within populations with 75% instead than among populations with 25%. This is in accordance with genetic diversity assessment in date palm genotypes of Sudan [74], and Tunis [77] by SSR markers. Also in Iran using scot [42], SSR and ISSR markers [78], by RAPD and ISSR markers [79], in Morocco by using isozyme markers [80].
The presence of genetic variability within populations is a condition for adaptation and evolutionary transition. Despite date palms are mostly vegetative propagated, most genetic variations observed are essentially caused by mutations, but in the other few cases through sexual reproduction, the meiosis and fertilization lead to immense genetic diversity. Moreover, the famer management had an important impact on this diversity.
However, the Nei analysis revealed low distance values between populations this is in line with AMOVA result which indicate the low diversity among populations with 25%. This finding can be explained by the same origin gene pool of Moroccan date palm. Indeed, Zehdi et al. [81] revealed a deep genetic structuring arising from two geographic gene pools, which means the existence of two cultivation origins of date palm, one in the east and other in the western. Morocco gene pool could belongs to the second region. In contrast, high genetic diversity observed within populations can be result of cloning process, seed reproduction, or natural mutation. This in agreement with Elmeer et al. [82] who reported high genetic diversity observed within the groups and the weak clustering of the cultivars suggested that they are not a result of a full cloning process.
Significant values of similarities were obtained mostly between Zagora and Goulmima populations, as illustrated in the UPGMA dendrogram (Fig. 4), where the two populations are in the same cluster. Sedra et al. [35] reported that most of the cultivars from Iraq were correlated with accessions already grown in Morocco. That agrees with our PCoA pattern which showed that the Tata population and the Iraqi varieties belong to the same group which has already been proven by the high genetic identity of 0.953, also in keeping with our UPGMA which revealed that the Iraqi and Moroccan varieties are in the same cluster that has been verified by similarity value of 0.918. Our results indicated the presence of higher genetic diversity in the Moroccan date palms oasis populations compared to the Iraqi and Moroccan date palms cultivars, which may be explained by intensive selection operations in breeding program [83].
Genetic variation partition in seven differentiated groups obtained with STRUCTURE regrouped Tinghir with Errachidia populations in the same cluster, and Goulmima in another one. While the latter is geographically closer to Errachidia than Tinghir. This can explained by human activities in exchanging disparate materials [84]. Tata population with Iraqi varieties grouped in the same cluster. This is in line with the PCoA result. Zagora population is sub-structured in three groups that confirm the high genetic diversity.
STRUCTURE and UPGMA cluster results showed that there is geo-graphical conservation of accessions. Genetic structure of date palms obtained in this study can be explained by distance geographical isolation, plant biological nature, and environmental conditions [84]. Furthermore, the history of cultivation may contribute to genotypes geographic conservation.
Zagora and Goulmima populations have the higher genetic diversity. Indeed, Zagora harbor 39 private alleles and Goulmima 23. These populations should be prioritized in conservation strategies.
Conclusion
This work provided a knowledge about genetic resources in the main distribution area of the Moroccan date palm. The SSR and DAMD markers showed adequate polymorphism and offered appropriate details for the genetic diversity assessment of Phoenix dactylifera L. Further, AMOVA of this plant showed a high degree of variation within populations.
Genetic diversity reported in our result is essential for evolutionary adaptation in climate change context and for date palm sustainable breeding purposes. Moreover, our results may be a significant guideline for the collection of core germplasm resources. So, we recommend in-situ and ex situ genetic resources management to assure the conservation of this diversity. The approach also must increase the local community awareness of the suitable farming techniques for Phoenix dactylifera L. preserving and conservation resources. Consequently, the sustainability of the oases, especially in the era of climate change, will be ensuring.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- AMOVA:
-
Analysis of the molecular variance
- DAMD:
-
Directed amplification of mini-satellite DNA
- DNA:
-
Deoxyribonucleic acid
- ISSR:
-
Inter simple sequence repeat
- PIC:
-
Polymorphic information content
- PCoA:
-
Principal coordinate analysis
- PCR:
-
Polymerase chain reaction
- RAPD:
-
Random amplified polymorphic DNA
- RFLP:
-
Restriction fragment length polymorphism
- Rp:
-
Resolving power
- SCoT:
-
Start codon targeted
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
References
Mohammed IA, Mohamed IA (2019) Validation of RAPD and ISSR markers used for sex determination in date palm grown under Sudan conditions. Adv Res Life Sci. 3(1):17–22. https://doi.org/10.2478/arls-2019-0005
Purayil FT, Robert GA, Gothandam KM, Kurup SS, Subramaniam S, Cheruth AJ (2018) Genetic variability in selected date palm (Phoenix dactylifera L.) cultivars of United Arab Emirates using ISSR and DAMD markers. 3 Biotech. 8(2):109. https://doi.org/10.1007/s13205-018-1108-3
Zango O et al (2017) Genetic diversity of Southeastern Nigerien date palms reveals a secondary structure within Western populations. Tree Genet Genomes. 13(4):75. https://doi.org/10.1007/s11295-017-1150-z
Ahmed MS, Nair KP, Khan MS, Algahtani A, Rehan M (2020) Evaluation of date seed (Phoenix dactylifera L.) oil as crop base stock for environment friendly industrial lubricants. Biomass Conv Bioref. https://doi.org/10.1007/s13399-020-00708-y
Al-Mssallem MQ, Alqurashi RM, Al-Khayri JM (2019) Bioactive compounds of date palm (Phoenix dactylifera L.). In bioactive compounds in underutilized fruits and nuts, H. N. Murthy et V. A. Bapat, Éd., in Reference Series in Phytochemistry. Springer International Publishing, Cham, pp 1–15. https://doi.org/10.1007/978-3-030-06120-3_6-1
Jamro MMR (2020) Production and quality of Date fruit in response to pollination methods. PAB. 9(3). https://doi.org/10.19045/bspab.2020.90184
Ibrahim KM (2010) The role of date palm tree in improvement of the environment. Acta Hortic. 882:777–778. https://doi.org/10.19045/bspab.2020.90184
Sedra MH (2015) Date Palm Status and Perspective in Morocco. In: Al-Khayri JM, Jain SM, Johnson DV (eds) Date Palm Genetic Resources and Utilization. Springer Netherlands, Dordrecht, pp 257–323. https://doi.org/10.1007/978-94-017-9694-1_8
Sedra MyH (2011) Development of New Moroccan Selected Date Palm Varieties Resistant to Bayoud and of Good Fruit Quality. In: Jain SM, Al-Khayri JM, Johnson DV (eds) Date Palm Biotechnology. Springer Netherlands, Dordrecht, pp 513–531. https://doi.org/10.1007/978-94-007-1318-5_24
FAOSTAT (2020) http://www.fao.org/faostat/en/#data/QC (consulté le 26 avril 2020)
Bedjaoui H, Benbouza H (2020) Assessment of phenotypic diversity of local Algerian date palm (Phoenix dactylifera L.) cultivars. J Saudi Soc Agric Sci. 19(1):65–75. https://doi.org/10.1016/j.jssas.2018.06.002
Elsafy M, Garkava-Gustavsson L, Mujaju C (2015) Phenotypic diversity of date palm cultivars ( Phoenix dactylifera L) from Sudan estimated by vegetative and fruit characteristics. Int J Biodivers. 2015:1–7. https://doi.org/10.1155/2015/610391
Khierallah HSM, Azhar HD (2016) Study of genetic diversity of iraqi date palms using some morphological markers. Int J Curr Microbiol App Sci. 5(3):317–327. https://doi.org/10.20546/ijcmas.2016.503.039
Simozrag A, Chala A, Djerouni A, Elmoncef Bentchikou M (2016) Phenotypic diversity of date palm cultivars (Phoenix dactylifera L.) from Algeria. Gayana Bot. 73(1):42–53. https://doi.org/10.4067/S0717-66432016000100006
Azeqour M, Majourhat K, Baaziz M (2001) Morphological variations and isoenzyme polymorphism of date palm clones from in vitro culture acclimatized and established on soil in South Morocco. p 10
Elhoumaizi M, Saaidi M, Oihabi A, Cilas C (2002) Phenotypic diversity of date-palm cultivars(Phoenix dactylifera L.) from Morocco. Genet Resour Crop Evol. 49:483–490. https://doi.org/10.1023/A:1020968513494
Al-Jibouri AAM, Adham KM (1990) Biochemical classification of date palm male cultivars. J Horticult Sci. 65(6):725–729. https://doi.org/10.1080/00221589.1990.11516114
Baaziz M, Saaidi M (1988) Preliminary identification of date palm cultivars by esterase isoenzymes and peroxidase activities. Can J Bot. 66(1):89–93. https://doi.org/10.1139/b88-013
Bendiab K, Baaziz M, Brakez Z, Sedra My H (1993) Correlation of isoenzyme polymorphism and Bayoud-disease resistance in date palm cultivars and progeny. Euphytica. 65(1):23–32. https://doi.org/10.1007/BF00022196
Jubrael JMS, Udupa SM, Baum M (2005) Assessment of AFLP-based genetic relationships among date palm (Phoenix dactylifera L.) varieties of Iraq. jashs. 130(3):442–447. https://doi.org/10.21273/JASHS.130.3.442
Khierallah et al (2011) Assessment of genetic diversity for some Iraqi date palms (Phoenix dactylifera L.) using amplified fragment length polymorphisms (AFLP) markers. Afr J Biotechnol. 10(47):9570–9576. https://doi.org/10.5897/AJB11.055
Adawy SS, Hussein EHA, Ismail SEME, El-Itriby HA (2005) Genomic diversity in date palm (Phoenix dactylifera L.) as revealed by AFLPs in comparison to RAPDs and ISSRs. Arab J Biotechnol. 8(1):99–114
El-Assar AM, Krueger RR, Devanand PS, Chao CCT (2005) Genetic Analysis of Egyptian Date (Phoenix dactylifera L.) Accessions Using AFLP Markers. Genet Resour Crop Evol. 52(5):601–607. https://doi.org/10.1007/s10722-004-0583-z
Rhouma Chatti S, Baraket G, Dakhlaoui Dkhil S, Zehdi Azouzi S, Trifi M (2011) Molecular research on the genetic diversity of Tunisian date palm (Phoenix dactylifera L.) using the random amplified microsatellite polymorphism (RAMPO) and amplified fragment length polymorphism (AFLP) methods. Afr J Biotechnol. 10(51):10352–10365. https://doi.org/10.5897/AJB10.2242
Diaz S, Pire C, Ferrer J, Bonete MJ (2003) Identification of Phoenix dactylifera L. varieties based on amplified fragment length polymorphism (AFLP) markers. Cell Mol Biol Letters. 8(4):891–900
Hela S, Salwa Z, Salem Ali OM, Abdelmajid R, Mohamed M, Mokhtar T (2004) Genetic polymorphism of plastid DNA in Tunisian date-palm germplasm (Phoenix dactylifera L.) detected with PCR-RFLP. Genet Resour Crop Evol. 51(5):479–487. https://doi.org/10.1023/B:GRES.0000024156.29139.23
Corniquel B, Mercier L (1994) Date palm (Phoenix dactylifera L.) cultivar identification by RFLP and RAPD. Plant Sci. 101(2):163–172. https://doi.org/10.1016/0168-9452(94)90252-6
Eissa EA, Abd El-Razek AB, El-Sharabasy SF, Rizk RM (2009) Morphological and molecular genetic characterization of soft date palm (Phoenix dactylifera L.) cultivars in Egypt. Egyptian J Genet Cytol. 38(2)
El Sharabasy SF, Soliman KA. Molecular analysis of date palm genetic diversity using random amplified polymorphic DNA (RAPD) and inter-simple sequence repeats (ISSRs). In Date Palm Biotechnology Protocols Volume II, J. M. Al-Khayri, S. M. Jain, et D. V. Johnson, Éd., in Methods in Molecular Biology, vol. 1638. New York, NY: Springer New York, 2017, p. 143–152. https://doi.org/10.1007/978-1-4939-7159-6_13
Kareem MAH, Al-Saadi AH, Naji HF (2018) Genetic diversity of iraqi date palm (Phoenix dactylifera L.) by using RAPD technique. J Univ Babylon. 26(1):114–131
Ahmed O, Chokri B, Noureddine D, Mohamed M, Mokhtar T (2009) Regeneration and molecular analysis of date palm (Phoenix dactylifera L.) plantlets using RAPD markers. Afr J Biotechnol. 8(5)
Trifi M, Rhouma A, Marrakchi M (2000) Phylogenetic relationships in Tunisian date-palm ( Phoenix dactylifera L.) germplasm collection using DNA amplification fingerprinting. Agronomie. 20(6):665–671. https://doi.org/10.1051/agro:2000158
AI-Moshileh AM, Motawei MI, AI-Wasel A, Abdel-Latif T (2004) Identification of some date palm (Phoenix dactylifera L.) cultivars in Saudi Arabia using RAPD fingerprints. J Agric Marine Sci 9(1):1–3
Bahraminejad A, Mohammadi-Nejad G (2015) Genetic diversity analysis of date palm (Phoenix dactylifera L.) genotypes using RAPD markers. ARRB. 5(1):41–47. https://doi.org/10.9734/ARRB/2015/11683
Sedra MH, Lashermes P, Trouslot P, Combes M-C (1998) Identification and genetic diversity analysis of date palm (Phoenix dactylifera L.) varieties from Morocco using RAPD markers. Euphytica. 103(1):75–82
Khierallah HSM, Al-Sammarraie SKI, Mohammed HI (2014) Molecular characterization of some iraqi date palm cultivars using RAPD and ISSR markers. J Asian Sci Res. 4(9):490-503
Sharifi M, Sheidai M, Koohdar F (2018) Genetic fingerprinting of date palm (Pheonix dactylifera L.) by using ISSR and cpDNA sequences. Indian J Genet. 78:507–514
Guettouchi A, Elshibli S, Haider N, Nabulsi I, Ykhlef N (2017) Molecular diversity in date palm (Phoenix dactylifera L.) cultivars from Algeria indicated by RAPD and ISSR polymorphisms. Plant Cell Biotechnol Mol Biol. 18(1-2):76–89
Mirbahar AA, Khan S, Markhand GS, Kauser N, Saeed R (2016) DNA fingerprinting of some pakistani date palm (Phoenix dactylifera L.) cultivars using ISSR markers. Pak J Bot. 48(5):2005–2010
Sabir JSM et al (2014) Characterization of ten date palm (Phoenix dactylifera L.) cultivars from Saudi Arabia using AFLP and ISSR markers. Comptes Rendus Biologies. 337(1):6–18. https://doi.org/10.1016/j.crvi.2013.11.003
Al-Qurainy F, Khan S, Nadeem M, Tarroum M (2015) SCoT marker for the assessment of genetic diversity in Saudi Arabian date palm cultivars. Pakistan J Bot. 47(2):637–643
Saboori S, Noormohammadi Z, Sheidai M, Marashi S (2020) SCoT molecular markers and genetic fingerprinting of date palm (Phoenix dactylifera L) cultivars. Genet Resour Crop Evol. 67(1):73–82. https://doi.org/10.1007/s10722-019-00854-x
Elmeer K, Mattat I (2015) Genetic diversity of Qatari date palm using SSR markers. Genet Mol Res. 14(1):1624–1635. https://doi.org/10.4238/2015.March.6.9
Yusuf AO et al (2015) Genetic diversity of Nigerian date palm (Phoenix dactylifera) germplasm based on microsatellite markers. IJBSBT. 7(1):121–132. https://doi.org/10.14257/ijbsbt.2015.7.1.12
Elsafy M, Zborowska A, Bryngelsson T, Fatih M, Mujaju C, Garkava-Gustavsson L (2016) Elucidating the genetic diversity of farmer cultivars of female date palms (Phoenix dactylifera L.) from Sudan by microsatellite markers. Genet Resour Crop Evol. 63(6):975–986. https://doi.org/10.1007/s10722-015-0294-7
Al-Faifi SA et al (2016) Development, characterization and use of genomic SSR markers for assessment of genetic diversity in some Saudi date palm (Phoenix dactylifera L.) cultivars. Electron J Biotechnol. 21:18–25. https://doi.org/10.1016/j.ejbt.2016.01.006
Racchi ML, Bove A, Turchi A, Bashir G, Battaglia M, Camussi A (2014) Genetic characterization of Libyan date palm resources by microsatellite markers. 3 Biotech. 4(1):21–32. https://doi.org/10.1007/s13205-013-0116-6
Racchi ML, Camussi A (2018) The date palms of Al Jufrah - Libya: a survey on genetic diversity of local varieties. J Agric Environ Int Dev. 112(1). https://doi.org/10.12895/jaeid.20181.776
Battesti V, Gros-Balthazard M, Ogéron C, Ivorra S, Terral J-F, Newton C (2018) Date Palm Agrobiodiversity (Phoenix dactylifera L.) in Siwa Oasis, Egypt: Combining Ethnography, Morphometry, and Genetics. Hum Ecol. 46(4):529–546. https://doi.org/10.1007/s10745-018-0006-y
Khierallah HSM, Bader SM, Baum M, Hamwieh A (2011) Genetic Diversity of Iraqi Date Palms Revealed By Microsatellite Polymorphism. J Amer Soc Hort Sci. 136(4):282–287. https://doi.org/10.21273/JASHS.136.4.282
Khierallah HSM, Bader SM, Hamwieh A, Baum M (2017) Date palm genetic diversity analysis using microsatellite polymorphism. In Date Palm Biotechnology Protocols Volume II, Al-Khayri JM, Jain SM, Johnson DV, Éd., in Methods in Molecular Biology. vol. 1638. New York, NY: Springer New York, p. 113‑124. https://doi.org/10.1007/978-1-4939-7159-6_11
N. Faqir (2016) Simple sequence repeat (SSR) markers show greater similarity among morphologically diverse Date palm (Phoenix dactylifera L) cultivars grown in Pakistan. PAB. 5(3). https://doi.org/10.19045/bspab.2016.50063
Jamil S, Shahzad R, Kanwal S, Yasmeen E, Rahman SU, Iqbal MZ (2020) DNA fingerprinting and population structure of date palm varieties grown in Punjab Pakistan using simple sequence repeat markers. Int J Agric Biol. 23(5):943-50
Bodian A, El Houmazi MA, Ndoye NK, Hasnaoui A, Nachtigall M, Wehling P (2012) Genetic diversity analysis of date palm (Phoenix dactylifera L.) cultivars from Figuig oasis (Morocco) using SSR markers. Int J Sci Adv Technol. 2(3):96-104
Marion Nachtigall AB (2014) Genetic diversity analysis of date palm (Phoenix dactylifera L.) cultivars from Morocco using SSR markers. J Biodivers Biopros Dev. 01(03). https://doi.org/10.4172/2376-0214.1000126
Faqir N et al (2019) Development of a single nucleotide polymorphism-based genetic identification key for date palm cultivars. Crop Sci. 59:12
Adjebeng-Danquah J, Manu-Aduening J, Asante IK, Agyare RY, Gracen V, Offei SK (2020) Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon. 6(1):e03154. https://doi.org/10.1016/j.heliyon.2019.e03154
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings Natl Acad Sci. 81(24):8014–8018. https://doi.org/10.1073/pnas.81.24.8014
Billotte N et al (2004) Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): characterization and utility across the genus Phoenix and in other palm genera. Mol Ecol Notes. 4(2):256–258. https://doi.org/10.1111/j.1471-8286.2004.00634.x
Smith JSC et al (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPS and pedigree. Theor Appl Genet. 95(1–2):163–173. https://doi.org/10.1007/s001220050544
Lamare A, Rao SR (2015) Efficacy of RAPD, ISSR and DAMD markers in assessment of genetic variability and population structure of wild Musa acuminata colla. Physiol Mol Biol Plants. 21(3):349–358. https://doi.org/10.1007/s12298-015-0295-1
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 98(1):107–112. https://doi.org/10.1007/s001220051046
Venkatesan J, Ramu V, Sethuraman T, Sivagnanam C, Doss G (2021) Molecular marker for characterization of traditional and hybrid derivatives of Eleusine coracana (L.) using ISSR marker. J Genet Eng Biotechnol. 19(1):178. https://doi.org/10.1186/s43141-021-00277-1
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 6(1):288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 76(10):5269–5273. https://doi.org/10.1073/pnas.76.10.5269
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet. 155(2):945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14(8):2611–2620
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292
Mirbahar AA, Markhand GS, Khan S, Abul-Soad AA (2014) Molecular characterization of some Pakistani date palm (Phoenix dactylifera L.) cultivars by RAPD markers. Pak J Bot. 46(2):619–625
Srivashtav V, Kapadia C, Mahatma M, Jha S, Ahmad S (2013) Genetic diversity analysis of date palm (Phoenix dactylifera L.) in the Kutchregion of India using RAPD and ISSR markers. Emir J Food Agric. 25(11):907. https://doi.org/10.9755/ejfa.v25i11.14325
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314
Chaluvadi SR, Khanam S, Aly MAM, Bennetzen JL (2014) Genetic diversity and population structure of native and introduced date palm (Phoenix dactylifera) germplasm in the United Arab Emirates. Tropical Plant Biol. 7(1):30–41. https://doi.org/10.1007/s12042-014-9135-7
Moussouni S, Pintaud J-C, Vigouroux Y, Bouguedoura N (2017) Diversity of Algerian oases date palm (Phoenix dactylifera L., Arecaceae): heterozygote excess and cryptic structure suggest farmer management had a major impact on diversity. PLoS ONE. 12(4):e0175232. https://doi.org/10.1371/journal.pone.0175232
Elshibli S, Korpelainen H (2008) Microsatellite markers reveal high genetic diversity in date palm (Phoenix dactylifera L.) germplasm from Sudan. Genetica. 134(2):251–260. https://doi.org/10.1007/s10709-007-9232-8
Al-Najm A et al (2017) Molecular variability and population structure of a core collection of date palm (Phoenix dactylifera L) cultivars from Australia and the Middle East. Aust J Crop Sci. 11(09):1106–1115. https://doi.org/10.21475/ajcs.17.11.09.pne517
Al-Najm A, Luo S, Ahmad NM, Trethowan R (2016) Molecular variability and genetic relationships of date palm ('Phoenix dactylifera’L.) cultivars based on inter-primer binding site (iPBS) markers. Aust J Crop Sci. 10(5):732
Zehdi S, Trifi M, Billotte N, Marrakchi M, Christophe Pintaud J (2005) Genetic diversity of Tunisian date palms (Phoenixdactylifera L.) revealed by nuclear microsatellite polymprohism: Genetic diversity of Tunisian date palms. Hereditas. 141(3):278–287. https://doi.org/10.1111/j.1601-5223.2004.01855.x
Babaei N, Shabanimofrad M (2018) Genetic diversity and population structure of selected Phoenix dactylifera L. Genotypes Iran. 9:9
Marsafari M, Mehrabi AA (2013) Molecular identification and genetic diversity of iranian date palm ('Phoenix dactylifera’L.) Cultivars using ISSR and RAPD markers. Aust J Crop Sci. 7(8):1160–1166
Fakir S (1992) Contribution a l’etude des ressources phytogenetiques chez le palmier dattier. Analyse du polymorphisme enzymatique et protéique. Doctoral dissertation, Paris 6
Zehdi-Azouzi S et al (2015) Genetic structure of the date palm ( Phoenix dactylifera ) in the Old World reveals a strong differentiation between eastern and western populations. Ann Bot. 116(1):101–112. https://doi.org/10.1093/aob/mcv068
khaled Elmeer et al (2019) Assessing genetic diversity of shishi date palm cultivars in Saudi Arabia and Qatar using microsatellite markers. Int J Hortic Sci Technol. 6(1). https://doi.org/10.22059/ijhst.2019.271662.267
Zehdi S, Sakka H, Rhouma A, Ould MSAMM, Trifi M (2004) Analysis of Tunisian date palm germplasm using simple sequence repeat primers. Afr J Biotechnol. 3(4):215–219. https://doi.org/10.5897/AJB2004.000-2040
Elshibli S, Korpelainen H (2009) Excess heterozygosity and scarce genetic differentiation in the populations of Phoenix dactylifera L.: human impact or ecological determinants. Plant Genet. Resour. 7(02):95–104. https://doi.org/10.1017/S1479262108060905
Acknowledgements
We would like to thank Dr. Chiquito Crasto from the Center for Biotechnology and Genomics, at Texas Tech University, Lubbock, TX, USA, for guidance, discussions, critical comments and valuable advice.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization, G.D., R.A., F.G., N.B., and M.I.; methodology, G.D., R.A., F.G., M.L., and M.I.; software, F.G., and M.I. validation, G.D., R.A.,N.B. and M.I.; formal analysis, F.G. and M.I.; investigation, D.I., R.M., J.E, M.I. and R.Z,; resources, G.D. and R.A.; data curation, F.G., and M.I.; writing—original draft preparation, M.I.; writing—review and editing, G.D., R.A. and M.F.; visualization, F.G. and M.I.; supervision, G.D., R.A., F.G., N.B; project administration, G.D., R.A.; funding acquisition, G.D., R.A., and D.I. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table
S1. Latitude and longitude of collected accessions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ibrahimi, M., Brhadda, N., Ziri, R. et al. Analysis of genetic diversity and population structure of Moroccan date palm (Phoenix dactylifera L.) using SSR and DAMD molecular markers. J Genet Eng Biotechnol 21, 66 (2023). https://doi.org/10.1186/s43141-023-00516-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-023-00516-7