Abstract
Background
An entomopathogenic nematode (EPN) was recovered by using Galleria baiting technique from the soils of marigold fields of Noida, Uttar Pradesh, India. Based on morphological, morphometrical and molecular characterizations, the isolated strain was identified as Steinernema abbasi and tagged as CS38. The isolated strain was conspecific to original description with minor deviations. Infective juveniles (IJs) of present strain were longer than original description.
Results
Molecular analysis was done using ITS1-5.8S-ITS2 and D2D3 regions. Pathogenicity and generative potential of the present strain CS38 were tested against larvae of 3 lepidopteran insect pests, namely, Galleria mellonella L., Helicoverpa armigera (Hb.) and Spodoptera litura (Fab.). Different concentrations of IJs/larva, viz. 25, 50, 100, and 200 IJs/larva, were used for bioassay trails. All experiments were repeated thrice to reach the optimum authenticity. Results of bioassays revealed that isolate CS38 was highly virulent against the 3 insect pests and caused (100%) mortality within 48 h under laboratory conditions. Generative potential of the studied S. abbasi CS38 was recorded high in G. mellonella (19 × 104 IJs/larva) at 100 IJs/larva concentration, followed by H. armigera (63.4 × 103 IJs/larva) and S. litura (60.5 × 103 IJs/larva).
Conclusions
Isolate CS38 is an indigenous, dominant and highly virulent strain that can be utilized as a biological control agent against the three studied insects. Moreover, it can be used for commercialization of the production of EPN-based biopesticide to be added under Integrated Pest Management in Indian agriculture system.
Similar content being viewed by others
Background
Agriculture has been facing distressing harm due to various factors worldwide and highly affected by attack of pests and diseases which decline the crop production. Insect pests are one of them causing huge yield loss (15–20%) of agricultural crops in India. The most common problem in the effective control of insect pests is that they develop resistance against chemical pesticides with time. In Integrated Pest Management (IPM), the biological control method is considered to suppress the population of insect pests in agricultural fields. Biopesticide is the effective potential measure that controls the pests by biological non-toxic means without causing environmental hazards. Entomopathogenic nematodes (EPNs) are among extraordinary biocontrol agents than others. They have been studied against various insect pests infesting agricultural crops in different biocontrol programs (Ehlers, 2001) and it has been proven that EPNs, belonging to the families Steinernematidae and Heterorhabditidae, are the most promising and highly effective biological control agents against many insect pests (Kaya and Gaugler, 1993), and they are harmless to vertebrate and plants (Burnell and Stock, 2000). They are found in various habitats from cultivated land to desert (Hominick et al., 1996). EPNs are the soil-inhabiting endoparasitoids, having a great potential to control both soil dwelling and above-ground insect pests (Kaya and Gaugler, 1993 and Ehlers, 2005). Therefore, to control a broad range of insect pests in cryptic and epigeal habitats, they can be used efficiently as a biocontrol agent against grubs, cutworms, crown borers, corn root worm, fungus gnats, thrips, cranefly, beetles, weevils, moths, butterflies, crickets, grasshoppers, and other insect pests. EPNs are effective as the highly recommended commercial products (e.g., Entonem), chemical insecticide (e.g., abamectin and thiamethoxam) that significantly decrease insect pest populations (Laznik et al. 2010). These nematodes are symbiotically associated with entomopathogenic bacteria belonging to the family enteriobacteriacae, which makes them lethal obligatory insect parasitoids that affect a variety of insects including their larval forms and kill them within a short period of time.
Infective juveniles (IJs), the only free-living stage of EPNs, has the ability to search the insect-hosts, enters them through natural openings (Poinar, 1990) or penetrates through thin sections of cuticle (Bedding and Molyneux, 1982) which releases its symbiotic bacteria into the haemolymph after entering the host. These symbionts are the primary agents which are responsible for killing the host within 24–72 h by septicaemia (Boemare and Akhurst, 1988). Two major genera Steinernema and Heterorhabditis are symbiotically associated with bacteria of genus Xenorhabdus and Photorhabdus, respectively. EPNs of both of these genera are used as biocontrol agents because they have the ability to find and kill even deep-seated insects (Bedding and Miller, 1981). Moreover, they are fast-acting, cost-effective and easy to handle. The EPNs are eco-friendly, safe to non-target organisms, have a great potential of reproduction (Kaya and Gaugler, 1993), wide host range, easy to mass produce, capable of resisting under unfavourable conditions (Askary and Ahmad, 2017), easy to apply, compatible with most of the insecticides (Chen, 2003) and sensitive to some insecticides (Laznik and Trdan, 2013a, b).
The greater wax moth, Galleria mellonella L., is a ubiquitous pest of honeybees and their hives. The cotton bollworm, Helicoverpa armigera (Hb.), and tobacco cutworm, Spodoptera litura (Fab.) are highly polyphagous and widely distributed insect pests.
The aim of the present study was to validate the EPN isolate Steinernema abbasi, on the basis of morpho-taxometrical and molecular characterizations. Furthermore, the pathogenicity and generative potential of the EPN isolate against 3 lepidopteran pests were studied.
Methods
Soil sampling
In 2017, a total of 23 soil samples were collected randomly by adopting the technique of Wallace (1971) from marigold fields of Noida (28° 32′ 7.8612″ N and 77° 23′ 27.7044″ E, 206 m above sea level), Uttar Pradesh, India, and brought to the Nematology Laboratory in a well labelled polythene bags for the isolation of EPNs. The characteristics of the positive soil sample were sandy loam with 6.5 pH.
Insect rearing and maintenance
In the present study, 3 lepidopteran insect pests, viz., G. mellonella, H. armigera and S. litura, were reared under laboratory conditions and utilised in experimental work. Larvae of G. mellonella were brought to the laboratory from Bio-control Lab, Sardar Vallabhbhai Patel University of Agriculture and Technology, Modipuram, and reared on semisynthetic diet as suggested by David and Kurup (1988). Eggs and larvae of H. armigera (National accession no. NBAII-MP-NOC-01) and S. litura (National accession no. NBAII-MP-NOC-02) were purchased from ICAR-National Bureau of Agriculturally Important Insects (NBAII), Bangalore. Larvae of H. armigera were reared under laboratory conditions on chickpea-based diet as suggested by Nagarkatti and Prakash (1974), modified by Kalia et al. (2001), while the larvae of S. litura were reared on fresh, properly washed and well-sterilized castor leaves. The 3rd to 4th larval instars of these insects, approximately same size and weight larvae, were used in bioassay experiments. However, the fully grown larvae have also been used for further implications such as isolation and mass production of nematodes.
Isolation and culture of nematodes
EPNs were isolated from the soil samples using the Galleria soil baiting technique (Bedding and Akhurst, 1975). Ten last instar larvae of G. mellonella were placed in a 250 ml sterilized polystyrene jar containing soil sample and kept in BOD at 27 ± 1 °C. For larval mortality, samples were checked daily up to 7 days. The cadavers of G. mellonella were collected from the jar and washed thrice with double-distilled water (DDW), disinfected with 0.1% sodium hypochlorite and transferred on White trap (White, 1927) for IJs. Of the total 23 soil samples collected, only one sample (4.35%) was found positive for EPNs. Koch’s postulate was performed for the confirmation of entomopathogenic nature of the nematodes and the isolated strain was designated as CS38. Isolated IJs were washed thrice with DDW, disinfected with 0.1% sodium hypochlorite and finally, stored in sterilized DDW into vented tissue culture flasks in BOD at 15 °C.
Morpho-taxometrical characterization
Fully grown larvae of G. mellonella were infected with isolated IJs (3rd stage) and adults (males and females) of first and second generations were obtained from cadavers by dissection on 3rd and 5th days after infection. In this study, 20 IJs, 15 males and 15 females of each generation were used for morphology and morphometry. Collected nematodes of different generations were killed separately with hot Ringer’s solution, fixed in TAF solution (2 ml triethanolamine, 7 ml formalin, 91 ml distilled water) (Courtney et al., 1955), dehydrated by Seinhorst method (Seinhorst, 1959) and finally kept in glycerol. Then, they were mounted onto a drop of glycerine on glass slides and coverslips were sealed with paraffin wax. Morphological observations were made using a light compound microscope (Magnus MLX) and phase contrast microscope (Nikon Eclipse 50i). Morphometric measurements were taken with the help of the inbuilt software of a phase contrast microscope (Nikon DS-L1).
Molecular characterization
For molecular studies, genomic DNA was extracted from freshly emerged IJs, using a DNeasy Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions and eluted DNA was stored at − 20 °C until further use. For PCR amplification, Internal Transcribed Spacers (ITS) and 28S ribosomal DNA (D2D3) were used. The Internal Transcribed Spacers regions (ITS1, 5.8S, ITS2) of the rDNA were amplified using primers 18S: 5′-TTGATTACGTCCCTGCCCTTT-3′ (forward) and 26S: 5′-TTTCACTCGCCGTTACTAAGG-3′ (reverse) (Vrain et al., 1992). The rDNA fragment containing D2D3 regions of 28S rDNA were amplified using primers (D2F): 5′-AGCGGAGGAAAAGAAACTAA-3′ (forward) and (D2R): 5′-TCGGAAGGAACCAGCTACTA-3′ (reverse) (Joyce et al., 1994).
The 30 μl PCR reaction mixture consisted of Dream Taq green PCR master mix (15 μl), DNA extract (3 μl), nuclease free distilled water (10 μl) and forward and reverse primers (1 μl of each). The cycling parameters were used as follows: for ITS: 1 cycle of 94 °C for 7 min, followed by 35 cycles of 94 °C for 1 min, 50 °C for 1 min, 72 °C for 1 min and a final extension at 72 °C for 10 min, and for D2D3 fragment of 28S rDNA: 1 cycle of 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 7 min.
PCR was followed by gel electrophoresis, in which the PCR products (5 μl) were analysed on 1% TAE (Tris–acetic acid–EDTA) buffered agarose gel (50 ml) stained with ethidium bromide (1 μl EtBr) at 70 volts for 30 min. The amplified PCR products were purified and then sequenced in both directions using ABI 3730 (48 capillary) electrophoresis instrument by Bioserve Pvt. Ltd. (Hyderabad, India). Obtained amplified forward and reverse sequences were aligned, edited, and assembled using BioEdit (Hall, 1999), and then, the sequencing results were submitted to GenBank with accession numbers MG198915 and MG198916 for ITS and D2D3 regions of Steinernema abbasi strain CS38 respectively.
Sequence alignment and phylogenetic analysis
The sequences were compared to other already available sequences in the GenBank using the Basic Local Alignment Search Tool (BLAST) of the National Centre for Biotechnology Information (NCBI). Based on nucleotide similarities, the related sequences were aligned using default CLUSTALW parameters in MEGA 6.0 (Tamura et al., 2013) and optimized manually in BioEdit (Hall, 1999). Pairwise distances of ITS and D2D3 regions between isolated strain CS38 and other Steinernema species from ‘bicornatum group’ were computed in MEGA 6.0. Codon positions included were 1st + 2nd + 3rd + Noncoding.
Phylogenetic trees were constructed by Maximum Parsimony (MP) using MEGA 6.0 with bootstrap analysis based on 10,000 replicates. Evolutionary distances were also computed using p-distance method in MEGA 6.0 and expressed in the units of base differences per site. All the characters were treated as equally weighted. Steinernema affine and S. scapterisci and S. nepalense were used as outgroups for ITS and D2D3 regions, respectively.
Bioassay for pathogenicity and generative potential
Freshly emerged IJs were used to perform all bioassay experiments to evaluate the pathogenicity and generative potential of S. abbasi CS38 against G. mellonella, H. armigera and S. litura as target hosts. For pathogenicity, bioassay trails were carried out in 6-well plates (well size 3.5 cm) lined with a double layer of Whatman Filter Paper No. 1. Four different concentrations of isolate, viz., 25, 50, 100 and 200 IJs, were prepared with a final volume of 400 μl, using DDW, and were poured into each well of well-plates with the help of micropipette. Ten replicates of the 3 insect larvae of the same size and weight were used for each concentration along with control (only DDW). Larvae were placed individually into each well of above prepared well plates and then, the plates were incubated at 28 ± 2 °C in BOD. Mortality rate was recorded after every 12 h of post infection period (PIP) till 100% mortality was observed. Dead larvae of target insects were transferred onto the modified White Trap (White, 1927) to observe the persistence of infection and for emergence of IJs. Each bioassay was placed separately and to reach the optimum authenticity, all experiments were repeated thrice.
For generative potential, 10 fully grown larvae of each insect (same size and weight) were infected with 100 IJs/larva and incubated at 27 ± 1 °C in BOD. Cadavers were transferred to the White Trap and emerged IJs were collected daily in tissue culture flask for progeny count, up to 20 days till the emergence stopped. Collected nematodes were quantified under stereomicroscope (Nikon SMZ 645) with the help of counting dish in 1 ml suspension.
Statistical analysis
The experimental data of larval mortality bioassay was analyzed statistically, using SPSS software (version 16.0) by applying Probit analysis. To evaluate the pathogenicity, LC50 and LT50 values were also computed at 95% confidence limit. Larval mortality was recorded in the form of percentage mortality and graphical presentations were made using excel. For generative potential, the total number of produced IJs/larva of the studied nematode was analysed by descriptive analysis and presented in number of IJ ± SE (range).
Results
Morpho-taxometrical characterization
The morpho-taxometrical characterizations of the different life stages (IJs, 1st and 2nd generation males and females) of the present isolate CS38 were compared to original described species S. abbasi (Elawad et al., 1997). Due to the presence of horn like cephalic armature in 3rd stage IJs, the present isolate CS38 was characterized as a member of “bicornatum” group and the morphological and taxometrical studies of different life stages revealed that it was conspecific to S. abbasi. Hence, it is named as the same. Light microscope images of S. abbasi CS38 are shown in Fig. 1. Body of IJs was observed larger in size when compared with IJs of already described species. Tail is attenuated, gradually tapering and dorsally curved at tip with slight ventral depression. Female body is robust, strongly curved and C-shaped. Cuticle with fine striae, oesophagus muscular with slightly swollen rounded basal bulb, gonads reflexed containing eggs, vulva with double flapped epiptygma. Tail short, conoid with pointed tip and ventral post anal swelling present in both generations. Male body is slender, ventrally curved and J-shaped upon fixation. Cuticle with fine transverse striae, oesophagus muscular with rounded basal bulb, nerve ring located above the basal bulb and excretory pore present anterior to nerve ring. Testis reflexed, spicule paired and golden dark yellow in colour. Ventrally curved gubernaculum observed boat-shaped and slightly swollen in the middle. Tail short and conoid with a bluntly rounded terminus. However, 2nd generation females and males are smaller in size than first generation. Spicules and gubernaculum of 2nd generation male are also shorter and thinner. Detailed morphometrics of all generation of present strain CS38 and comparative morphometrics of all generations of CS38 with S. abbasi are presented in Tables 1 and 2, respectively.
Light microscope image of Steinernema abbasi CS38 drawn to scale at 400X. a–c: 1st generation female, a- anterior region, b- vulva region, c- posterior region. d–e: 1stgeneration male, d- anterior region, e- posterior region. f–h: 2nd generation female, f- anterior region, g- vulva region, h- posterior region. i–j: 2nd-generation male, i- anterior region, j- posterior region. k–l: Infective juveniles, k- anterior region, l- posterior region
Molecular characterization
For molecular studies, Internal Transcribed Spacer (ITS) and D2D3 region of rDNA were analysed. In the ITS sequence of rDNA, the present strain CS38 did not show any variation with the already described S. abbasi (AY230158). The sequence length of ITS in S. abbasi CS38 was 739 bp with ITS-1 268 bp, 5.8S 157 bp and ITS-2 314 bp same as found in original description. The ITS nucleotide composition of CS38 was A = 175 bp, C = 111 bp, G = 162, T = 291 bp (Table 3). The ITS sequence of CS38 was separated from the other related species of “bicornatum” group by 33 to 191 bp. The pairwise distance matrix is presented in Table 4. The sequence of the D2D3 region was 883 bp and its nucleotide composition was A = 225 bp, C = 146 bp, G = 266 bp, T = 246 bp (Table 5). The D2D3 sequences of CS38 were separated from the other related species of “bicornatum” group by 138 to 208 bp (Table 6). Pairwise distances of ITS and D2D3 regions of rDNA showed 100% similarity and 0 total character difference between present strain and original description.
Phylogenetic analysis
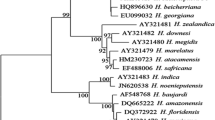
For phylogenetic analysis of ITS and D2D3 regions, the phylogenetic trees were constructed by the Maximum Parsimony Method, using Subtree-Pruning-Regrafting (SPR) algorithm. Both of these regions were too conservative among closely related species to resolve the phylogenetic relations. For ITS region, phylogenetic analysis involved 15 nucleotide sequences and the relationships among those 15 Steinernema species, obtained with MP Tree length = 757; Consistency index = 0.665094; Retention index = 0.742754 and Composite index = 0.533762 (0.494001). For D2D3 region, phylogenetic analysis involved 14 nucleotide sequences and the relationship between those 14 Steinernema species were obtained with MP Tree length = 266; Consistency index = 0.630137; Retention index = 0.736156; Composite index = 0.511988 (0.463879). Phylogenetic analysis of the “bicornatum group” showed a clear monophyly of the group formed by the isolates CS38 and original S. abbasi isolates (Figs. 2 and 3). All positions containing gaps and missing data were eliminated in both ITS and D2D3 regions.
Phylogenetic relationships in the “bicornatum” group of Steinernema based on analysis of ITS rDNA regions. Steinernema affine were used as outgroup taxa. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches. Branch lengths indicate evolutionary distances and are expressed in the units of number of base differences per site
Phylogenetic relationships in the “bicornatum” group of Steinernema based on analysis of D2D3 regions. Steinernema scapterisci and Steinernema nepalense were used as outgroup taxa. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches. Branch lengths indicate evolutionary distances and are expressed in the units of number of base differences per site
Bioassay for pathogenicity and generative potential
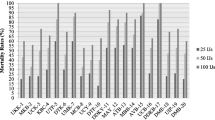
To test the pathogenicity, different IJ concentrations were applied to all target host larvae and different parameters were measured. The data revealed that the present strain CS38 was highly virulent against all the studied insects. Mortality was initiated in all the target insects after 24 h of post infection period (PIP) and recorded as 80, 80, 90 and 100% mortality in G. mellonella, followed by 30, 50, 50 and 60% mortality in H. armigera, and 0, 10, 10 and 20% mortality in S. litura, with 25, 50, 100 and 200 IJs/larva concentrations. At 36 h of PIP, 100% mortality was recorded in G. mellonella with all concentrations, while, in H. armigera and S. litura 100% mortality was observed only in 50, 100 and 200 IJs/larva concentrations. In 25 IJs/larva, complete mortality was observed at 48 h of PIP in H. armigera and S. litura. In control group, no mortality was observed during the experiment even after 60 h. Graphical presentations of percentage mortality in larvae of G. mellonella, H. armigera and S. litura are shown in Fig. 4a–c. Lethal concentration (LC50) was calculated at 24 and 36 h PIP and showed the requirement of 7.806 IJs/larva and 86.032 IJs/larva of the isolate at 24 h to kill the half population of G. mellonella and H. armigera, respectively, which were only 7.67 IJs/larva and 16.202 IJs/larva at 36 h in H. armigera and S. litura, respectively. The lowest LT50 was found at 100 IJs/larva concentration recorded as 19.4 h in G. mellonella, while in H. armigera, the LT50 values were 26.96, 23.17 and 24.32 h at 25, 200 and combined IJs/larva, respectively. However, in S. litura LT50 values were 26.53, 26.53 and 25.74 h at concentrations of 50, 100 and 200 IJs/larva, respectively.
a Percentage mortality in larvae of Galleria mellonella infected with different doses of Steinernema abbasi isolate CS38. b Percentage mortality in larvae of Helicoverpa armigera infected with different doses of Steinernema abbasi isolate CS38. c Percentage mortality in larvae of Spodoptera litura infected with different doses of Steinernema abbasi isolate CS38
To evaluate the generative potential of present isolate, the cadavers of target hosts, infected with 100 IJs/larva concentration, were used for progeny production. The highest progeny count at 100 IJs/larva concentration was recorded as 24.9 × 104, 92 × 103 and 90 × 103 IJs/larva, while the lowest progeny count was 14.8 × 104, 37 × 103 and 31 × 103 IJs/larva in G. mellonella, H. armigera and S. litura, respectively. The mean IJs/larva production count was 19 × 104 IJs/larva in case of G. mellonella 63.4 × 103 IJs/larva in H. armigera and 60.5 × 103 IJs/larva in S. litura (Fig. 5).
Discussion
This is the first valid report of the existence of S. abbasi in Noida, Uttar Pradesh, India. Based on the morpho-taxometrical and molecular studies, the present isolate CS38 was identified as Steinernema abbasi (Elawad et al., 1997). Morphologically, the body of IJs was thin, elongated and larger in size than already described species. Intra-individual variability in the ITS sequence was not found in molecular studies. In the D2D3 sequence of rDNA, there was also no difference between CS38 and the original S. abbasi (AF331890). For ITS region, sequences of S. abbasi formed a monophyletic group with S. yirgalemense (Nguyen et al., 2004) and this pair was sister to the pair of S. biddulphi (Cimen et al., 2016), S. pakistanense (Fayyaz et al., 2001) and S. bifurcatum (Fayyaz et al., 2014). The phylogenetic analysis also showed that the alignment resulted in 1633 characters, of which 289 were constant, 579 variable characters, 375 characters were parsimony uninformative and 390 characters were parsimony informative. A total of 589 positions were present in final dataset. For D2D3 region, S. abbasi CS38 formed a monophyletic group with S. bifurcatum (Fayyaz et al., 2014), S. yirgalemense (Nguyen et al., 2004), S. pakistanense (Fayyaz et al., 2001) and S. biddulphi (Cimen et al., 2016) (Fig. 3). The phylogenetic analysis showed that the alignment resulted in 1309 characters, of which 547 constant, 377 variable characters, 188 parsimony uninformative and 197 characters are parsimony informative. A total of 555 positions were present in the final dataset.
Efficacy of the S. abbasi CS38 was tested on larvae of all the studied insects, i.e., G. mellonella, H. armigera and S. litura, and was found to be excellent biocontrol agent against all 3 target lepidopteran pests under the laboratory conditions. Bhat et al. (2015) also tested 3 isolates of Steinernema sp. against G. mellonella and recorded 100% mortality after 60 h of PIP. Istkhar et al. (2016) observed that the mortality initiated in 24 h of PIP and one strain of Steinernema showed 100% mortality within 60 h in all concentrations, but in case of second strain of Steinernema, the complete mortality was not observed even after 72 h of PIP in 25 and 50 IJs/larva concentrations. Chaubey et al. (2019) studied 3 isolates of S. abbasi and observed that the mortality initiated in larvae of G. mellonella, H. armigera and S. litura only after 12 h of PIP and complete mortality was recorded within 48 h of infection. Gorashi et al. (2014) recorded a complete mortality rate with the same species after 96 and 192 h of PIP in H. armigera and S. litura, respectively. These virulence alterations showed by different species or strains of EPNs against insect pests depend on some factors such as locality and climate, type of insect host, foraging strategy, penetration rate, bacterium complex, generative potential, and other abiotic and biotic factors (Kaya and Gaugler, 1993). IJs are also attracted by roots damaged by insect larvae, showing a chemotactic response to volatile compounds and influenced by different factors and their interactions, which is a strain-specific characteristic of EPN (Laznik and Trdan, 2013a, b). Chaubey et al. (2019) observed the differences in generative potential of different strains of S. abbasi against larvae of the 3 insect pests and recorded a high progeny production in case of G. mellonella. Patil et al. (2020) also noted the difference in generative potential of 3 strains of Steinernema sp. against larval and pupal stages of Mythimna separata (oriental armyworm). The variations found in generative potential of different species or stains of EPNs might be due to host-specificity, larval stage of insect, size of larvae, concentration of nematode and other factors. In the present study, S. abbasi CS38 produced significantly more progeny in G. mellonella than H. armigera and S. litura.
Conclusions
Based on the findings, the present isolate S. abbasi CS38 was characterized as highly virulent to the larvae of G. mellonella, H. armigera and S. litura, which shows that the isolate has good biocontrol potential on insect pests. In comparison to all target insect pest, the rate of generative potential of isolate was high in G. mellonella. The above findings concluded that S. abbasi can be utilized as a good biocontrol agent against all three insect pests. Further studies are still needed before recommending the strain to be applied.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EPN:
-
Entomopathogenic nematode
- IJs:
-
Infective juveniles
- DDW:
-
Double-distilled water
- TAF:
-
Triethanolamine formalin
- ITS:
-
Internal Transcribed Spacer
- PCR:
-
Polymerase chain reaction
- PIP:
-
Post infection period
- LC:
-
Lethal concentration
- LT:
-
Lethal time
- IPM:
-
Integrated Pest Management
References
Askary TH, Ahmad MA (2017) Entomopathogenic nematodes: mass production, formulation and application. In: Abd-Elgawad MM, Askary TH, Coupland J (eds) Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes. CABI International, UK, pp 261–286
Bedding RA, Akhurst RJ (1975) A simple technique for the detection of insect paristic rhabditid nematodes in soil. Nematologica. 21(1):109–110
Bedding RA, Miller LA (1981) Disinfesting blackcurrant cuttings of Synanthedon tipuliformis, using the insect parasitic nematode, Neoaplectana bibionis. Environmental Entomology 10(4):449–453
Bedding RA, Molyneux AS (1982) Penetration of insect cuticle by infective juveniles of Heterorhabditis spp. (Heterorhabditidae: Nematoda). Nematologica 28(3):354–359
Bhat AH, Istkhar A, Chaubey AK (2015) Pathogenicity and reproductive potential of Steinernema sp. isolated from the soils of Baghpat and Bulandshahr districts of Uttar Pradesh. In: Ahmad F (ed) Proceedings of 11th JK Science Congress. Publisher: University of Kashmir, Srinagar, J&K, pp z1–z8
Boemare NE, Akhurst RJ (1988) Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J General Microbiol 134(3):751–761
Burnell AM, Stock SP (2000) Heterorhabditis, Steinernema and their bacterial symbionts - lethal pathogens of insects. Nematology 2(1):31–42
Chaubey AK, Aasha BAH (2019) Notes on Steinernema abbasi (Rhabditida: Steinernematidae) strains and virulence tests against lepidopteran and coleopterans pests. J Entomol Zool Stud 7:954–964
Chen S (2003) Infectivity and persistence of entomopathogenic nematodes used to control the cabbage root fly Delia radicum. Ph. D. Thesis, University of Ghent, Belgium, p 135
Cimen H, Půža V, NermuŤ J, Hatting J, Ramakuwela T, Hazir S (2016) Steinernema biddulphi n. sp., a new entomopathogenic nematode (Nematoda: Steinernematidae) from South Africa. J Nematol 48(3):148–158
Courtney WD, Polley D, Miller VL (1955) TAF, an improved fixative in nematode technique. Plant Dis Reporter 39(7):570–571
David H, Kurup NK (1988) Techniques for mass production of Sturmiopsis inferens. In: David H, Easwaramoorthy E (eds) Biocontrol technology for sugarcane pest management. Sugarcane Breeding Institute, Coimbatore, India, pp 87–92
Ehlers RU (2001) Mass production of entomopathogenic nematodes for plant protection. Appl Microbiol Biotechnol 56:623–633
Ehlers RU (2005) Forum on safety and regulation. In: Grewal PS, Ehlers RU, Shapiro Ilan DI (eds) Nematodes as Biocontrol Agents. CABI Publishing, Wallingford, UK, pp 107–114
Elawad S, Ahmad W, Reid AP (1997) Steinernema abbasi sp. n. (Nematoda: Steinernematidae) from the Sultanate of Oman. Fundamental and Appl Nematol 20(5):435–442
Fayyaz S, Anis M, Reid AP, Rowe J, Maqbool MA (2001) Steinernema pakistanense sp. n. (Rhabditida: Steinernematidae) from Pakistan. Int J Nematol 11(1):124–133
Fayyaz S, Yan X, Qiu L, Han R, Gulsher M, Khanum TA, Javed S (2014) A new entomopathogenic nematode, Steinernema bifurcatum n. sp. (Rhabditida: Steinernematidae) from Punjab, Pakistan. Nematology 16(7):821–836
Gorashi NE, Tripathi M, Kalia V, Gujar GT (2014) Identification and characterization of the Sudanese Bacillus thuringiensis and related bacterial strains for their efficacy against Helicoverpa armigera and Tribolium castaneum. NISCAIR Online Periodicals Repository, pp 637–649
Hall TA (1999) BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/ NT. Nucleic Acids Symposium Series 41:95–98
Hominick WM, Alexander PR, David AB, Bernard RB (1996) Entomopathogenic Nematodes: Biodiversity, geographical distribution and the convention on biological diversity. Biocontrol Sci Technol 6:317–332
Istkhar BAH, Aasha B, Panwar A, Chaubey AK (2016) A report on dose dependent pathogenicity of populations of Steinernema surkhetense along with Steinernema abbasi isolates of Indian origin using laboratory host Galleria mellonella. J Experimental Zool India. 19:1393–1398
Joyce SA, Reid A, Driver F, Curran J (1994) Application of polymerase chain reaction (PCR) methods to the identification of entomopathogenic nematodes. In: Burnell AM, Ehlers R-U, Masson JP (eds) COST 812 Biotechnology: Genetics of entomopathogenic nematode-bacterium complexes, Proceedings of Symposium & Workshop, St. Patrick's College, Maynooth, Co. Kildare, Ireland. European Commission, DG XII, Luxembourg, pp 178–187
Kalia V, Chaudhari S, Gujar GT (2001) Changes in haemolymph constituents of American bollworm, Helicoverpa armigera (Hübner), infected with nucleopolyhedrovirus. Indian J Experimental biol 39(11):1123–1129
Kaya HK, Gaugler R (1993) Entomopathogenic nematodes. Ecofriendly Pest Management for Food Security 38:181–206
Laznik Ž, Tóth T, Lakatos T, Vidrih M, Trdan S (2010) Control of the Colorado potato beetle (Leptinotarsa decemlineata [Say]) on potato under field conditions: a comparison of the efficacy of foliar application of two strains of Steinernema feltiae (Filipjev) and spraying with thiametoxam. J Plant Dis Protection 117(3):129–135
Laznik Ž, Trdan S (2013a) The influence of insecticides on the viability of entomopathogenic nematodes (Rhabditida: Steinernematidae and Heterorhabditidae) under laboratory conditions. Pest Management Sci 70(5):784–789
Laznik Ž, Trdan S (2013b) An investigation on the chemotactic responses of different entomopathogenic nematode strains to mechanically damaged maize root volatile compounds. Experimental Parasitol 134(3):349–355
Nagarkatti S, Prakash A (1974) Rearing Heliothis armigera (Hubn.) on an artificial diet. Technical Bulletin Common Wealth Institute of Biological Control, Bangalore, India, pp 17–169
Nguyen KB, Tesfamariam M, Gozel U, Gaugler R, Byron JA (2004) Steinernema yirgalemense n. sp. (Rhabditida: Steinernematidae) from Ethiopia. Nematology 6(6):839–856
Patil J, Vijayakumar R, Linga V, Sivakumar G (2020) Susceptibility of Oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae) larvae and pupae to native entomopathogenic nematodes. J Appl Entomol. 00:1–8. https://doi.org/10.1111/jen.12786
Poinar GO (1990) Taxonomy and biology of Steinernematidae and Heterorhabditidae. Pg. 23–61 in R. Gaugler and H. K. Kaya, eds. Entomopathogenic Nematodes in Biological Control. CRC Press, Boca Raton
Seinhorst JW (1959) A rapid method for the transfer of nematodes from fixative to anhydrous glycerin. Nematologica 4(1):67–69
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12):2725–2729
Vrain T, Wakarchuk D, Levesque A, Hamilton R (1992) Intraspecific rDNA restriction fragment length polymorphism in the Xiphinema americanum group. Fundamental Appl Nematol 15(6):563–573
Wallace HR (1971) In: Zukerman RARBM, Mai WM (eds) Plant Parasitic Nematodes. Pp. 257–80 in Abiotic influences in the soil environment. Academic, New York: New York
White GF (1927) A method for obtaining infective nematode larvae from cultures. Science 66:302–303
Acknowledgements
The authors are thankful to the Head of the Department of Zoology, Chaudhary Charan Singh University, Meerut, for providing necessary laboratory facilities for conducting the experiments.
Funding
There are no funding sources for this manuscript.
Author information
Authors and Affiliations
Contributions
The study was designed by all authors. H and AR calculated the morphometric measurements. H and AHB performed the molecular analysis. Interpretation of the data, manuscript writing and reference settings were done by the first author (H). Errors and grammatical mistakes in manuscript were removed and edited by AKC. Then, final manuscript was read and approved by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Heena, Rana, A., Bhat, A.H. et al. Morpho-taxometrical and molecular characterization of Steinernema abbasi (Nematoda: Steinernematidae) and its pathogenicity and generative potential against lepidopteran pests. Egypt J Biol Pest Control 31, 21 (2021). https://doi.org/10.1186/s41938-020-00359-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-020-00359-1