Abstract
Background
Wolbachia, obligate intracellular bacteria, infect the majority of arthropods, including many mosquito species of medical importance. Some Wolbachia strains interfere with the development of Plasmodium parasites in female Anopheles, a major vector of malaria. The use of Wolbachia as a means to block malaria transmission is an emerging vector control strategy in highly endemic areas. Hence, identification of native Wolbachia strains in areas where malaria transmission is low may uncover a particular Wolbachia strain capable of Plasmodium interference. This study aims to identify native Wolbachia strains in female Anopheles spp. that are predominant in a low-malaria transmission area in mainland Southeast Asia.
Methods
Following a 2-year survey of malaria vectors in Umphang Valley of Tak Province, Thailand, DNA extracts of female An. minimus, An. peditaeniatus, and An. maculatus were subjected to amplification of the conserved region of the 16S rRNA-encoding gene. The DNA sequences of the amplicons were phylogenetically compared with those of known Wolbachia strains.
Results
Among three Anopheles spp., amplification was detected in only the DNA samples from An. minimus. The DNA sequencing of amplicons revealed 100% similarity to Wolbachia pipientis, confirming the specificity of amplification. The Wolbachia-positive An. minimus samples were devoid of Plasmodium 18S rRNA amplification. The phylogenetic trees indicate a close relationship with Wolbachia strains in subgroup B.
Conclusion
To the best of our knowledge, the data presented herein provide the first molecular evidence of a Wolbachia strain in An. minimus, hereinafter named wAnmi, in a low-malaria transmission area in the Umphang Valley of western Thailand. Further biological characterization is required to examine its potential for malaria transmission control in the field.
Graphical Abstract

Similar content being viewed by others
Background
Malaria is a vector-borne parasitic disease caused by Plasmodium species. Ongoing malaria control programs significantly decreased morbidity and mortality in Africa and Asia between 2010 and 2018 [1]; however, many parts of the border regions, especially in Southeast Asia, are still malaria-endemic areas [1, 2]. Given that Anopheles mosquitoes are malaria vectors, one of the effective strategies to control malaria transmission relies on the use of insecticides, including indoor residual spraying and the use of insecticide-treated mosquito nets. Accordingly, resistance to insecticides has emerged as a biological threat to malaria control and elimination efforts in endemic areas, including many regions located in forest-mountain landscapes along the Thailand-Myanmar [3] and Thailand-Laos PDR borders [4]. Although widespread insecticide resistance has increased globally in many malaria-endemic regions, there was no evidence of an association between insecticide resistance and malaria burden [5]. However, a meta-analysis-based model of malaria transmission predicted that insecticide resistance potentially increased malaria incidence in part because of the decrement of mosquito mortality [6], challenging global malaria eradication. Thus, an effective alternative to insecticides is needed.
Wolbachia is an intracellular bacterium that naturally infects the majority of insect species [7]. Wolbachia bacteria reside in the cytoplasmic vacuoles of various types of insect somatic and germ cells, allowing maternal transmission to their progeny. Wolbachia is capable of manipulating host reproduction through cytoplasmic incompatibility [8, 9], in which Wolbachia-infected eggs form viable offspring and noninfected eggs do not. Moreover, the infected progeny tend to sexually develop into females, while unfertilized eggs develop into females [10, 11], leading to widespread Wolbachia infection in an insect population [12, 13], including mosquito species of medical importance. Since Wolbachia is reportedly capable of suppressing pathogen development and blocking disease transmission, the use of Wolbachia has been proposed as a mean of controlling transmission of pathogenic viruses causing dengue virus [14, 15], West Nile virus [16], yellow fever virus, and Chikungunya virus [17].
Given failures in the detection of Wolbachia in Anopheles mosquitoes, it was initially hypothesized that Anopheles mosquitoes are refractory to Wolbachia infection [18,19,20,21]. However, a study demonstrated that the Aedes albopictus-specific Wolbachia strain AlbB (wAlbB) could infect laboratory-reared An. stephensi and suppress the development of P. falciparum within female Anopheles mosquitoes [22]. In concordance with the laboratory study, Wolbachia infections were observed in natural populations of An. gambiae and An. coluzzii, two major vectors in malaria-endemic regions of Burkina Faso [23, 24]. Interestingly, researchers phylogenetically identified Anopheles-infecting Wolbachia as a new arthropod-specific subgroup named wAnga [23]. Previous reports have shown evidence of natural Wolbachia endosymbiosis in other Anopheles species as well as its effects on Plasmodium development. In the high-malaria transmission area of Burkina Faso, a field study showed that natural infection with the Wolbachia strain wAnga in blood-fed An. coluzzii females was negatively correlated with Plasmodium development [25]. Based on a mathematical model, natural Wolbachia infection potentially blocks malaria transmission from vector to human [25]. Furthermore, infection by the Wolbachia strain wAnga-Mali in An. gambiae was associated with a reduced prevalence and intensity of sporozoite infection in field-collected females in Mali [26]. Altogether, studies strongly suggest that Anopheles mosquitoes are permissible to Wolbachia endosymbiosis and that some strains of Wolbachia are capable of interfering with the development of Plasmodium parasites in female Anopheles. Thus, the release of laboratory-reared, Wolbachia-infected Anopheles mosquitoes to replace the wild Anopheles population is a potential strategy to block malaria transmission. Hence, identification of native Wolbachia strains in areas where malaria transmission is low may uncover a particular Wolbachia strain capable of interfering with Plasmodium development in Anopheles.
In Thailand, only one survey of Wolbachia in mosquitoes was conducted to amplify the filamenting temperature-sensitive mutant Z (ftsz) and Wolbachia surface protein (wsp) genes. All 23 mosquito species in the genera Aedes, Culex, and Mansonia were positive for the ftsz and wsp genes, whereas none of the 19 Anopheles species were positive [18]. Failure to detect Wolbachia-specific genes in Anopheles spp. was consistent with the results of studies in European, African, and American specimens [19, 20]. Nevertheless, detection of the Wolbachia 16S rRNA region was accomplished. The W-Spec primers were designed to specifically amplify a 438-bp sequence at the 3ʹ region of the 16S rRNA gene in Wolbachia [27]. The W-Spec primers allowed the detection of Wolbachia in temperate North American arthropods, including the family Culicidae but excluding other mosquito families. Subsequently, Baldini et al. reported the first evidence of Wolbachia in the reproductive organs of male and female An. gambiae, a major malaria vector in sub-Saharan Africa. In the same DNA samples, the W-Spec primer-based PCR was able to amplify the 16S rRNA fragment, whereas Wolbachia-specific surface protein and fructose-biphosphate aldolase-based PCR failed [23], implying good sensitivity of the W-Spec primers. Moreover, Shaw et al. further improved the sensitivity of W-Spec primer-based PCR by using nested primers (16SNF and 16SNR). The use of nested PCR allowed the detection of Wolbachia in An. coluzzii [25], An. gambiae in Mali [26], and An. arabiensis in Tanzania [28]. Additional studies were able to amplify the Wolbachia 16S rRNA fragment in DNA samples extracted from head-thorax or thorax-abdomen, implying the possibility of Wolbachia infection in nonreproductive organs [22, 29]. Collectively, Wolbachia infection in somatic and germ cells can be detected using nested PCR, which amplifies the conserved region of the Wolbachia 16S rRNA gene. Considering the availability of DNA extracts from major Anopheles species obtained during a 2-year survey of malaria vectors [30] and An. minimus is the important malaria vector carrying P. vivax sporozoites [31], this study aims to identify native Wolbachia strains in female Anopheles spp. that are predominant in a low-transmission area in Umphang Valley, located near the Thailand-Myanmar border of mainland Southeast Asia.
Methods
Biosafety for using biological samples of mosquitoes
The protocol for the use of DNA samples extracted from Anopheles mosquitoes was approved by the Siriraj Safety Risk Management Taskforce, Faculty of Medicine Siriraj Hospital, Mahidol University (SI2020-010). In accordance with the guidelines for ethics in animal use, this study submitted the DNA extraction protocol and sampling details to the Siriraj Animal Care and Use Committee, Faculty of Medicine Siriraj Hospital, Mahidol University (COA 012/2563), and received permission.
Collection and identification of Anopheles species
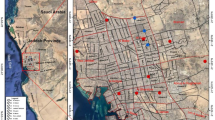
Since this study is an extension of a previously published report, we did not collect and identify Anopheles species. For detailed collection methods, we highly recommend reading the original article [30]. The collection site was located in Ban Nong Luang village (16° 04ʹ 36.3ʺ N 98° 45ʹ 8.0ʺ E), Umphang District of Tak Province, located in western Thailand (Fig. 1a). The village is located approximately 4 km from the border of Kayin state, Myanmar. Mosquitoes were captured for five consecutive nights every 2 months from February 2015 to December 2016. The standard mosquito landing collection procedure followed that in a previous report [32]. Briefly, mosquito capture methods included indoor human landing, outdoor human landing, and cattle-baited outdoor collections. For human landing, mosquitoes were collected in a 6-h period: 18:00-00:00 h and 00:00-06:00 h. In indoor human landing, a volunteer was sitting inside the house. When mosquitoes landed on the lower part of the legs, they were collected using aspiration. The collection was performed continuously for 45 min, followed by a 15-min resting period. To collect mosquitoes outdoors, another volunteer stayed outside the house located 30 m away from the same house. For the cattle-baited outdoor collections, an adult cow was covered with a two-layered cotton bed net. To prevent mosquito biting, the inner layer enclosed the ground, while the outer layer was above the ground, allowing entry of mosquitoes into the net. Interlayer-residing mosquitoes were collected at the end of each 45-min collection period. Mosquitoes were identified following a standard dichotomous key [33], and Mansonia mosquito were morphologically identified according to the key characteristics [34]. Individual mosquitoes were placed in a 1.5-ml tube, frozen in liquid nitrogen, and stored at − 80 °C. For molecular identification of Anopheles spp. and Wolbachia, DNA from individual mosquitoes was extracted from the head and thorax and subjected to the multiplex allele-specific polymerase chain reaction assays and nested PCR, respectively. There were four assays of multiplex allele-specific PCR with regard to the Dirus Complex (An. dirus, An. cracens, An. scanloni, An. baimaii, and An. nemophilous) [35], the Minimus Complex and related species (An. minimus, An. harrisoni, An. aconitus, An. varuna and An. pampanai) [36], the Maculatus Group (An. maculatus, An. sawadwongporni, An. pseudowillmori, An. dravidicus and An. rampae (former Form K)) [37], and the Hyrcanus Group (An. argyropus, An. crawfordi, An. nigerrimus, An. nitidus, An. paraliae, An. peditaeniatus, An. pursati, and An. sinensis) [38]. A total of 731 DNA samples were obtained from An. minimus (n = 401, 55%), An. peditaeniatus (n = 200, 27%), and An. maculatus (n = 130, 18%) (Fig. 1b). To pool the DNA from each mosquito species for analysis, 2 μL of 10–11 DNA samples was mixed in the same tube. There were 40, 20, and 13 DNA sample pools from An. minimus, An. peditaeniatus, and An. maculatus, respectively (Fig. 1b).
Collection site of Anopheles spp. a Google map of mainland Southeast Asia (left) and the location in which the Anopheles spp. were collected (right side). The collection site was located at 16° 04ʹ 36.3ʺ N 98° 45ʹ 8.0ʺ E (red pins) in Umphang Valley of Tak Province in western Thailand. The yellow arrow indicates the Thailand-Myanmar border. Scale bars are 500 and 5 km. b Anopheles spp. in the subgenera Cellia and Anopheles and the total number of DNA samples included in this study
Amplification of the Wolbachia-specific 16S rRNA coding region
To amplify a conserved region of the Wolbachia 16S rDNA-encoding gene, W-SpecF and W-SpecR primers were used in the initial standard PCR, and 16SNF and 16SNR primers were used in the nested PCR, following a previous report [27] (Fig. 2a). Primer sequences are shown in Table 1. Samples were prepared with a total volume of 10 µL, which was composed of 0.5 μM of each primer and 1 μL of DNA template and AccuStart™ II Gel Track PCR SuperMix (Quantabio, Beverly, MA, USA). Amplification was performed with DNA template denaturation at 95 °C for 3 min; followed by 35 cycles of DNA denaturation at 95 °C for 15 s, primer annealing at 50 °C for 25 s, and DNA extension at 72 °C for 30 s; and final extension at 72 °C for 5 min. To visualize the DNA bands, the PCR products were mixed with ViSafe Red (Vivantis Technologies Sdn. Bhd., Selangor Darul Ehsan, Malaysia) and subsequently electrophoresed in 2% agarose gel in 1× TAE buffer at a voltage of 100 V for 45–50 min. The ViSafe Red-intercalated, double-stranded DNA sequences were then exposed to UV light (Molecular Imager® Gel Doc™ XR System, Bio-Rad Laboratories, Inc., Hercules, CA) for visualization. The length of the amplicon yielded from the initial PCR was approximately 438 bp in length. Subsequently, 1 µL of the initial PCR product was used as template for the nested PCR, in which 16SNF and 16SNR primers to bind to the internal sequence of W-SpecF and W-SpecR were added (Fig. 2a, lower panel). The thermal cycles included initial denaturation at 95 °C for 3 min; followed by 35 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 25 s and extension at 72 °C for 30 s; with a final extension at 72 °C for 5 min. The length of the amplicon yielded from the nested PCR was approximately 412 bp in length. To confirm specificity, the 412-bp amplicons were purified from the agarose gel and sequenced by an ABI 3730XL DNA Analyzer (Bionics, Seoul, South Korea). The 16SNF and 16SNR primers were used as DNA sequencing primers. Since Mansonia uniformis and M. indiana are naturally infected with Wolbachia [18, 21] in Thailand, DNA extracts of Mansonia mosquitoes were used as the positive control. PCR without DNA template was used as the negative control.
Amplification of the Wolbachia 16S rRNA-encoding gene. a Schematic diagram showing two-step PCR, including standard and nested PCR. In the standard PCR, W-SpecF and W-SpecR primers (blue colored arrows in upper panel) attached to the 3ʹ region of the Wolbachia 16S rRNA-encoding gene, amplifying a 438-bp fragment. In the nested PCR, the 438-bp amplicons generated from the regular PCR were used as templates. The 16SNF and 16SNR primers attached the internal sequence of the 438-bp fragment, generating a 412-bp PCR product. b A representative image of the 438-bp amplicons obtained from the standard PCR. Two and three representative pools of DNA extracts of Anopheles are shown. c A representative image of amplicons derived from the standard PCR (438 bp) and nested PCR (412 bp). For the standard PCR, templates were obtained from the DNA extracts from An. minimus pool numbers 21 (P21), 32 (P32), and 36 (P36). DNA from Mansonia spp. was used as the positive control (P), while absent template DNA was used as the negative control (NTC). In the nested PCR, all the samples from the standard PCR were used as templates. The PCR products were analyzed with electrophoresis in a 2% agarose gel. Lane M: the DNA ladder electrophoresed simultaneously with the PCR product to determine amplicon size
Quantitative amplification of Wolbachia 16S rRNA and Plasmodium 18S rRNA region
Level of Wolbachia 16S rRNA- and Plasmodium 18S rRNA-coding DNA sequence was examined in the same DNA sample as the nested PCR using quantitative real-time PCR (qPCR). The use of W-Specf and W16S primers in qPCR had > 100% efficiency in amplification and detected the Wolbachia 16S rRNA-coding region at concentrations lower than combination of W-Specf and W-Specr did [26, 39]. Thus, this study deployed the W-Specf and W16S primers. Detection of Plasmodium 18S rRNA and An. minimus ITS2A region was performed using the primer set published by Shaw et al. [25] and Eamet al. [40]. Primer sets are shown in Table 1. Luna® Universal qPCR Master Mix (New England BioLabs) was used, and the primer concentration was 250 nM for each primer. After an initial denaturation at 95 °C for 1 min, thermal cycles were as follows: denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 30 s (CFX96™ Real-Time System and C1000™ Thermal Cycler, Bio-Rad). The amplicons having melting temperatures similar to the positive control were interpreted as “detectable,” whereas amplicons that have different melting temperature or cycle threshold (Ct) values > 40 were regarded as “undetectable” [26]. To compare the amount of gDNA template of Wolbachia and Plasmodium spp., the level of Wolbachia 16S rRNA- and Plasmodium 18S rRNA-coding DNA sequence was normalized with the level of the An. minimus ITS2A region. Levels of Wolbachia and Plasmodium spp. in a given sample were compared using the 2−∆∆CT method based on assumption of 100% qPCR efficiency and were shown as the relative level of Wolbachia [41]. Gene expression analyses were carried out in triplicate for each sample. The gDNA of Mansonia spp. or Plasmodium falciparum strain K1 [42] were used as positive control, respectively. Nuclease-free water was set as the no template control. Melt cure analysis was performed at the end of amplification.
Bioinformatics
The obtained sequences of the Wolbachia 16S rRNA fragment were edited and assembled using BioEdit Sequence Alignment Editor (version 7.2.5). DNA sequences were deposited in GenBank (accession nos. MT449018 and MT449019). To identify similar sequences, the GenBank database was searched with BLASTN [43]. For sequence alignment, the following sequences of the 16S rRNA-encoding gene of Wolbachia subgroup B were obtained: Wolbachia strain wNo from Drosophila simulans (CP003883.1), strain wAlbB from Ae. albopictus (KX155506.1), and wPip from Culex quinquefasciatus (AM999887.1). To analyze nucleotide substitution, the multiple sequence alignment was performed using MSAViewer.
Phylogenetic analysis
The conserved region of the Wolbachia 16S rRNA-encoding gene was phylogenetically analyzed using Molecular Evolutionary Genetics Analysis (MEGA) software version 10.0 [44] and NGPhylogeny.fr [45]. Wolbachia 16S rRNA sequences of other strains belonging to subgroups A, B, C, D, E, F, H, and Anopheles-specific subgroups were retrieved from GenBank for analysis (Table 2). Rooted and unrooted phylogenetic trees were analyzed based on the neighbor-joining method. Rickettsia montanensis was used as a non-Wolbachia outgroup for rooted tree analysis.
Data analysis
Statistical analyses and graph generation were performed using GraphPad Prism software version 5.0 (GraphPad Software, Inc., San Diego, CA, USA). Statistically significant differences were identified using the non-parametric Mann-Whitney test. A p value < 0.05 was regarded as being statistically significant.
Results
Amplification of the Wolbachia 16S rRNA-encoding gene from field-captured Anopheles species
In the initial PCR, the W-SpecF and W-SpecR primers specifically bound to a conserved region at the 3ʹ end of the Wolbachia 16S rRNA-encoding gene, generating an amplicon with an approximate length of 438 bp (Fig. 2a, upper panel). A representative image of agarose gel electrophoresis shows low-intensity DNA bands of between 400 and 500 bp, amplified from DNA pools of An. minimus, An. maculatus, and An. peditaeniatus (Fig. 2b). Among 73 DNA sample pools, 14 pools yielded 438-bp amplicons: 10 pools of An. minimus, 3 pools of An. peditaeniatus, and 1 pool of An. maculatus. Preliminary data of two pools of An. aconitus containing seven DNA samples in each pool yielded 538-bp amplicons in one pool (additional file 1). The W-Spec-based PCR product obtained from the 15 DNA sample pools from the initial runs was subsequently used as template in the nested PCR using the 16SN primer (Fig. 1a, lower panel). The 16SNF and 16SNR primers yielded amplicons from An. minimus pool numbers 21, 32, and 36 (Fig. 2c and Additional file 1: Fig. S1). Given that the size and intensity of the amplicon obtained from An. minimus pool numbers 36 and 40 were similar to that of the no template control or smaller than that of the positive control, respectively (Fig. 2c), we therefore interpreted this as a negative result. No amplification was observed in An. peditaeniatus, An. maculatus, or An. aconitus in the nested PCR (Additional file 1).
To confirm whether the yielded amplicons were the conserved region of Wolbachia 16S rRNA, the PCR products derived from DNA pool numbers 21 (P21) and 32 (P32) from An. minimus were subjected to DNA sequencing using 16SNF and 16SNR primers. The GenBank database was searched for similar sequences using the BLASTN program. DNA sequencing of P21 and P32 yielded 326 nucleotides, and all 326 nucleotides of P21 and P32 were aligned with 97 and 100 sequences, respectively, of Wolbachia pipientis 16S rRNA (query coverage = 100). P21 and P32 were 100% and 99.39% identical to all 16S rRNA sequences of W. pipientis, respectively. The BLASTN and MSAViewer results are provided as an Additional file 2. Hereafter, we referred to the Wolbachia strain identified in An. minimus as wAnmi. The place and pool number were tagged; wAnmi_UmpP21 and wAnmi_UmpP32 represent Wolbachia from An. minimus isolated from Umphang Valley and from pool numbers 21 and 32, respectively.
Correlation of Wolbachia and Plasmodium infection in Anopheles minimus
Levels of Wolbachia and Plasmodium in the DNA sample of An. minimus were calculated based on the level of the respective 16S and 18S rRNA amplification using quantitative real-time PCR. Among 21 individual samples of pool number 21 (n = 10) and 32 (n = 11), five samples exhibited amplification of the Wolbachia 16S rRNA region (orange dots in Fig. 3a). The amplicons were detected at the cycle threshold of 32–39 and had the same melting temperature. After normalization with levels of the An. minimus ITS2A region, relative levels of Wolbachia 16S rRNA varied among the five samples: 7–776 fold differences (Fig. 3a). To examine correlation between Wolbachia and Plasmodium in the individual An. minimus, we deployed the heatmap to indicate the relative level of both microorganisms in the individual An. minimus. All Wolbachia-positive samples (n = 5) exhibited undetectable levels of Plasmodium 18S rRNA (Fig. 3b). By contrast, Plasmodium-positive An. minimus (n = 6) had undetectable levels of Wolbachia, implying negative correlation between Wolbachia and Plasmodium (Fig. 3b). However, the level and prevalence of Plasmodium in An. minimus samples were not statistically significantly different between the two groups: the undetectable and detectable Wolbachia (p = 0.71, Fig. 3c).
Correlation of Wolbachia and Plasmodium infection in An. minimus. Levels of Wolbachia 16S rRNA and Plasmodium 18S rRNA were examined using quantitative PCR. DNA levels in individual samples were normalized with that of An. minimus ITS2A. Levels of each DNA were relative to a reference sample, which was set as 1. a Relative levels of Wolbachia (X axis) and Plasmodium (Y axis) in the individual samples of An. minimus pool numbers 21 and 32 were plotted. Five DNA samples were detectable for Wolbachia 16S rRNA (orange dots), while the others were undetectable (blue dots). b Heatmap compares relative levels of Wolbachia 16S rRNA and Plasmodium 18S rRNA in all 21 An. minimus samples. High to low DNA levels were displayed as red to green, respectively. Numbers are relative levels calculated using the 2−∆∆CT method. W: Wolbachia 16S rRNA-coding region; P: plasmodium 18S rRNA-coding region. c Level and prevalence of Plasmodium in the An. minimus DNA samples with and without Wolbachia (blue and orange dots, respectively)
Phylogenetic analysis
To assess the relationship of wAnmi_UmpP21 and wAnmi_UmpP32 with other known Wolbachia strains, we assembled a phylogenetic tree to determine genetic similarity and heterogeneity based on the conserved region of the Wolbachia 16S rRNA-encoding gene (Fig. 2a, c), a heritable region in prokaryotes. To validate the output data, we employed rooted and unrooted phylogenetic inference methods. Based on an assumption of a common ancestral path, the rooted maximum likelihood phylogram illustrated that both wAnmi_UmpP21 and wAnmi_UmpP32 were genetically related to Wolbachia subgroup B (Fig. 4). wAnm_UmpP21 was in the same cluster as the wPip strain from Cx. quinquefasciatus and the wAlbB strain from Ae. albopictus. In contrast, wAnmi_UmpP32 was closely related to wAnga isolated from An. gambiae in Burkina Faso (wAnga_BF) and wAnga isolated from An. arabiensis in Tanzania (wAnga_TZ140) (Fig. 4). To reveal the extent of genetic similarity between the Wolbachia members in subgroup B, we aligned the Wolbachia 16S rRNA-conserved regions of the wNo, wAlbB, wPip, wAnmi_UmpP21, and wAnmi_UmpP32 strains (Fig. 5). One hundred percent similarity was observed among the wAlbB, wPip, and wAnmi_UmpP21 strains. However, wAnmi_UmpP32 had two single nucleotide polymorphisms at two locations, implying genetic variation in Wolbachia in wild An. minimus in Umphang Valley. In agreement with the rooted maximum likelihood phylogenetic tree, excluding the assumption of a common ancestor, the unrooted, bifurcating phylogenetic tree revealed that wAnmi_UmpP21 and wAnmi_UmpP32 clustered in the leaf node of Wolbachia subgroup B, confirming a close genetic relationship (Fig. 6).
Rooted phylogenetic tree of the Wolbachia strain identified in An. minimus collected from Umphang Valley compared to other Wolbachia strains. The conserved region of the Wolbachia 16S rRNA-encoding DNA sequence obtained from the identified Wolbachia strains (blue letters) was phylogenetically compared with those in Wolbachia subgroups A, B, C, D, E, F, and H (black letters) and Anopheles-specific subgroups (orange letters) using MEGA software version 10. Rickettsia montanensis was used as the reference outgroup. A tree scale of 0.01 corresponds to inferred evolutionary changes. Details of the DNA sequences retrieved from GenBank are shown in Table 2
Multiple sequence alignment of the conserved region of the Wolbachia 16S rRNA gene. The sequence of the conserved region of the Wolbachia 16S rRNA gene was compared to those of wNo in Drosophila simulans (CP003883.1), wAlbB in Aedes albopictus (KX155506.1), and wPip in Culex quinquefasciatus (AM999887.1), members of subgroup B. DNA sequences were aligned with the wAnmi_UmpP21 and wAnmi_UmpP32 strains identified in the present study. The consensus sequence was illustrated using MSAViewer
Unrooted phylogenetic tree of the Wolbachia strain identified in An. minimus collected from Umphang Valley compared to other Wolbachia strains. The conserved region of the Wolbachia 16S rRNA-encoding DNA sequence obtained from the identified Wolbachia strains (blue letters) was phylogenetically compared with those of Wolbachia subgroups A, B, C, D, E, F, and H (black letters) and Anopheles-specific subgroups (orange letters) using NGPhylogeny.fr. Rickettsia montanensis was used as the reference outgroup. A tree scale of 0.01 corresponds to inferred evolutionary changes. Details of the DNA sequences retrieved from GenBank are shown in Table 2
Discussion
The data presented herein are preliminary evidence of native Wolbachia in An. minimus, a major malaria vector in an endemic area of mainland Southeast Asia. The identified Wolbachia in An. minimus, herein called wAnmi, was phylogenetically clustered in subgroup B, similar to wAlbB, which has been associated with the suppression of development of P. falciparum in An. stephensi [22]. Along the Thailand-Myanmar border, malaria transmission continues in many areas [46, 47], including the Umphang District of Tak Province in western Thailand. Most collection sites were in the Ban Nong Luang Village of Umphang District located in a valley primarily surrounded by forested mountains. Since some villagers engage in hunting-gathering and agriculture in areas near the forest (Fig. 1a), the risk of malaria infection in individuals is high. In the Umphang Valley, there are reportedly seven putative malaria vectors: An. minimus, An. dirus, An. baimaii, An. sawadwongporni, An. maculatus, An. pseudowillmori, and An. aconitus [30]. In the Thasongyang District northern Umphang Valley, female An. minimus, An. maculatus, An. annularis, and An. barbirostris have been shown to carry P. vivax sporozoites, confirming their role in malaria transmission [31]. Regarding the number of Anopheles in the Umphang Valley, An. minimus was the most abundant (> 50%), followed by the potential malaria vectors An. peditaeniatus (~ 20%) and An. maculatus (~ 10%) [30]. Hence, this study included samples from An. minimus, An. peditaeniatus, and An. maculatus, the major malaria vectors, for the detection of Wolbachia. In addition, we examined 14 DNA samples of An. aconitus samples as a preliminary data.
In Thailand, only one survey of Wolbachia in mosquitoes was conducted to amplify the filamenting temperature-sensitive mutant Z (ftsz) and Wolbachia surface protein (wsp) genes. All 23 mosquito species in the genera Aedes, Culex, and Mansonia were positive for the ftsz and wsp genes, whereas none of the 19 Anopheles species were positive [18]. Failure to detect Wolbachia-specific genes in Anopheles spp. was consistent with the results of studies in European, African, and American specimens [19, 20]. Nevertheless, detection of the Wolbachia 16S rRNA region was accomplished. The W-Spec primers were designed to specifically amplify a 438-bp sequence at the 3ʹ region of the 16S rRNA gene in Wolbachia [27]. The W-Spec primers allowed the detection of Wolbachia in temperate North American arthropods, including the family Culicidae but excluding other mosquito families. Subsequently, Baldini et al. reported the first evidence of Wolbachia in the reproductive organs of male and female An. gambiae, a major malaria vector in sub-Saharan Africa. In the same DNA samples, the W-Spec primer-based PCR was able to amplify the 16S rRNA fragment, whereas Wolbachia-specific surface protein and fructose-biphosphate aldolase-based PCR failed [23], implying good sensitivity of the W-Spec primers. Moreover, Shaw et al. further improved the sensitivity of W-Spec primer-based PCR by using nested primers (16SNF and 16SNR). The use of nested PCR allowed the detection of Wolbachia in An. coluzzii [25], An. gambiae in Mali [26], and An. arabiensis in Tanzania [28]. Additional studies were able to amplify the Wolbachia 16S rRNA fragment in DNA samples extracted from head-thorax or thorax-abdomen, implying the possibility of Wolbachia infection in nonreproductive organs [22, 29]. Collectively, Wolbachia infection in somatic and germ cells can be detected using nested PCR, which amplifies the conserved region of the Wolbachia 16S rRNA gene.
Nested PCR is regarded as a highly sensitive tool for detecting targets of interest that are present in very low amounts. We sometimes failed to amplify the 438-bp fragment using W-SpecF and W-SpecR in the initial PCR; however, there were 412-bp amplicons observed in the nested PCR [25], implying good sensitivity of the nested PCR. As such, false-negative results may occur in cases of low-intensity Wolbachia infection because the quantity of the target of interest is below the limit of nested PCR. Given that DNA samples were extracted from the head and thorax of female Anopheles, detection failure in nested PCR is possibly because of low-intensity infection or the reproductive organ specificity of Wolbachia. Therefore, assays with high sensitivity, such as quantitative PCR, may aid in the detection of low-intensity Wolbachia infection [39, 48, 49]. Moreover, DNA preparation from the whole body of mosquitoes ensures the inclusion of Wolbachia strains that specifically infect germ cells.
The high sensitivity of nested PCR may cause low specificity, especially when primers bind to the conserved region of a common gene. Since Wolbachia is capable of infecting the majority of insect species [50], and the W-Spec primers amplify the conserved region of the Wolbachia 16S rRNA-coding gene, false-positive results due to environmental contamination from other insects may occur. In our study, 438-bp DNA amplicons with low-fluorescence intensity were present in the initial PCR, but we failed to reamplify these amplicons in the subsequent nested PCR using the 16SNF and 16SNR primers, suggesting the possibility of nonspecific amplification in the initial run. Moreover, if the environmental contamination is at an extremely low level in the initial run and cannot be detected, the subsequent runs will be able to amplify, owing to a sufficient amount of template. As shown in Fig. 2c, the DNA band could be observed in the no template control. To minimize environmental contamination, PCR preparations were performed in a clean hood for the initial and nested PCR. Despite great care in the pre-PCR steps, we sometimes observed DNA bands in the negative control lane. Thus, DNA sequencing of the PCR product was necessary to confirm Wolbachia-specific amplification. Importantly, given the possible environmental contamination in the previous survey of Anopheles spp., a new field study of Wolbachia in A. minimus needs to be conducted in the same area. To address environmental contamination, the DNA probe- or antibody-based microscopic imaging of Wolbachia will be employed to validate the PCR-based findings. We are now undertaking the field study in the Umphang Valley. Because the additional work may take some time, we will report the finding in a new study.
To the best of our knowledge, the present data are the first preliminary evidence of native Wolbachia in An. minimus. However, this study has limitations. First, the presence of the Wolbachia 16S rRNA gene in DNA samples from Anopheles is not direct evidence of natural Wolbachia infection because environmental contamination during mosquito capture, DNA extraction, and PCR preparation is possible. Intracellular localization of Wolbachia in Anopheles spp. is a more definitive indicator of Wolbachia infection than sequencing. This could be done by using in situ hybridization [25, 51]. Moreover, given the availability of the remaining DNA samples obtained during a previous 2-year survey [30], this study included the DNA samples regardless of the sample size. Thus, the prevalence of Wolbachia in Anopheles spp. in Umphang Valley could not be statistically estimated. Finally, all mosquitoes were collected from the same area located in the Ban Nong Luang village, and only Wolbachia subgroup B was identified in An. minimus. By contrast, Sawasdichai et al. could molecularly detect the high diversity of Wolbachia in An. minimus and An. maculatus collected from different villages [39]. Thus, the Wolbachia subgroup B in An. minimus may represent a subpopulation of Wolbachia, and undetectable amplification of Wolbachia in An. maculatus, An. peditaenitus, and An. aconitus did not indicate that these species are refractory to Wolbachia infection. Detection of Wolbachia in more diverse areas will address this issue. Collectively, identification of the Wolbachia strain in Anopheles spp. requires further confirmation, in which high-sensitivity assays, such as fluorescent in situ hybridization [25] and quantitative PCR [39], whole mosquitoes, and more diverse areas will be included.
Wolbachia has been under investigation for its potential application in blocking malaria transmission. In a recent report, An. gambiae mosquitoes were naturally infected with Wolbachia at different levels, and those infected with a high level of Wolbachia were likely devoid of Plasmodium development [26]. In agreement with the study in An. gambiae, the relative level of Wolbachia varied among An. minimus examined in our study. The likelihood of Plasmodium inhibition was observed in the Wolbachia-deteced An. minimus; however, low sample numbers of An. minimus having Wolbachia 16S rRNA resulted in a non-significant difference in the prevalence and level of Plasmodium. Thus, more field-isolated An. minimus need to be included to provide a definitive tendency. As proof of concept, field trials in Australia demonstrated that the release of laboratory-reared mosquitoes infected with Wolbachia resulted in the rapid spread of Wolbachia among wild uninfected mosquito populations [15]. Population invasion by a particular Wolbachia strain depends on the level of cytoplasmic incompatibility, host fitness (survival, fecundity and fertility), and vertical transmission. Therefore, the following issues need to be assessed: the potential of the native Wolbachia identified in An. minimus to render resistance to Plasmodium parasites and interfere with malaria transmission, its ability to cause cytoplasmic incompatibility, and its effects on host fitness.
Conclusion
To the best of our knowledge, the data presented herein are the first molecular evidence of a Wolbachia strain in An. minimus, named wAnmi, in a low-malaria transmission area in the Umphang Valley of western Thailand. Further biological characterization is required to examine its potential as a malaria transmission control strategy in the field.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional information files. The sequences of Wolbachia 16S rRNA fragments generated in the present study were submitted to GenBank under the accession numbers MT449018 and MT449019 and are retrievable.
Abbreviations
- An.:
-
Genus Anopheles
- 16S rRNA:
-
Small subunit 16 of ribosomal ribonucleic acid
- 16SNF:
-
Small subunit 16 of ribosomal ribonucleic acid-nested forward primer
- 16SNR:
-
Small subunit 16 of ribosomal ribonucleic acid-nested reverse primer
- Ump:
-
Umphang
- W-SpecF:
-
Wolbachia-specific forward primer
- W-SpecR:
-
Wolbachia-specific reverse primer
References
Organization WH. World malaria report 2019. 2019. https://www.who.int/news-room/feature-stories/detail/world-malaria-report-2019.
Wongsrichanalai C, Kurdova-Mintcheva R, Palmer K. Current malaria situation in Asia-Oceania. Methods Mol Biol. 2019;2013:45–56. https://doi.org/10.1007/978-1-4939-9550-9_3.
Chaumeau V, Cerqueira D, Zadrozny J, Kittiphanakun P, Andolina C, Chareonviriyaphap T, et al. Insecticide resistance in malaria vectors along the Thailand-Myanmar border. Parasit Vectors. 2017;10:165. https://doi.org/10.1186/s13071-017-2102-z.
Sumarnrote A, Overgaard HJ, Marasri N, Fustec B, Thanispong K, Chareonviriyaphap T, et al. Status of insecticide resistance in Anopheles mosquitoes in Ubon Ratchathani province Northeastern Thailand. Malar J. 2017;16:299. https://doi.org/10.1186/s12936-017-1948-z.
Kleinschmidt I, Bradley J, Knox TB, Mnzava AP, Kafy HT, Mbogo C, et al. Implications of insecticide resistance for malaria vector control with long-lasting insecticidal nets: a WHO-coordinated, prospective, international, observational cohort study. Lancet Infect Dis. 2018;18:640–9. https://doi.org/10.1016/S1473-3099(18)30172-5.
Churcher TS, Lissenden N, Griffin JT, Worrall E, Ranson H. The impact of pyrethroid resistance on the efficacy and effectiveness of bednets for malaria control in Africa. Elife. 2016;5:e16090. https://doi.org/10.7554/eLife.16090.
Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7:e38544. https://doi.org/10.1371/journal.pone.0038544.
Breeuwer JA, Werren JH. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature. 1990;346:558–60. https://doi.org/10.1038/346558a0.
Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–51. https://doi.org/10.1038/nrmicro1969.
Weeks AR, Breeuwer JA. Wolbachia-induced parthenogenesis in a genus of phytophagous mites. Proc Biol Sci. 2001;268:2245–51. https://doi.org/10.1098/rspb.2001.1797.
Negri I, Pellecchia M, Mazzoglio PJ, Patetta A, Alma A. Feminizing Wolbachia in Zyginidia pullula (Insecta, Hemiptera), a leafhopper with an XX/X0 sex-determination system. Proc Biol Sci. 2006;273:2409–16. https://doi.org/10.1098/rspb.2006.3592.
O’Neill SL, Karr TL. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature. 1990;348:178–80. https://doi.org/10.1038/348178a0.
Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–2. https://doi.org/10.1038/353440a0.
Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–3. https://doi.org/10.1038/nature10355.
Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–7. https://doi.org/10.1038/nature10356.
Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE. 2010;5:e11977. https://doi.org/10.1371/journal.pone.0011977.
van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, et al. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis. 2012;6:e1892. https://doi.org/10.1371/journal.pntd.0001892.
Kittayapong P, Baisley KJ, Baimai V, O’Neill SL. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J Med Entomol. 2000;37:340–5. https://doi.org/10.1093/jmedent/37.3.340.
Ricci I, Cancrini G, Gabrielli S, D’Amelio S, Favi G. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J Med Entomol. 2002;39:562–7. https://doi.org/10.1603/0022-2585-39.4.562.
Rasgon JL, Scott TW. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J Med Entomol. 2004;41:255–7. https://doi.org/10.1603/0022-2585-41.2.255.
Wiwatanaratanabutr I. Geographic distribution of wolbachial infections in mosquitoes from Thailand. J Invertebr Pathol. 2013;114:337–40. https://doi.org/10.1016/j.jip.2013.04.011.
Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–51. https://doi.org/10.1126/science.1236192.
Baldini F, Segata N, Pompon J, Marcenac P, Shaw WR, Dabire RK, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985. https://doi.org/10.1038/ncomms4985.
Buck M, Nilsson LK, Brunius C, Dabire RK, Hopkins R, Terenius O. Bacterial associations reveal spatial population dynamics in Anopheles gambiae mosquitoes. Sci Rep. 2016;6:22806. https://doi.org/10.1038/srep22806.
Shaw WR, Marcenac P, Childs LM, Buckee CO, Baldini F, Sawadogo SP, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772. https://doi.org/10.1038/ncomms11772.
Gomes FM, Hixson BL, Tyner MDW, Ramirez JL, Canepa GE, Alves ESTL, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci USA. 2017;114(47):12566–71. https://doi.org/10.1073/pnas.1716181114.
Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–85. https://doi.org/10.1098/rspb.2000.1139.
Baldini F, Rouge J, Kreppel K, Mkandawile G, Mapua SA, Sikulu-Lord M, et al. First report of natural Wolbachia infection in the malaria mosquito Anopheles arabiensis in Tanzania. Parasit Vectors. 2018;11:635. https://doi.org/10.1186/s13071-018-3249-y.
Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. https://doi.org/10.1371/journal.ppat.1002043.
Tananchai C, Pattanakul M, Nararak J, Sinou V, Manguin S, Chareonviriyaphap T. Diversity and biting patterns of Anopheles species in a malaria endemic area, Umphang Valley, Tak Province, western Thailand. Acta Trop. 2019;190:183–92. https://doi.org/10.1016/j.actatropica.2018.11.009.
Sriwichai P, Samung Y, Sumruayphol S, Kiattibutr K, Kumpitak C, Payakkapol A, et al. Natural human Plasmodium infections in major Anopheles mosquitoes in western Thailand. Parasit Vectors. 2016;9:17. https://doi.org/10.1186/s13071-016-1295-x.
Sungvornyothin S, Kongmee M, Muenvorn V, Polsomboon S, Bangs MJ, Prabaripai A, et al. Seasonal abundance and bloodfeeding activity of Anopheles dirus sensu lato in western Thailand. J Am Mosq Control Assoc. 2009;25:425–30. https://doi.org/10.2987/09-5907.1.
Rattanarithikul R, Harrison BA, Harbach RE, Panthusiri P, Coleman RE, Panthusiri P. Illustrated keys to the mosquitoes of Thailand. IV. Anopheles. Southeast Asian J Trop Med Public Health. 2006;37(Suppl 2):1–128. https://www.ncbi.nlm.nih.gov/pubmed/17262930.
Rattanarithikul R, Panthusiri P. Illustrated keys to the medically important mosquitos of Thailand. Southeast Asian J Trop Med Public Health. 1994;25(Suppl 1):1–66. https://www.ncbi.nlm.nih.gov/pubmed/7831585.
Walton C, Handley JM, Kuvangkadilok C, Collins FH, Harbach RE, Baimai V, et al. Identification of five species of the Anopheles dirus complex from Thailand, using allele-specific polymerase chain reaction. Med Vet Entomol. 1999;13:24–32. https://doi.org/10.1046/j.1365-2915.1999.00142.x.
Garros C, Koekemoer LL, Coetzee M, Coosemans M, Manguin S. A single multiplex assay to identify major malaria vectors within the African Anopheles funestus and the Oriental An. minimus groups. Am J Trop Med Hyg. 2004;70:583–90. https://www.ncbi.nlm.nih.gov/pubmed/15210996.
Walton C, Somboon P, O’Loughlin SM, Zhang S, Harbach RE, Linton YM, et al. Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infect Genet Evol. 2007;7:93–102. https://doi.org/10.1016/j.meegid.2006.05.001.
Hempolchom C, Otsuka Y, Baimai V, Thongsahuan S, Saeung A, Taai K, et al. Development of a multiplex PCR assay for the identification of eight species members of the Thai Hyrcanus Group (Diptera: Culicidae). Appl Entamol Zool. 2013;48:469–76. https://doi.org/10.1007/s13355-013-0207-1.
Sawasdichai S, Chaumeau V, Dah T, Kulabkeeree T, Kajeechiwa L, Phanaphadungtham M, et al. Detection of diverse Wolbachia 16S rRNA sequences at low titers from malaria vectors in Kayin state, Myanmar. Wellcome Open Res. 2019;4:11. https://doi.org/10.12688/wellcomeopenres.15005.4.
Eamkum P, Sungvornyothin S, Kritpetcharat O, Daduang J, Lek-Uthai U, Charerntanyarak L, et al. A single-round multiplex PCR assay for the identification of Anopheles minimus related species infected with Plasmodium falciparum and Plasmodium vivax. Parasitol Int. 2014;63:442–9. https://doi.org/10.1016/j.parint.2013.11.001.
Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinform Biomath. 2013;3:71-85. https://www.ncbi.nlm.nih.gov/pubmed/25558171.
Kulkeaw K, Ketprasit N, Tungtrongchitr A, Palasuwan D. A simple monochromatic flow cytometric assay for assessment of intraerythrocytic development of Plasmodium falciparum. Malar J. 2020;19:74. https://doi.org/10.1186/s12936-020-03156-1.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. https://doi.org/10.1093/nar/25.17.3389.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. https://doi.org/10.1093/molbev/msy096.
Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, et al. NGPhylogenyfr.: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47:W260–5. https://doi.org/10.1093/nar/gkz303.
Lawpoolsri S, Sattabongkot J, Sirichaisinthop J, Cui L, Kiattibutr K, Rachaphaew N, et al. Epidemiological profiles of recurrent malaria episodes in an endemic area along the Thailand-Myanmar border: a prospective cohort study. Malar J. 2019;18:124. https://doi.org/10.1186/s12936-019-2763-5.
Saita S, Pan-Ngum W, Phuanukoonnon S, Sriwichai P, Silawan T, White LJ, et al. Human population movement and behavioural patterns in malaria hotspots on the Thai-Myanmar border: implications for malaria elimination. Malar J. 2019;18:64. https://doi.org/10.1186/s12936-019-2704-3.
Jeffries CL, Lawrence GG, Golovko G, Kristan M, Orsborne J, Spence K, et al. Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa. Wellcome Open Res. 2018;3:113. https://doi.org/10.12688/wellcomeopenres.14765.2.
Niang EHA, Bassene H, Makoundou P, Fenollar F, Weill M, Mediannikov O. First report of natural Wolbachia infection in wild Anopheles funestus population in Senegal. Malar J. 2018;17:408. https://doi.org/10.1186/s12936-018-2559-z.
Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc Biol Sci. 2015;282:20150249. https://doi.org/10.1098/rspb.2015.0249.
Zouache K, Voronin D, Tran-Van V, Mousson L, Failloux AB, Mavingui P. Persistent Wolbachia and cultivable bacteria infection in the reproductive and somatic tissues of the mosquito vector Aedes albopictus. PLoS ONE. 2009;4:e6388. https://doi.org/10.1371/journal.pone.0006388.
Acknowledgements
The authors acknowledge the staff of the District Vector-Borne Disease Control Unit (VBDU 9.3.3) in Umphang District, Tak Province, and students and staff of the Department of Entomology, Faculty of Agriculture, Kasetsart University, for the collection, morphological identification, DNA extraction, and molecular typing of mosquito species. We thank Prof. Dr. Sirirchit Wongkumchai and Achinya Phuakrod of the Department of Parasitology, Faculty of Siriraj Hospital Mahidol University, for providing DNA samples of Masonia. The authors gratefully acknowledge the receipt of a Research Career Development Grant from the Thailand Science Research and Innovation (grant no. [IO] RSA6280102) and a grant from the Siriraj Research Fund, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (grant no. [IO] R016233004).
Funding
Funding was provided by the Research Career Development Grant from Thailand Science Research and Innovation, no. RSA6280102, and the Siriraj Research Fund, Faculty of Medicine Siriraj Hospital, Mahidol University, no. R016233004.
Author information
Authors and Affiliations
Contributions
NT performed the experiments and analyzed and interpreted the data; PR analyzed and interpreted the data and wrote the manuscript; CT and TC performed the experiments; KK designed the study, analyzed and interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Results of the standard PCR.
Additional file 2:
BLASTN result of P21. BLASTN result of P32.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tongkrajang, N., Ruenchit, P., Tananchai, C. et al. Molecular identification of native Wolbachia pipientis in Anopheles minimus in a low-malaria transmission area of Umphang Valley along the Thailand-Myanmar border. Parasites Vectors 13, 579 (2020). https://doi.org/10.1186/s13071-020-04459-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04459-7