Abstract
Background
Masked palm civets are known to play an important role in the transmission of some zoonotic pathogens. However, the distribution and zoonotic potential of Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium spp. in these animals remain unclear.
Methods
A total of 889 fecal specimens were collected in this study from farmed masked palm civets in Hainan, Guangdong, Jiangxi and Chongqing, southern China, and analyzed for these pathogens by nested PCR and DNA sequencing.
Results
Altogether, 474 (53.3%), 34 (3.8%) and 1 (0.1%) specimens were positive for E. bieneusi, G. duodenalis and Cryptosporidium sp., respectively. Sequence analysis revealed the presence of 11 novel E. bieneusi genotypes named as PL1–PL11 and two known genotypes Peru8 and J, with PL1 and PL2 accounting for 90% of E. bieneusi infections. Phylogenetically, PL4, PL5, PL9, PL10 and PL11 were clustered into Group 1, while PL1, PL2, PL3, PL6, PL7 and PL8 were clustered into Group 2. Assemblage B (n = 33) and concurrence of B and D (n = 1) were identified among G. duodenalis-positive animals. Further multilocus genotyping of assemblage B has revealed that all 13 multilocus genotypes in civets formed a cluster related to those from humans. The Cryptosporidium isolate from one civet was identified to be genetically related to the Cryptosporidium bamboo rat genotype II.
Conclusions
To the best of our knowledge, this first report of enteric protists in farmed masked palm civets suggests that these animals might be potential reservoirs of zoonotic E. bieneusi and G. duodenalis genotypes.

Similar content being viewed by others
Background
Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium spp. are enteric pathogens in humans and various wild and domestic animals, causing diarrhea and other gastrointestinal symptoms. Humans can be infected by these pathogens through contact of contaminated fomites or ingestion of contaminated food or water (food-borne or water-borne transmission) [1,2,3].
The identification of genetic diversity in these pathogens has accelerated in recent years. Thus far, nearly 500 E. bieneusi genotypes have been identified, belonging to 11 phylogenetic groups with different host preferences, including the zoonotic Group 1 and host-adapted Groups 2–11 [2]. Similarly, eight distinct G. duodenalis assemblages of A–H with different host ranges have been identified by genetic characterization [4]. Among them, assemblages A and B are the most common causes of human giardiasis [5]. There are also nearly 40 recognized Cryptosporidium species and at least as many genotypes of unknown species status, most of which have host preference [3]. Recently, high-resolution multilocus genotyping (MLG) tools have been employed to elucidate the genetic heterogeneity of these pathogens in humans and animals [6,7,8].
Masked palm civets (Paguma larvata), belonging to the order Carnivora and family Viverridae, are wild mammals distributed widely in Asia [9]. In southern China, they are raised as new farm animals, as their meat is considered a culinary delicacy. In 2003, masked palm civets gained attention due to their potential involvement in the outbreak of severe acute respiratory syndrome (SARS), which originated from southern China and spread to over 30 countries [10]. In addition, results of other studies have suggested that they may play a role in the transmission of other zoonotic pathogens such as Salmonella enterica, Campylobacter spp., Bartonella henselae and Toxoplasma gondii [11,12,13,14]. Thus far, the occurrence and zoonotic potential of E. bieneusi, G. duodenalis and Cryptosporidium spp. in masked palm civets remain unclear.
This study was conducted to examine the prevalence, genetic identity and zoonotic potential of E. bieneusi, G. duodenalis and Cryptosporidium spp. in farmed masked palm civets in southern China.
Methods
Specimens
From April 2018 to March 2019, a total of 889 fecal specimens were collected from masked palm civets on four commercial farms in Hainan, Guangdong, Jiangxi and Chongqing, southern China (Fig. 1). The management of animals on all four farms was similar, with the farm in Jiangxi being the largest in scale and having the longest history of over 20 years. The sanitary conditions of farms in Hainan and Chongqing were poor compared with the other two farms. Masked palm civets were kept in groups of 2–6 animals per cage, with some interactions with those in neighboring cages. Fresh fecal droppings from civets were collected on the ground under each cage, with one fecal specimen being collected from each cage for this study. The age of animals was divided into three groups: < 1 year (n = 469); 1–2 years (n = 129); and > 2 years (n = 291). All animals sampled in this study were clinically normal without obvious signs of diarrhea. Specimens were stored at 4 °C in 2.5% potassium dichromate before DNA extraction.
DNA extraction
Prior to genomic DNA extraction, potassium dichromate was removed by washing 500 μl of fecal suspension three times with distilled water by centrifugation at 2000×g for 10 min. DNA was extracted from the sediment using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) as described [15]. The extracted DNA was stored at − 20 °C until being used in PCR analyses.
PCR amplification
Enterocytozoon bieneusi was identified by nested PCR amplification of the internal transcribed spacer (ITS) of the rRNA gene [16]. Giardia duodenalis was detected by nested PCR targeting the triosephosphate isomerase (tpi), β-giardin (bg), and glutamate dehydrogenase (gdh) genes [17]. Cryptosporidium spp. were detected by PCR and sequence analyses of the small subunit (SSU) rRNA and further characterized by sequence analysis of the 60-kDa glycoprotein (gp60), 70-kDa heat-shock protein (hsp70) and actin genes [18,19,20,21]. Two replicates were used in PCR analysis of each target for each specimen. DNA of genotype PtEb IX from dogs, assemblage E from cattle, and Cryptosporidium bovis from cattle were used as positive controls in PCR analysis of E. bieneusi, G. duodenalis and Cryptosporidium spp., respectively, while reagent-grade water was used as the negative control.
Sequence analysis
All positive secondary PCR products were sequenced bi-directionally on an ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). To determine the genetic identity of E. bieneusi, G. duodenalis and Cryptosporidium spp., sequences obtained were assembled using ChromasPro 2.1.6. (http://technelysium.com.au/ChromasPro.html), edited using BioEdit 7.1 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and aligned with each other and reference sequences downloaded from GenBank using ClustalX 2.0.11 (http://clustal.org). Genotypes or subtypes of these pathogens were named according to the established nomenclature system [22, 23]. Maximum likelihood (ML) trees were constructed to infer the phylogenetic relationships among species or genotypes of these pathogens using MEGA 7.0.14 (http://www.megasoftware.net/). The general time-reversible model was used in substitution rate calculations and 1000 replicates were used in bootstrapping analysis.
Statistical analysis
The Chi-square test was used to compare differences in the prevalence of pathogens among geographical locations or age groups. Differences were considered significant at P < 0.05.
Nucleotide accession numbers
Representative nucleotide sequences generated in this study were deposited in the GenBank database under the accession numbers MT497888–MT497900 (E. bieneusi), MT487560–MT487588 (G. duodenalis), MT487589–MT487591 and MT779807 (Cryptosporidium sp.).
Results
Occurrence of E. bieneusi, G. duodenalis and Cryptosporidium sp
Occurrence of the three pathogens was determined based on the PCR positivity of at least one PCR replicate. Of the 889 fecal specimens collected from masked palm civets, 474 (53.3%) were positive for E. bieneusi, with infection rates ranging from 35.2% (172/489) to 86.1% (180/209) among the four farms sampled. Infection rates on the farms in Hainan (86.1%) and Chongqing (85.9%) were significantly higher than in Guangdong (46.2%; χ2 = 56.407 and 32.149, respectively; P < 0.0001) and Jiangxi (35.2%; χ2 = 152.05 and 76.11, respectively; P < 0.0001) (Table 1). By age, E. bieneusi infection rates in animals of < 1 year (61.0%; χ2 = 29.110, P < 0.0001) and 1–2 years (53.5%; χ2 = 5.734, P = 0.0166) were significantly higher than in animals of > 2 years (40.9%) (Table 1).
For G. duodenalis, 34 (3.8%) of the 889 specimens were positive based on the PCR positivity at any one of the three genetic loci. The highest infection rate was 13.2% (14/106) on the farm in Guangdong, followed by Chongqing (8.2%, 7/85), Hainan (3.8%, 8/209) and Jiangxi (1.0%, 5/489). The infection rate on the farm in Guangdong was significantly higher than in Hainan (χ2 = 9.525, P = 0.002) and Jiangxi (χ2 = 37.993, P < 0.0001) (Table 1). Additionally, animals of 1–2 years (10.9%) had significantly higher prevalence of G. duodenalis than those of < 1 year (2.8%; χ2 = 15.324, P < 0.0001) and > 2 years (2.4%; χ2 = 13.427, P = 0.0002) (Table 1).
Among the 889 specimens analyzed, only one (0.1%) from the farm in Hainan was positive for Cryptosporidium sp. This civet was co-infected with E. bieneusi. In addition, co-infection of E. bieneusi and G. duodenalis was detected in 31 other animals, with a significantly higher occurrence of co-infection of the two pathogens on the farm in Guangdong (12.3%) than in Hainan (2.9%; χ2 = 10.949, P = 0.0009) and Jiangxi (1.0%; χ2 = 33.793, P < 0.0001). By age, a significantly higher co-infection rate was detected in civets aged 1–2 years (10.1%) than < 1 year (2.6%; χ2 = 14.278, P = 0.0002) and > 2 years (2.1%; χ2 = 13.296, P = 0.0003) (Table 1).
Enterocytozoon bieneusi genotypes
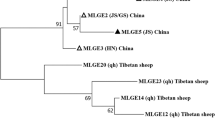
Sequence analysis of the ITS PCR products was successful for 457 of 474 E. bieneusi-positive specimens. The remaining 17 specimens generated sequences with underlying signals of mixed nucleotides, probably due to concurrence of more than one E. bieneusi genotype in each specimen. Altogether, 13 E. bieneusi genotypes were detected, including two known ones (Peru8 and J) and 11 novel genotypes (named as PL1–PL11) (Table 1). The latter were identified based on nucleotide sequence differences from known E. bieneusi genotypes. The ITS sequences of Peru8 in Group 1 and genotype J in Group 2 were identical to the GenBank reference sequences AY371283 and MF592787, respectively. Among the 11 novel E. bieneusi genotypes, PL4, PL5, PL9, PL10 and PL11 were phylogenetically placed in Group 1, while PL1, PL2, PL3, PL6, PL7 and PL8 formed a clade named as Group 2-like, which was mostly related to Group 2 (Fig. 2).
Phylogenetic relationship among Enterocytozoon bieneusi genotypes based on the maximum-likelihood analysis of the internal transcribed spacer of the rRNA gene. Bootstrap values greater than 50% from 1000 replicates are shown on the branches. Known and novel genotypes identified in this study are indicated by blue and red triangles, respectively
PL1 (53.0% or 242/457) and PL2 (37.0% or 169/457) were the two most prevalent genotypes (Table 1). PL1 was the dominant genotype on the three farms in Hainan, Chongqing and Jiangxi, while PL2 was the most common one on the farm in Guangdong. The highest genetic diversity was observed on the farm in Jiangxi, with 2 known and 8 novel genotypes. Several unique genotypes were detected on some of the farms, including PL4, PL5, PL6, PL9, PL10, PL11 on the farm in Jiangxi, PL7 and PL8 on the farm in Guangdong, and PL3 on the farm in Chongqing.
Giardia duodenalis assemblages
Among the 34 G. duodenalis-positive specimens, assemblage B was the dominant genotype, being detected in 33 specimens. One specimen, however, had the concurrence of assemblages B and D. By genetic locus, 13 sequence types were present among the 28 assemblage B-positive specimens at the tpi locus, including 3 known ones and 10 new ones. The three known sequence types, i.e. MB8 (n = 8), MB7 (n = 3) and B14 (n = 1), were identical to GenBank reference sequences KF679746, KF679745 and KF679737, respectively. In contrast, the new sequence types B-PL01 to B-PL10 had 1 to 6 single nucleotide polymorphisms (SNPs) compared with the GenBank reference sequence KF679746.
At the bg locus, one known and two new sequence types were obtained among the 16 assemblage B-positive specimens. Bb-3 (n = 5) was identical to the GenBank reference sequence KJ888976, while the new sequence types of B-PL11 and B-PL12 had one SNP compared with the GenBank reference sequence KJ888976.
At the gdh locus, 10 new sequence types (B-PL13 ~ B-PL22) were detected among the 22 assemblage B-positive specimens, with 1–4 SNPs compared with the GenBank reference sequence LC430575.
Multilocus genotypes of G. duodenalis assemblage B
Of the 33 assemblage B-positive specimens, 13 were positive by PCR at all three loci, forming 13 MLGs (Civet-MLG-B1 to Civet-MLG-B13) based on the concatenated sequences of the gdh, bg and tpi loci. Phylogenetic analysis of these MLGs was conducted together with those from previous studies of assemblage B in humans and various animals [24,25,26,27,28]. It showed that all 13 MLGs from civets in this study formed a cluster, which was a sister group with MLGs from humans in Sweden (Fig. 3).
Phylogeny of multilocus genotypes (MLGs) of Giardia duodenalis assemblage B from this and previous reports based on the maximum-likelihood analysis of concatenated sequences of the gdh, bg and tpi loci. Bootstrap values greater than 50% from 1000 replicates are shown on the branches. MLGs identified in this study are indicated by red triangles
Cryptosporidium genotype
For the Cryptosporidium-positive specimen SCAU12558, DNA sequencing of PCR products of the SSU rRNA, gp60 and hsp70 genes revealed that it was phylogenetically related to the Cryptosporidium bamboo rat genotype II (Fig. 4). The SSU rRNA sequence generated had one SNP and three nucleotide insertions compared with the sequence from the bamboo rat genotype II (GenBank: MK731962). The gp60 sequence generated had a maximum nucleotide identity of 83.0% to the bamboo rat genotype II (GenBank: MK731966), with extensive sequence differences in the non-repeat regions. The hsp70 sequence generated had 17 SNPs and 1 nucleotide deletion with the bamboo rat genotype II (GenBank: MK731969), mostly over the repeat region at the 3′-end of the gene. Due to the absence of the actin gene sequence from the bamboo rat genotype II in the GenBank database, the actin sequence of SCAU12558 was found to be most similar to the Cryptosporidium ferret genotype (GenBank: MF411076) with 12 SNPs.
Phylogeny of Cryptosporidium spp. based on the maximum-likelihood analyses of the small subunit rRNA (a), 60-kDa glycoprotein (b), 70-kDa heat-shock protein (c) and actin (d) genes. Bootstrap values greater than 50% from 1000 replicates are shown on the branches. The Cryptosporidium-positive specimen detected in this study is indicated by red triangle
Discussion
In this study, to the best of our knowledge, we report for the first time the occurrence and genetic identity of E. bieneusi, G. duodenalis and Cryptosporidium sp. in masked palm civets, with infection rates of 53.3%, 3.8% and 0.1%, respectively. Within the family Viverridae, several small carnivores genetically related to masked palm civets have been examined for these pathogens without much success. For example, a recent study in Spain reported the occurrence of an unknown Cryptosporidium genotype in one of six genets (Genetta genetta) examined for Cryptosporidium spp. and G. duodenalis [29]. Another study in the Philippines examined an Asian palm civet (Paradoxurus hermaphroditus) for G. duodenalis but did not detect this pathogen [30].
Results of the present study have revealed some differences in infection rates of E. bieneusi and G. duodenalis among farms and age groups. Significantly higher infection rates of E. bieneusi were observed on farms in Hainan (86.1%) and Chongqing (85.9%) than in Guangdong (46.2%) and Jiangxi (35.2%). This was likely due to the poor sanitary conditions of the two farms in Hainan and Chongqing, as the housing and management of animals were similar among the four farms. By age, a higher prevalence of E. bieneusi was obtained from young civets of < 1 year. This is in agreement with previous findings of age-associated occurrence of E. bieneusi infections in other farmed wild animals such as macaques, boars and squirrels [31,32,33]. In contrast, a significantly higher infection rate of G. duodenalis was observed on the farm in Guangdong, and masked palm civets of 1–2 years had the highest infection rate of G. duodenalis, suggesting that E. bieneusi and G. duodenalis are transmitted differently in these animals.
Results of the sequence analysis have demonstrated a high genetic diversity of E. bieneusi in masked palm civets. Altogether, 13 E. bieneusi ITS genotypes were identified in this study, with two novel genotypes PL1 and PL2 accounting for about 90% E. bieneusi infections in civets. These two ITS genotypes together with several other novel ones formed a clade related to Group 2, thus might be civet-adapted E. bieneusi genotypes. In comparison, the remaining novel genotypes PL4, PL5, PL9, PL10 and PL11 were phylogenetically clustered into the zoonotic Group 1. In addition, one civet was found positive for Peru8, which is a common zoonotic genotype in humans [2]. Thus, masked palm civets could carry human-pathogenic E. bieneusi.
High genetic heterogeneity of E. bieneusi was observed in masked palm civets on a large farm in Jiangxi. This farm is the largest among the four study farms and has been established for over 20 years. It has 10 of the 13 E. bieneusi genotypes identified in the study, including all six ITS genotypes (Peru8, PL4, 5, 9, 10 and 11) of the zoonotic Group 1. One explanation for the high heterogeneity is that genetic exchange might have occurred among ancestral types on this farm during the past 20 years [34]. The long history of the farm and high genotype diversity have probably facilitated the genetic exchange among E. bieneusi isolates and emergence of novel genotypes in farmed masked palm civets.
The zoonotic assemblage B appears to be the dominant G. duodenalis genotype in masked palm civets. In this study, all G. duodenalis-positive civets harbored assemblage B, with only one having concurrent infection of assemblages B and D. Assemblage B is the most common human-pathogenic genotype in both industrialized and developing countries [4, 35]. In animals, assemblage B has been recently reported as the dominant G. duodenalis genotype in farmed macaques, horses, donkeys, rabbits, chinchillas and bamboo rats, mostly in China [24, 36,37,38,39,40,41,42,43]. Within assemblage B found in this study, a high genetic heterogeneity was observed at the three genetic loci examined. Of the known subtypes, MB7 and MB8 at the tpi locus and Bb-3 at the bg locus were reported in non-human primates [27, 44], while B14 at the tpi locus was detected in urban wastewater previously [45]. In addition, phylogenetic analysis of the concatenated sequences from the three loci showed that the MLGs of assemblage B in civets formed a separate cluster, which confirms the previous suggestion on the likely occurrence of host adaptation within assemblage B [24, 27]. Nevertheless, the assemblage B MLGs from civets were genetically related to several MLGs from humans, indicating that they could have zoonotic potential.
The Cryptosporidium isolate from masked palm civets was found to be genetically related to the Cryptosporidium bamboo rat genotype II from bamboo rats in south-central China based on sequence analysis of the SSU rRNA, gp60 and hsp70 genes [46]. As only one fecal specimen was positive for Cryptosporidium, whether it is a native parasite of masked palm civets remains unclear. Further studies are needed to understand its host range.
Conclusions
This report of the occurrence and identity of E. bieneusi, G. duodenalis and Cryptosporidium bamboo rat genotype II in farmed masked palm civets in southern China extends our knowledge on the genetic diversity and transmission of these pathogens in farmed wild animals. The presence of zoonotic E. bieneusi genotypes and G. duodenalis assemblage B in masked palm civets suggests that these animals might be potential reservoirs for human infections with these pathogens. Further studies involving extensive sampling of animals as well as farmworkers are needed to elucidate the role of newly domesticated wild animals in the epidemiology of human microsporidiosis, giardiasis and cryptosporidiosis.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. Representative nucleotide sequences generated in this study were deposited in the GenBank database under the accession numbers MT487560-MT487591, MT497888-MT497900 and MT779807.
Abbreviations
- PCR:
-
polymerase chain reaction
- ITS:
-
internal transcribed spacer
- tpi :
-
triosephosphate isomerase
- bg :
-
β-giardin
- gdh :
-
glutamate dehydrogenase
- SSU rRNA:
-
the small subunit rRNA
- gp60 :
-
60-kDa glycoprotein gene
- hsp70 :
-
70-kDa heat shock protein gene
- ML:
-
maximum likelihood
- SNPs:
-
single nucleotide polymorphisms
- MLGs:
-
multilocus genotypes
References
Ryan U, Hijjawi N, Feng Y, Xiao L. Giardia: an under-reported foodborne parasite. Int J Parasitol. 2019;49:1–11.
Li W, Feng Y, Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019;35:436–51.
Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011.
Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–40.
Caccio SM, Lalle M, Svard SG. Host specificity in the Giardia duodenalis species complex. Infect Genet Evol. 2018;66:335–45.
Gatei W, Hart CA, Gilman RH, Das P, Cama V, Xiao L. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J Eukaryot Microbiol. 2006;53:S43–8.
Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–31.
Feng Y, Li N, Dearen T, Lobo ML, Matos O, Cama V, et al. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl Environ Microbiol. 2011;77:4822–8.
Inoue T, Kaneko Y, Yamazaki K, Anezaki T, Yachimori S, Ochiai K, et al. Genetic population structure of the masked palm civet Paguma larvata, (Carnivora: Viverridae) in Japan, revealed from analysis of newly identified compound microsatellites. Conserv Genet. 2012;13:1095–107.
Guan Y, Zheng B, He Y, Liu X, Zhuang Z, Cheung C, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–8.
Hou G, Zhao J, Zhou H, Rong G. Seroprevalence and genetic characterization of Toxoplasma gondii in masked palm civet (Paguma larvata) in Hainan province, tropical China. Acta Trop. 2016;162:103–6.
Lee K, Iwata T, Nakadai A, Kato T, Hayama S, Taniguchi T, et al. Prevalence of Salmonella, Yersinia and Campylobacter spp. in feral raccoons (Procyon lotor) and masked palm civets (Paguma larvata) in Japan. Zoonoses Public Hlth. 2011;58:424–31.
Sato S, Kabeya H, Shigematsu Y, Sentsui H, Une Y, Minami M, et al. Small Indian mongooses and masked palm civets serve as new reservoirs of Bartonella henselae and potential sources of infection for humans. Clin Microbiol Infect. 2013;19:1181–7.
Wicker LV, Canfield PJ, Higgins DP. Potential pathogens reported in species of the family Viverridae and their implications for human and animal health. Zoonoses Public Health. 2017;64:75–93.
Jiang J, Alderisio KA, Singh A, Xiao L. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microbiol. 2005;71:1135–41.
Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003;69:4495–501.
Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80.
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–83.
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–7.
Sulaiman IM, Lal AA, Xiao L. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J Parasitol. 2002;88:388–94.
Sulaiman IM, Morgan UM, Thompson RC, Lal AA, Xiao L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl Environ Microbiol. 2000;66:2385–91.
Santin M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56:34–8.
Xiao L, Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32.
Chen L, Zhao J, Li N, Guo Y, Feng Y, Feng Y, et al. Genotypes and public health potential of Enterocytozoon bieneusi and Giardia duodenalis in crab-eating macaques. Parasit Vectors. 2019;12:254.
Qi M, Yu F, Li S, Wang H, Luo N, Huang J, et al. Multilocus genotyping of potentially zoonotic Giardia duodenalis in pet chinchillas (Chinchilla lanigera) in China. Vet Parasitol. 2015;208:113–7.
Minetti C, Lamden K, Durband C, Cheesbrough J, Fox A, Wastling JM. Determination of Giardia duodenalis assemblages and multi-locus genotypes in patients with sporadic giardiasis from England. Parasit Vectors. 2015;8:444.
Karim MR, Wang R, Yu F, Li T, Dong H, Li D, et al. Multi-locus analysis of Giardia duodenalis from nonhuman primates kept in zoos in China: geographical segregation and host-adaptation of assemblage B isolates. Infect Genet Evol. 2015;30:82–8.
Xu H, Jin Y, Wu W, Li P, Wang L, Li N, et al. Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai. China. Parasit Vectors. 2016;9:121.
Mateo M, de Mingo MH, de Lucio A, Morales L, Balseiro A, Espi A, et al. Occurrence and molecular genotyping of Giardia duodenalis and Cryptosporidium spp. in wild mesocarnivores in Spain. Vet Parasitol. 2017;235:86–93.
Velante NAP, Oronan RB, Reyes MF, Divina BP. Giardia duodenalis in captive tigers (Panthera tigris), palawan bearcats (Arctictis binturong whitei) and Asian palm civet (Paradoxurus hermaphroditus) at a wildlife facility in Manila, Philippines. Iran J Parasitol. 2017;12:348–54.
Feng S, Jia T, Huang J, Fan Y, Chang H, Han S, et al. Identification of Enterocytozoon bieneusi and Cryptosporidium spp. in farmed wild boars (Sus scrofa) in Beijing, China. Infect Genet Evol. 2020;80:104231.
Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2014;63:132–7.
Deng L, Li W, Yu X, Gong C, Liu X, Zhong Z, et al. First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS ONE. 2016;11:e0163605.
Li W, Xiao L. Multilocus sequence typing and population genetic analysis of Enterocytozoon bieneusi: host specificity and its impacts on public health. Front Genet. 2019;10:307.
Ryan U, Caccio SM. Zoonotic potential of Giardia. Int J Parasitol. 2013;43:943–56.
Ma X, Wang Y, Zhang H, Wu H, Zhao G. First report of Giardia duodenalis infection in bamboo rats. Parasit Vectors. 2018;11:520.
Li F, Wang R, Guo Y, Li N, Feng Y, Xiao L. Zoonotic potential of Enterocytozoon bieneusi and Giardia duodenalis in horses and donkeys in northern China. Parasitol Res. 2020;119:1101–8.
Zhang X, Qi M, Jing B, Yu F, Wu Y, Chang Y, et al. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in rabbits in Xinjiang, China. J Eukaryot Microbiol. 2018;65:854–9.
Jiang J, Ma JG, Zhang NZ, Xu P, Hou G, Zhao Q, et al. Prevalence and risk factors of Giardia duodenalis in domestic rabbbits (Oryctolagus cuniculus) in Jilin and Liaoning province, northeastern China. J Infect Public Health. 2018;11:723–6.
Gherman CM, Kalmar Z, Gyorke A, Mircean V. Occurrence of Giardia duodenalis assemblages in farmed long-tailed chinchillas Chinchilla lanigera (Rodentia) from Romania. Parasit Vectors. 2018;11:86.
Zhang X, Zhang F, Li F, Hou J, Zheng W, Du S, et al. The presence of Giardia intestinalis in donkeys, Equus asinus, in China. Parasit Vectors. 2017;10:3.
Deng L, Li W, Zhong Z, Liu X, Chai Y, Luo X, et al. Prevalence and molecular characterization of Giardia intestinalis in racehorses from the Sichuan province of southwestern China. PLoS ONE. 2017;12:e0189728.
Debenham JJ, Tysnes K, Khunger S, Robertson LJ. Occurrence of Giardia, Cryptosporidium, and Entamoeba in wild rhesus macaques (Macaca mulatta) living in urban and semi-rural north-west India. Int J Parasitol Parasites Wildl. 2017;6:29–34.
Karim MR, Zhang S, Jian F, Li J, Zhou C, Zhang L, et al. Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int J Parasitol. 2014;44:1039–47.
Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. 2012;6:e1809.
Wei Z, Liu Q, Zhao W, Jiang X, Zhang Y, Zhao A, et al. Prevalence and diversity of Cryptosporidium spp. in bamboo rats (Rhizomys sinensis) in south central China. Int J Parasitol Parasites Wildl. 2019;9:312–6.
Acknowledgements
We would like to thank the farm owners or managers for the assistance in sample collection from masked palm civets.
Funding
This study was supported in part by the National Natural Science Foundation of China (31820103014, U1901208), 111 Project (D20008), Innovation Team Project of Guangdong University (2019KCXTD001), and Projects of COVID-19 Control and Prevention of the Department of Education of Guangdong Province (2020KZDZX1033).
Author information
Authors and Affiliations
Contributions
NL, YF and LX conceived and designed the experiments. ZY, XW and RY performed the experiments. ZY, XH and YG analyzed the data. ZY, LX and NL wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This research was approved by the Ethics Committee of the South China Agricultural University. Masked palm civets sampled were handled following the Animal Ethics Procedures and Guidelines of the People’s Republic of China. Specimen collection was conducted with the permission of the owners or farm managers.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yu, Z., Wen, X., Huang, X. et al. Molecular characterization and zoonotic potential of Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium sp. in farmed masked palm civets (Paguma larvata) in southern China. Parasites Vectors 13, 403 (2020). https://doi.org/10.1186/s13071-020-04274-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04274-0