Abstract
Background
Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi are important zoonotic protists in humans and animals around the world, including nonhuman primates (NHPs). However, the prevalence, genetic identity and zoonotic potential of these pathogens in wild NHPs remain largely unclear.
Methods
A total of 348 fecal samples were collected from wild NHPs at four locations in Yunnan, southwestern China, and analyzed for these pathogens using nested PCR targeting various genetic loci and DNA sequence analysis of the PCR products. The zoonotic potential of the pathogens was assessed by comparing the genetic identity of the pathogens in these animals with that previously reported in humans.
Results
Altogether, two (0.6%), 25 (7.2%) and 30 (8.6%) samples were positive for Cryptosporidium sp., G. duodenalis and E. bieneusi, respectively. The Cryptosporidium sp. identified belonged to C. parvum subtype IIdA20G1. Both assemblages A (n = 3) and B (n = 22) were identified among G. duodenalis-positive animals. Five genotypes in zoonotic Group 1 were identified within E. bieneusi, including Type IV (n = 13), D (n = 7), Peru8 (n = 6), MMR86 (n = 2) and HNFS01 (n = 2). All genotypes and subtypes identified are known human pathogens or phylogenetically related to them.
Conclusions
Data from this study suggest a common occurrence of zoonotic genotypes of G. duodenalis and E. bieneusi in wild NHPs in southwestern China.
Graphical Abstract

Similar content being viewed by others
Background
Cryptosporidiosis, giardiasis and microsporidiosis are three emerging zoonotic diseases caused by Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi, respectively, that have been reported in humans as well as domestic and wild animals. Humans can be infected with these pathogens through the ingestion of contaminated food or water, causing diarrhea in immunocompetent individuals and significant mortality in immunocompromised ones [1,2,3]. To date, over 40 Cryptosporidium species and 100 genotypes have been recognized, including Cryptosporidium parvum, which has a broad host range [4]. Similarly, at least eight distinct assemblages (i.e., A–H) are known in G. duodenalis [5]. Among these, assemblages A and B have a broad host range and as such can be transmitted between humans and animals, whereas assemblages C–H are host specific [6]. For E. bieneusi, the nearly 500 genotypes identified thus far have been classified into 11 major groups, with Group 1 genotypes having a broad host range [7].
Nonhuman primates (NHPs), due to their high genetic homology to humans, are considered potential sources of zoonotic parasites in humans [8]. Data from several studies indicate that captive and farmed NHPs are reservoirs of Cryptosporidium spp., G. duodenalis and E. bieneusi, with the most prevalent genotypes of the pathogens also detected in humans [9]. Compared with captive NHPs in zoos or breeding facilities, free-range NHPs are in more contact with humans and can thus potentially transmit pathogens to humans [9].
Yunnan Province in China is home to many wild NHPs [10]. The high biodiversity and lifestyle of native ethnic groups facilitate the contact of people with wild animals [11]. For example, in Yunnan Golden Monkey National Park in northern Yunnan, snub-nosed monkeys (Rhinopithecus bieti) are in frequent contact with local Tibetan residents [12]. In the Jizu Mountains and Shibao Mountains of western Yunnan, rhesus macaques (Macaca mulatta) are in close contact with monks living in temples and with pilgrims and tourists visiting the mountains [13]. In Xishuangbanna Primeval Forest Park, which is a famous tropical rainforest reserve in southern Yunnan, tourists are encouraged to feed and interact with NHPs [13].
In a small-scale survey of animals, captive NHPs from zoos and research laboratories in Yunnan Province were shown to be infected with several genotypes of E. bieneusi [14]. A few studies on the genetic identity of Cryptosporidium spp., G. duodenalis and E. bieneusi in NHPs have also been conducted in China with captive animals [8, 9, 15,16,17,18,19,20,21]. There is a need for molecular characterizations of these pathogens in wild NHPs. In the study presented here, four tourist attractions in Yunnan Province home to many wild NHPs were examined for the prevalence, genetic identity and public health potential of Cryptosporidium spp., G. duodenalis and E. bieneusi in these animals.
Methods
Sample collection
A total of 348 fecal samples from rhesus macaques (M. mulatta, n = 320), snub-nosed monkeys (R. bieti, n = 20), and Assamese macaques (Macaca assamensis, n = 8) at four tourist attractions in Yunnan Province, China were collected from June 2019 to January 2021 (Table 1; Fig. 1). As there are only a limited number of Assamese macaques in Xishuangbanna Primeval Forest Park, to better protect them, they are kept in an isolated area that is separate from those frequented by rhesus macaques. The sampling size was strictly controlled not to exceed one third of the estimated total number of NHPs at each sampling location. Each sample was from one animal and consisted of a fresh fecal dropping collected from the ground. All monkeys appeared to be clinically normal without obvious signs of diarrhea. The samples were stored at 4 °C in 2.5% potassium dichromate prior to DNA extraction and PCR analysis.
DNA extraction and PCR
Before genomic DNA extraction, approximately 300 mg of fecal material was washed with distilled water by centrifugation (2000 g, 10 min). DNA was extracted from the pellet using a FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) as previously described [22]. The extracted DNA was stored at – 20 °C until being analyzed by PCR.
PCR amplification
Cryptosporidium spp. were detected and genotyped by PCR and sequence analyses of the small subunit (SSU) rRNA gene [23]. The C. parvum detected was further subtyped by sequence analysis of the 60-kDa glycoprotein (gp60) gene [24]. In contrast, G. duodenalis was detected and genotyped by PCR and sequence analyses of the β-giardin (bg), triosephosphate isomerase (tpi) and glutamate dehydrogenase (gdh) genes [25,26,27]. Enterocytozoon bieneusi in the extracted DNA was detected and genotyped by PCR and sequence analyses of a rRNA fragment containing the entire internal transcribed spacer (ITS) [28]. Each sample was subjected to PCR analysis with two technical replicates at each genetic locus. DNA preparations of Cryptosporidium tyzzeri from mice, assemblage G from mice and genotype PtEb IX from dogs were used as positive controls in the PCR analysis for Cryptosporidium spp., G. duodenalis and E. bieneusi, respectively, whereas reagent-grade water was used as the negative control.
Sequence analysis
All positive secondary PCR products were sequenced by Sangon Biotech (Shanghai, China) bidirectionally on an ABI 3730 Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA). The raw sequences obtained were assembled using ChromasPro 1.33 (http://technelysium.com.au/ChromasPro.html), edited using BioEdit 7.1 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and aligned with reference sequences from GenBank using ClustalX 2.1 (http://clustal.org). Genotypes and subtypes of the pathogens were named according to the established nomenclature [29, 30]. A maximum likelihood (ML) tree was constructed to evaluate the phylogenetic relationships among genotypes of E. bieneusi using MEGA 7.0.14 (http://www.megasoftware.net/) with substitution rates calculated with the general time-reversible model. The reliability of the tree was assessed using bootstrap analysis with 1000 replicates.
Statistical analysis
The Chi-square test was used to compare differences in infection rates of pathogens between geographical locations. Differences were considered to be significant at P < 0.05.
Results
Occurrence and genetic identity of Cryptosporidium sp.
Of the 348 fecal samples analyzed, only two (0.6%) samples from rhesus macaques in Xishuangbanna Primeval Forest Park were positive for Cryptosporidium (Table 2). Sequence analysis of the SSU rRNA PCR products identified both as C. parvum. One sample, SCAU39270, was successfully subtyped at the gp60 locus as IIdA20G1, while the expected gp60 PCR product in the other C. parvum-positive sample (SCAU39236) could not be generated.
Occurrence and assemblages of G. duodenalis
In nested PCR analyses of the three genetic loci, 25 (7.2%) of the 348 fecal samples were positive for G. duodenalis, with infection rates ranging from 2.8% (4/141) to 15.5% (11/71) among the four sampling locations (Table 2). Among these, the infection rate was significantly higher among samples collected at Jizu Mountains (15.5%) than among those collected at Xishuangbanna Primeval Forest Park (2.8%; χ2 = 11.504, P = 0.0006). DNA sequence analysis of the bg, tpi and gdh PCR products revealed the presence of assemblages A (n = 3) and B (n = 22) in those monkeys. By animal species, only M. mulatta and R. bieti samples were positive for G. duodenalis (Table 2), with R. bieti only positive for assemblage B and M. mulatta positive for both assemblages A and B. The assemblage A was identified as A1 and A5 subtypes at the bg and tpi loci and as A5 subtype at the gdh locus; thus, all belonged to the AI subassemblage.
Occurrence and genotypes of E. bieneusi
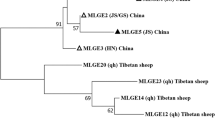
Of the 348 fecal samples collected from wild NHPs, 30 (8.6%) were positive for E. bieneusi in the PCR analysis of the ITS, with infection rates ranging from 1.7% (2/116) to 17.0% (24/141) among the four locations (Table 2). The infection rate at Xishuangbanna Primeval Forest Park (17.0%) was significantly higher than at Jizu Mountains (5.6%; χ2 = 5.341, P = 0.0208) and Shibao Mountains (1.7%; χ2 = 16.377, P = 0.00005). Five E. bieneusi genotypes were detected in these animals, including Type IV (n = 13), D (n = 7), Peru8 (n = 6), MMR86 (n = 2) and HNFS01 (n = 2). The sequences of Type IV, D, Peru8, MMR86 and HNFS01 were identical to GenBank sequences JX683801(from humans), MK982516 (from giraffes), KF305584 (from rhesus macaques), MN399818 (from humans) and MK947105 (from flying squirrels), respectively. In the phylogenetic analysis, all were placed in zoonotic Group 1 (Fig. 2).
Phylogenetic relationship of Enterocytozoon bieneusi genotypes from wild nonhuman primates in Yunnan Province, China based on a maximum-likelihood analysis of sequences of the ribosomal internal transcribed spacer. Bootstrap values > 50% from 1000 replicate analysis are shown on nodes. Representative sequences obtained from this study are indicated with red triangles. The major hosts of the genotypes are indicated with black animal symbols
By animal species, 8.4% (27/320) M. mulatta and 37.5% (3/8) M. assamensis sampled were positive for E. bieneusi. Among these, all five subtypes (Type IV, Peru8, D, MMR86 and HNFS01) were detected in M. mulatta and two subtypes (D and Peru8) were found in M. assamensis (Table 2). None of the 20 R. bieti samples examined were positive for E. bieneusi.
Coinfection of enteric pathogens
Among five samples from M. mulatta at Xishuangbanna Primeval Forest Park, one C. parvum-positive sample had concurrence of E. bieneusi genotype D, two samples (one with Peru8 and another with Type IV) had concurrence of assemblage B of G. duodenalis and two samples had concurrence of two E. bieneusi genotypes based on inconsistent sequencing results of the two PCR replicates (one with both Type IV and Peru8 and another with both Peru8 and D).
Discussion
This is the first report of the occurrence of Cryptosporidium sp., G. duodenalis and E. bieneusi in wild NHPs in Yunnan Province, China. These pathogens were identified in 0.6, 7.2 and 8.6% of fecal samples, respectively. The occurrence rates of these pathogens are lower than those found in captive, laboratory and zoo NHPs in previous investigations, which reported infection rates ranging from 9.1% to 46.7% for Cryptosporidium spp. [8, 15, 31], 8.5% to 32.3% for G. duodenalis [8, 19, 32,33,34] and 11.4% to 46.2% for E. bieneusi [8, 14, 18, 19, 21, 35,36,37,38]. This difference was expected as the congregation of susceptible animals in an enclosed environment could facilitate the transmission of enteric pathogens on farms and in captivity.
The zoonotic IIdA20G1 subtype of C. parvum is the only Cryptosporidium identified in the present study. Thus far, three Cryptosporidium species, C. hominis, C. parvum and C. muris, have been identified as common ones in NHPs in China [15, 39]. Unlike IIa subtypes that are prevalent in many industrialized countries, IId subtypes are the most common C. parvum in China [39]. A previous study also found the IIdA15G2R1 and IIdA19G1 subtypes in NHPs in China [15, 36] and, more recently, the IIdA20G1 subtype was detected in cattle and deer in several areas in China, causing an outbreak of cryptosporidiosis in pre-weaned calves [40,41,42,43]. Therefore, the occurrence of this emerging subtype in NHPs indicates an expansion of host range of this emerging subtype.
Similar to other studies in NHPs, the zoonotic assemblage B was identified as the dominant genotype of G. duodenalis in M. mulatta and R. bieti in this study [8, 19, 20, 44, 45]. Assemblage B is the most common pathogenic genotype in humans in both industrialized and developing countries [5, 44]. The assemblage A identified in NHPs in the present study appears to belong to AI subassemblage, which is common in animals but nevertheless has been found in humans [30]. Thus, wild NHPs could serve as potential reservoirs for zoonotic G. duodenalis.
The five E. bieneusi genotypes detected in NHPs in Yunnan Province all belong to Group 1, which contains most of the zoonotic genotypes of major public health concern [7]. Among these, Type IV, D and Peru8 are known human and NHP pathogens in many countries [19, 46,47,48]. To our knowledge, our study is the first to report genotypes MMR86 and HNFS01 in NHPs in China. MMR86 was initially seen in humans in Myanmar, which borders Yunnan province, sampled in the present study [49], while HNFS01 was recently identified in flying squirrels in Henan, China (GenBank accession number MK947105). Previous studies on E. bieneusi in captive and farmed NHPs also detected mostly Group 1 genotypes [14, 18, 21, 35, 38, 50].
Conclusions
In summary, the results of the present study show a common occurrence of zoonotic G. duodenalis and E. bieneusi genotypes in wild NHPs, which in turn represents a significant public health concern as tourists are coming into increasing contact with NHPs. In response, public education programs on the infective potential of pathogens from NHPs and the utility of hygiene in disease prevention should be implemented to inform park personnel and tourists as part of the One Health approach to the prevention and control of Cryptosporidium spp., G. duodenalis and E. bieneusi infections.
Availability of data and materials
Representative nucleotide sequences generated in the study were deposited in GenBank under the accession numbers OM212450, OM212451, OM212347, OM212012-OM212021, OM221354-OM221360.
Abbreviations
- bg :
-
β-Giardin gene
- gdh :
-
Glutamate dehydrogenase gene
- gp60 :
-
60 KDa glycoprotein gene
- ITS:
-
Internal transcribed spacer
- NHPs:
-
Nonhuman primates
- SSU rRNA:
-
Small subunit rRNA gene
- tpi :
-
Triosephosphate isomerase gene
References
Ryan U, Hijjawi N, Feng Y, Xiao L. Giardia: an under-reported foodborne parasite. Int J Parasitol. 2019;49:1–11.
Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen X, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis. 2015;15:85–94.
Li W, Feng Y, Xiao L. Enterocytozoon bieneusi. Trends Parasitol. 2022;38:95–6.
Feng Y, Ryan U, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011.
Cai W, Ryan U, Xiao L, Feng Y. Zoonotic giardiasis: an update. Parasitol Res. 2021;120:4199–218.
Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80.
Li W, Feng Y, Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019;35:436–51.
Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg Infect Dis. 2012;18:1640–3.
Karim MR, Zhang S, Jian F, Li J, Zhou C, Zhang L, et al. Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int J Parasitol. 2014;44:1039–47.
Garber PA. Why China is important in advancing the field of primatology. Zool Res. 2018;39:241–3.
Afonso E, Fu R, Dupaix A, Goydadin AC, Yu Z, Callou C, et al. Feeding sites promoting wildlife-related tourism might highly expose the endangered Yunnan snub-nosed monkey (Rhinopithecus bieti) to parasite transmission. Sci Rep. 2021;11:15817.
Huang Z, Scott MB, Li Y, Ren G, Xiang Z, Cui L, et al. Black-and-white snub-nosed monkey (Rhinopithecus bieti) feeding behavior in a degraded forest fragment: clues to a stressed population. Primates. 2017;58:517–24.
Liu Z, Qian X, Hong M, Zhang J, Li D, Wang T, et al. Global view on virus infection in non-human primates and implications for public health and wildlife conservation. Zool Res. 2021;42:626–32.
Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, et al. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl Environ Microb. 2014;80:1893–8.
Chen L, Hu S, Jiang W, Zhao J, Li N, Guo Y, et al. Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques. Parasit Vectors. 2019;12:350.
Guo Y, Li N, Feng Y, Xiao L. Zoonotic parasites in farmed exotic animals in China: implications to public health. Int J Parasitol Parasites Wildl. 2021;14:241–7.
Liu X, Xie N, Li W, Zhou Z, Zhong Z, Shen L, et al. Emergence of Cryptosporidium hominis monkey genotype II and novel subtype family Ik in the squirrel monkey (Saimiri sciureus) in China. PLoS ONE. 2015;10:e0141450.
Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2014;63:132–7.
Chen L, Zhao J, Li N, Guo Y, Feng Y, Feng Y, et al. Genotypes and public health potential of Enterocytozoon bieneusi and Giardia duodenalis in crab-eating macaques. Parasit Vectors. 2019;12:254.
Karim MR, Wang R, Yu F, Li T, Dong H, Li D, et al. Multi-locus analysis of Giardia duodenalis from nonhuman primates kept in zoos in China: Geographical segregation and host-adaptation of assemblage B isolates. Infect Genet Evol. 2015;30:82–8.
Yu F, Wu Y, Li T, Cao J, Wang J, Hu S, et al. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet Res. 2017;13:158.
Jiang J, Alderisio KA, Singh A, Xiao L. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microbiol. 2005;71:1135–41.
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microb. 1999;65:1578–83.
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41:2744–7.
Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–31.
Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Caccio SM. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol. 2005;35:207–13.
Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–52.
Sulaiman IM, Fayer R, Lal AA, Trout JM, Schaefer FW, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003;69:4495–501.
Santin M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56:34–8.
Xiao L, Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32.
Feng Y. Cryptosporidium in wild placental mammals. Exp Parasitol. 2010;124:128–37.
Johnston AR, Gillespie TR, Rwego IB, McLachlan TL, Kent AD, Goldberg TL. Molecular epidemiology of cross-species Giardia duodenalis transmission in western Uganda. PLoS Negl Trop Dis. 2010;4:e683.
Beck R, Sprong H, Bata I, Lucinger S, Pozio E, Caccio SM. Prevalence and molecular typing of Giardia spp. in captive mammals at the zoo of Zagreb, Croatia. Vet Parasitol. 2011;175:40–6.
Debenham JJ, Tysnes K, Khunger S, Robertson LJ. Occurrence of Giardia, Cryptosporidium, and Entamoeba in wild rhesus macaques (Macaca mulatta) living in urban and semi-rural North-West India. Int J Parasitol Parasites Wildl. 2017;6:29–34.
Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS ONE. 2015;10:e0117991.
Du S, Zhao G, Shao J, Fang Y, Tian G, Zhang L, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-Human primates in Qinling Mountains. Korean J Parasitol. 2015;53:395–402.
Li W, Deng L, Yu X, Zhong Z, Wang Q, Liu X, et al. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasit Vectors. 2016;9:395.
Zhong Z, Li W, Deng L, Song Y, Wu K, Tian Y, et al. Multilocus genotyping of Enterocytozoon bieneusi derived from nonhuman primates in southwest China. PLoS ONE. 2017;12:e0176926.
Feng Y, Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front Microbiol. 2017;8:1701.
Tao W, Li Y, Yang H, Song M, Lu Y, Li W. Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from northeast China: public health concerns. J Parasitol. 2018;104:10–7.
Xie F, Zhang Z, Zhao A, Jing B, Qi M, Wang R. Molecular characterization of Cryptosporidium and Enterocytozoon bieneusi in Pere David’s deer (Elaphurus davidianus) from Shishou, China. Int J Parasitol Parasites Wildl. 2019;10:184–7.
Xue N, Liu F, Tao W, Zhao Q, Qiu H, Hu Y, et al. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi in Longjiang Wagyu cattle in Northeastern China. Microb Pathog. 2020;149:104526.
Zhang Z, Su D, Meng X, Liang R, Wang W, Li N, et al. Cryptosporidiosis outbreak caused by Cryptosporidium parvum subtype IIdA20G1 in neonatal calves. Transbound Emerg Dis. 2021;00:1–8.
Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–40.
Levecke B, Geldhof P, Claerebout E, Dorny P, Vercammen F, Caccio SM, et al. Molecular characterisation of Giardia duodenalis in captive non-human primates reveals mixed assemblage A and B infections and novel polymorphisms. Int J Parasitol. 2009;39:1595–601.
Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. 2012;2012:981424.
Santin M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. 2011;90:363–71.
Wang SS, Wang RJ, Fan XC, Liu TL, Zhang LX, Zhao GH. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. 2018;183:142–52.
Shen Y, Gong B, Liu X, Wu Y, Yang F, Xu J, et al. First identification and genotyping of Enterocytozoon bieneusi in humans in Myanmar. BMC Microbiol. 2020;20:10.
Yang H, Lin Y, Li Y, Song M, Lu Y, Li W. Molecular characterization of Enterocytozoon bieneusi isolates in laboratory macaques in north China: zoonotic concerns. Parasitol Res. 2017;116:2877–82.
Acknowledgements
We thank the Yunnan Golden Monkey National Park staff and Xishuangbanna Primeval Forest Park staff for their assistance in sample collection in Yunnan Province, China.
Funding
This work was supported by the National Key R&D Program of China (2018YFC1602504), National Natural Science Foundation of China (U1901208), Applied Basic Research Program of Yunnan Province (2018FD069), 111 Project (D20008), and Innovation Team Project of Guangdong University (2019KCXTD001).
Author information
Authors and Affiliations
Contributions
FS and NL conceived and designed the experiments; FS and SS performed the experiments; YW, LF, YG and YF analyzed the data; FS, LX and NL wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The collection of fecal samples in the study was approved by the local government. The experimental protocol was reviewed by the Research Ethics Committee of South China Agricultural University. The animals were handled following the Animal Ethics Procedure and Guidelines of the People’s Republic of China. The sampling did not involve the capture of these free-range animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shu, F., Song, S., Wei, Y. et al. High zoonotic potential of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in wild nonhuman primates from Yunnan Province, China. Parasites Vectors 15, 85 (2022). https://doi.org/10.1186/s13071-022-05217-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05217-7