Abstract
Background
Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi are the main causal pathogens of gastrointestinal disease. However, there are limited reports about the prevalence of these organisms in captive Eurasian wild boars worldwide. Therefore, we examined the occurrence and identified the species/assemblages/genotypes of these pathogens in captive Eurasian wild boars, and estimated the zoonotic potential.
Findings
Of 357 fecal samples collected from captive Eurasian wild boars in China, 155 (43.4%) were infected with Cryptosporidium, G. duodenalis and/or E. bieneusi. The infection rates significantly differed in different areas, but did not differ between wild boars kept indoors and outdoors. Three (0.8%), 11 (3.1%) and 147 (41.2%) fecal samples were positive for Cryptosporidium, G. duodenalis and E. bieneusi, respectively. Sequence analysis of SSU rRNA gene revealed that all of the Cryptosporidium strains belonged to C. scrofarum. Based on the sequence analysis of the β-giardia gene of G. duodenalis, assemblages E and A were characterized. Fourteen E. bieneusi genotypes comprising five novel (WildBoar 7–11) and eight known (EbpC, F, CHG19, CHC5, PigEBITS5, D, RWSH4, SC02) genotypes were identified by ITS sequencing. EbpC was the most frequent genotype, detected in 85 specimens. Phylogenetic analysis revealed that all 14 genotypes belonged to Group 1.
Conclusions
This first report on the occurrence of Cryptosporidium, G. duodenalis and E. bieneusi in captive wild boars in China indicates that the presence of zoonotic species/assemblages/genotypes poses a threat to public health. The findings suggest that wild boars could be a significant source of human infection and water pollution.
Similar content being viewed by others
Background
Cryptosporidiosis, giardiasis and microsporidiosis are emerging infectious diseases that are mainly caused by the pathogens Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi, respectively [1]. Humans and animals infected with these protozoan parasites show acute or chronic diarrhea or other symptoms [2]. Currently, over 30 Cryptosporidium species are considered valid, and most of the Cryptosporidium species, genotypes, or subtypes are host-adapted [3, 4]. Cryptosporidium suis and C. scrofarum, previously known as pig genotype I and pig genotype II, are the dominant species infecting pigs [5], although C. muris, C. tyzzeri, C. parvum, C. felis, C. hominis, C. andersoni and C. meleagridis have also been observed in swine [6]. Given that all of these species were frequently or occasionally found in human infections and have zoonotic potential, they represent a significant public health risk [3, 4]. Giardia duodenalis, also known as G. intestinalis, or G. lamblia is comprised of at least 8 assemblages (assemblages A-H). Assemblages A and B have broad host specificities, having been found in humans and various mammals, while assemblages C-H have strong host specificities and narrow host ranges and assemblages A-F have been identified in pigs [7–9]. Enterocytozoon bieneusi is the most common microsporidian species infecting humans. To date, over 200 genotypes have been identified and have been divided into eight groups (Group 1–8) based on phylogenetic analysis; most of the genotypes belonging to Group 1 have zoonotic potential [10]. At least 60 E. bieneusi genotypes have been characterized in swine to date [11]. Cryptosporidium spp., G. duodenalis and E. bieneusi are considered to be primarily food-borne and water-borne parasites, posing an invisible threat to public health [12].
Eurasian wild boars (Sus scrofa) and domestic pigs (Sus scrofa domesticus) belong to the same species (S. scrofa), suggesting that they could share the same pathogens, with high potential for transmission between them [13]. Although the presence of Cryptosporidium spp. and E. bieneusi has been reported in domestic pigs, no survey on the occurrence of G. duodenalis in swine of China has been conducted. Domestic pigs in Chongqing, Shaanxi, Shanghai, Heilongjiang, Henan, Jiangsu and Taiwan were found to be infected with Cryptosporidium spp., with the infection rates ranging from 3.3 to 55.8%, and C. suis, C. scrofarum, C. andersoni and C. tyzzeri were the species identified [14–20]. Several reports have revealed the existence of E. bieneusi in domestic pigs in China, and over 20 genotypes have been identified, including CS-1, CS-3, CS-4, CS-6, CS-9, CS-10, EbpA, EbpB, EbpC, EbpD, Henan-I, Henan-IV, G, D, H, O, LW1, CHN1, CHN7, CHN8, CHN9, CHN10, EBITS3, PigEBITS5 and HLJ-I to HLJ-IV, with most of the identified genotypes confirmed to be zoonotic [21–25].
Wild boars have a worldwide distribution. They not only provide meat for human beings but are also widely used in scientific research. Unfortunately, wild boars are readily exposed and susceptible to parasites such as helminths and/or protozoa. To date, there is no published report on the occurrence of Cryptosporidium spp., G. duodenalis and E. bieneusi in wild boars in China. Therefore, the aim of this study was to identify the species/assemblages/genotypes using molecular characterization. Moreover, the role of wild boars as a potential reservoir of protozoa for other animals and human beings was estimated.
Methods
Sample collection
During 2014 to 2015, a total of 357 fecal samples were collected from captive wild boars in four sites of Sichuan province, including 239, 60, 50 and 8 specimens collected from Aba, Mingshan, Qionglai and Hanyuan, respectively. Among them, 308 and 49 were from wild boars kept indoors and outdoors, respectively. During specimen collection, we only gathered the top layer of the feces to ensure no contamination of the samples. The specimens were stored in centrifuge tubes containing 2.5% potassium dichromate and then placed in containers filled with ice packs and transported to the laboratory immediately.
DNA extraction and nested PCR amplification
Before extracting DNA, the fecal samples were washed with distilled water until the potassium dichromate was removed. Subsequently, genomic DNA was extracted from approximately 200 mg of semi-purified product using the E.Z.N.A Tool DNA Kit (D4015–02; Omega Bio-Tek Inc., Norcross, GA, USA) following the manufacturer’s instructions. DNA samples were stored in 200 μl of the kit’s Solution Buffer at -20 °C until use.
Cryptosporidium spp., G. duodenalis and E. bieneusi were identified by nested PCR amplification of the small subunit (SSU) rRNA gene, β-giardin (bg), and internal transcribed spacer (ITS) genes, respectively. The primers and annealing temperatures were previously reported [26]. For Cryptosporidium spp. and E. bieneusi, the annealing temperature was 55 °C for both primary and secondary PCR amplification. For G. duodenalis detection, the annealing temperatures of 65 °C and 55 °C were used in the primary and secondary PCR amplification, respectively. The secondary PCR products were visualized by staining with Golden View following 1% agarose gel electrophoresis.
Data analysis
The amplicons of the expected size were sent to Invitrogen (Shanghai, China) for sequencing. To ensure sequence accuracy, two-directional sequencing methods were used. To determine Cryptosporidium species, G. duodenalis assemblages and E. bieneusi genotypes, the sequences obtained in this study were aligned with sequences downloaded from the GenBank database via BLAST analysis (http://blast.ncbi.nlm.nih.gov) and using ClustalX software. For E. bieneusi, phylogenetic analysis of ITS sequences was performed using Mega software [27], and neighbor-joining phylogenetic analysis of the aligned E. bieneusi sequences was utilized to assess genetic clustering of genotypes. A total of 1,000 replicates were used for bootstrap analysis.
The infection rates between animals in different areas and with different farming modes (indoor or outdoor) were compared using the Chi-square test. A P-value < 0.05 was considered to indicate a significant difference. All tests were conducted using SPSS version 17.0 software (SPSS Inc., Chicago, IL, USA).
Nucleotide sequence GenBank accession numbers
All of the nucleotide sequences of SSU rRNA and the β-giardin gene from wild boars obtained in this study were deposited in the GenBank database under the accession numbers KU668893, KU668897 and KU668898 for Cryptosporidium, and KU668880, KU668883–KU668892 for G. duodenalis. The representative E. bieneusi sequences were deposited in the GenBank database under the accession numbers KX670577–KX670590.
Results and discussion
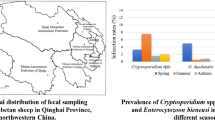
In our study, mixed infection was detected in six specimens, and all cases involved the combination of G. duodenalis and E. bieneusi. Considering all three pathogens evaluated in this study, a total of 155 individuals were infected, for an overall infection rate of 43.4%. Studies on these three pathogens in China have thus far focused on wildlife in captivity, companion animals, domestic animals and wastewater [26, 28, 29]. Here, we provide the first report of the prevalence and genetic characteristics of Cryptosporidium, G. duodenalis and E. bieneusi in captive wild boars in China, with infection rates of 0.8%, 3.1% and 41.2%, respectively, suggesting that the presence of E. bieneusi is generally more common. Considering the pathogens together, the infection rates were 35.6% (85/239), 60% (36/60), 66% (33/50) and 12.5% (1/8) in Aba, Mingshan, Qionglai and Hanyuan, respectively. A significant difference was observed between different areas (χ 2 = 26.207, df = 3, P = 0.001), which confirms the results of a study conducted on E. bieneusi infection in farmed foxes in northern China [30]. However, in the previous study, the prevalence was found to be highly associated with the farming mode, whereas in the present study, the infection rates were not significantly different (χ 2 = 0.156, df = 1, P > 0.05) between the wild boars kept indoors (135/308; 43.8%) and those kept outdoors (20/49; 40.8%) [30].
For Cryptosporidium spp., three Cryptosporidium-positive fecal samples were detected in Qionglai (n = 2) and Hanyuan (n = 1), and identified as C. scrofarum (Tables 1, 2). To date, infection of Cryptosporidium in wild boars has mainly been reported in European countries, with a prevalence of 7.6–16.7% in Spain, 16.5–16.9% in the Czech Republic, 18.2% in Austria, 0–8.5% in Poland and 5.4% in the Slovak Republic [6, 31–34]. Furthermore, C. scrofarum, C. suis, C. parvum, and mixed infection of C. scrofarum with C. suis have been identified in Europe, and C. scrofarum was the predominant Cryptosporidium species, followed by C. suis (Table 3). No other Cryptosporidium species that can infect domestic pigs has been detected in wild boars thus far. In our study, only three wild boars (0.8%) were found to be infected with Cryptosporidium, suggesting a much lower infection rate compared to those reported elsewhere, and only C. scrofarum was identified. Collectively, these findings suggest that wild boars might mainly harbor C. scrofarum and C. suis, which are both porcine-specific species, having been reported in domestic pigs worldwide [33]. Cryptosporidium scrofarum is recognized as a zoonotic species, and has been detected in humans, domestic animals, and even source water [20, 35, 36]. Thus, wild boars can serve as an environmental reservoir of Cryptosporidium transmitted to animals, humans and water.
We found a prevalence of 3.1% for G. duodenalis (Table 1), which is higher than that detected in wild boars in Poland (0%), Croatia (1.4%), and Spain (1.3%) [31, 34, 37]. In contrast, the prevalence in domestic pigs has been reported to vary from 1 to 66.4% (Table 3) [38–42]. Reports on G. duodenalis infection in wild boars are limited, and only one wild boar isolate was successfully identified using a molecular method, which turned out to be a part of assemblage A. In our sample, 11 wild boars were found to be infected with G. duodenalis. Specifically, two were infected with assemblage A and nine were infected with assemblage E (Table 2). Based on epidemiological data, assemblages A-F have been found in domestic pigs [7]. Among them, assemblages A and E were both found in domestic pigs and wild boars, and the latter was the most prevalent genotype, suggesting the possibility of inter-species transmission; however, the species of origin is not clear at present. A recent study conducted in Rio de Janeiro, Brazil detected that 15 people were infected with assemblage E, which further demonstrated that humans can be infected with assemblage E through an anthropozoonotic cycle [43]. Thus, the dominant assemblage E, and assemblage A detected in our study could be transmitted to humans through an anthropozoonotic transmission cycle.
In the present study, a total of 147 specimens were found to be E. bieneusi-positive, forming 14 E. bieneusi genotypes comprising five novel genotypes (WildBoar 7–11) and eight known genotypes (EbpC, F, CHG19, CHC5, PigEBITS5, D, RWSH4 and SC02), as identified by ITS sequencing analysis (Tables 1, 2). The most frequent genotype was EbpC, followed by genotype F (Table 2). The first report of wild boar E. bieneusi infection was in Poland, but this was confirmed by chromotrope 2R and fluorescence in situ hybridization analyses, and the genotype was not characterized [44]. We could only found one published paper reporting the prevalence and genetic characterization of E. bieneusi in wild boars: 11 genotypes were found in Central Europe (D, EbpA, EbpC, G, Henan-I, and WildBoar1–6), and EbpA was the most frequently detected microsporidian, which is different from our results [13]. Furthermore, the infection rate of the previous study was only 7.17% (33/460), which is markedly lower than that detected in our study (41.2%) [13]. These differences between the two studies may be due to the different geographic areas and living conditions. The novel genotypes (WildBoar 1–11) identified in wild boars of China may indicate that E. bieneusi in wild boars has relatively higher genetic variability. The genotypes EbpC, D, and PigEBITS5 previously identified in domestic pigs were also detected in wild boars. However, ten genotypes (F, CHG19, CHC5, RWSH4, SC02, WildBoar 7–11) have only been detected in wild boars to date. Thus, further studies in swine are needed to determine whether or not these ten genotypes have the capacity to infect domestic pigs, and if the genotypes found in domestic pigs can infect wild boars. This is the first report of these ten genotypes in wild boars, which may indicate that the wild boar is a new host for E. bieneusi of these genotypes.
Based on the phylogenetic analysis of the ITS gene, we found that all of the 14 E. bieneusi genotypes identified in our study belong to Group 1 (Fig. 1), which suggests their zoonotic potential. Among them, the human-pathogenic genotype EbpC was the most prevalent, which has been frequently found in hospitalized children in Shanghai, HIV+ and HIV- people in Henan, wastewater in four cities, and pigs in multiple cities in China [1, 21, 23–25, 45, 46]. These studies suggest that pigs might be a source of human microsporidiosis and water pollution. Moreover, EbpC may represent the main cause of human microsporidiosis in China [21, 23–25]. Our results confirm this possibility by demonstrating the potential for the zoonotic transmission of E. bieneusi. Furthermore, the identification of five novel genotypes has broadened the recognized genotypes and suggests high genetic variability among E. bieneusi.
Phylogenetic relationships of ITS nucleotide sequences of the Enterocytozoon bieneusi genotypes identified in this study and other reported genotypes. The phylogeny was inferred by a neighbor-joining analysis. Bootstrap values were obtained using 1,000 pseudo-replicates and those greater than > 50% are shown on nodes. The genotypes identified in this study are marked by outlined triangles and the novel genotypes are marked by filled triangles
Conclusions
We provide the first report on the presence of Cryptosporidium, G. duodenalis and E. bieneusi in captive wild boars from China, and identified C. scrofarum, assemblages A/E, and 14 E. bieneusi genotypes. Most of the species/assemblages/genotypes identified have been detected in humans. Our results revealed that wild boars and domestic pigs can share the same pathogens, and that wild boars could be an important source of animal and human cryptosporidiosis, giardiasis and microsporidiosis, as well as water contamination. Thus, measures should be taken to control the possible transmission. Furthermore, more molecular epidemiological surveys of Cryptosporidium, G. duodenalis and E. bieneusi in wild boars, humans, animals and water samples are needed to better elucidate the transmission risk and mode.
Abbreviations
- Bg:
-
β-giardin
- ITS:
-
Internal transcribed spacer
- SSU rRNA:
-
The small subunit rRNA
References
Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in Wastewater. PLoS Negl Trop Dis. 2012;6(9):e1809.
Li W, Li Y, Song M, Lu Y, Yang J, Tao W, et al. Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet Parasitol. 2015;208(3-4):125–34.
Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals - a One Health approach to prophylaxis. Parasite Immunol. 2016;38(9):535–47.
Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124(1):80–9.
Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141(13):1667–85.
García-Presedo I, Pedraza-Díaz S, González-Warleta M, Mezo M, Gómez-Bautista M, Ortega-Mora LM, et al. Presence of Cryptosporidium scrofarum, C. suis and C. parvum subtypes IIaA16G2R1 and IIaA13G1R1 in Eurasian wild boars (Sus scrofa). Vet Parasitol. 2013;196(3–4):497–502.
Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24(1):110–40.
Minetti C, Taweenan W, Hogg R, Featherstone C, Randle N, Latham SM, et al. Occurrence and diversity of Giardia duodenalis sssemblages in livestock in the UK. Transbound Emerg Dis. 2014;61(6):60–7.
Farzan A, Parrington L, Coklin T, Cook A, Pintar K, Pollari F, et al. Detection and characterization of Giardia duodenalis and Cryptosporidium spp. on swine farms in Ontario, Canada. Foodborne Pathog Dis. 2011;8(11):1207–13.
Wan Q, Xiao L, Zhang X, Li Y, Lu Y, Song M, et al. Clonal evolution of Enterocytozoon bieneusi populations in swine and genetic differentiation in subpopulations between isolates from swine and humans. PLoS Negl Trop Dis. 2016;10(8):e0004966.
Fiuza VR, Oliveira FC, Fayer R, Santín M. First report of Enterocytozoon bieneusi in pigs in Brazil. Parasitol Int. 2015;64(4):18–23.
Ye J, Xiao L, Jian L, Huang W, Amer SE, Guo Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2014;63(1):132–7.
Němejc K, Sak B, Květoňová D, Hanzal V, Janiszewski P, Forejtek P, et al. Prevalence and diversity of Encephalitozoon spp. and Enterocytozoon bieneusi in wild boars (Sus scrofa) in Central Europe. Parasitol Res. 2014;113(2):761–7.
Hsu BM, Wun HY, Hsu CL. Detection and species identification of Cryptosporidium from Taiwan feeding animals. J Parasitol. 2008;94(1):252–6.
Yin JH, Yuan ZY, Cai HX, Shen YJ, Jiang YY, Zhang J, et al. Age-related infection with Cryptosporidium species and genotype in pigs in China. Biomed Environ Sci. 2013;26(6):492–5.
Lin Q, Wang XY, Chen JW, Ling D, Zhao GH. Cryptosporidium suis infection in post-weaned and adult pigs in Shaanxi province, northwestern China. Korean J Parasitol. 2015;53(1):113–7.
Chen Z, Mi R, Yu H, Shi Y, Yan H, Chen Y, et al. Prevalence of Cryptosporidium spp. in pigs in Shanghai, China. Vet Parasitol. 2011;181(2–4):113–9.
Zhang W, Yang F, Liu A, Wang R, Zhang L, Shen Y, et al. Prevalence and genetic characterizations of Cryptosporidium spp. in pre-weaned and post-weaned piglets in Heilongjiang province, China. PLoS One. 2013;8(7):e67564.
Wang R, Qiu S, Jian F, Zhang S, Shen Y, Zhang L, et al. Prevalence and molecular identification of Cryptosporidium spp. in pigs in Henan, China. Parasitol Res. 2010;107(6):1489–94.
Yin J, Shen Y, Yuan Z, Lu W, Xu Y, Cao J. Prevalence of the Cryptosporidium pig genotype II in pigs from the Yangtze River Delta, China. PLoS One. 2010;6(6):e20738.
Li W, Li Y, Li W, Yang J, Song M, Diao R, et al. Genotypes of Enterocytozoon bieneusi in livestock in China: high prevalence and zoonotic potential. PLoS One. 2014;9(5):e97623.
Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. 2011;49(5):2006–8.
Zhao W, Zhang W, Yang F, Cao J, Liu H, Yang D, et al. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl Environ Microbiol. 2014;80(12):3699–707.
Qiang W, Lin Y, Mao Y, Yang Y, Qiao L, Zhang S, et al. High prevalence and widespread distribution of zoonotic Enterocytozoon bieneusi genotypes in swine in Northeast China: Implications for Public Health. J Eukaryot Microbiol. 2015;63(2):162–70.
Li W, Wei T, Jiang Y, Diao R, Yang J, Xiao L. Genotypic distribution and phylogenetic characterization of Enterocytozoon bieneusi in diarrheic chickens and pigs in multiple cities, China: potential zoonotic transmission. PLoS One. 2014;9(9):e108279.
Li J, Qi M, Chang Y, Wang R, Li T, Dong H, et al. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou Zoo, China. J Eukaryot Microbiol. 2015;62(6):833–9.
Molecular evolutionary Genetics Analysis. http://www.megasoftware.net/. Accessed 5 Sep 2016.
Peng XQ, Tian GR, Ren GJ, Yu ZQ, Lok JB, Zhang LX, et al. Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infect Genet Evol. 2016;41:26–31.
Ma J, Feng Y, Hu Y, Villegas EN, Xiao L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J Water Health. 2016;14(3):411–23.
Zhang XX, Cong W, Lou ZL, Ma JG, Zheng WB, Yao QX, et al. Prevalence, risk factors and multilocus genotyping of Enterocytozoon bieneusi in farmed foxes (Vulpes lagopus). Northern China Parasit Vectors. 2015;9(1):1–7.
Němejc K, Sak B, Květoňová D, Hanzal V, Janiszewski P, Forejtek P, et al. Cryptosporidium suis and Cryptosporidium scrofarum in Eurasian wild boars (Sus scrofa) in Central Europe. Vet Parasitol. 2013;197(3-4):504–8.
Castro-Hermida JA, García-Presedo I, González-Warleta M, Mezo M. Prevalence of Cryptosporidium and Giardia in roe deer (Capreolus capreolus) and wild boars (Sus scrofa) in Galicia (NW, Spain). Vet Parasitol. 2011;179(1–3):216–9.
Němejc K, Sak B, Květoňová D, Hanzal V, Jeníková M, Kváč M. The first report on Cryptosporidium suis and Cryptosporidium pig genotype II in Eurasian wild boars (Sus scrofa) (Czech Republic). Vet Parasitol. 2012;184(2–4):122–5.
Paziewska A, Bednarska M, Niewegłowski H, Karbowiak G, Bajer A. Distribution of Cryptosporidium and Giardia spp. in selected species of protected and game mammals from North-Eastern Poland. Ann Agric Environ Med. 2007;14(2):265–70.
Kvác M, Kvetonová D, Sak B, Ditrich O. Cryptosporidium pig genotype II in immunocompetent man. Emerg Infect Dis. 2009;15(6):982–3.
Xiao S, Wei A, Chen Z, Zhang D, Yu J, Yang M. Occurrences and genotypes of Cryptosporidium oocysts in river network of southern-eastern China. Parasitol Res. 2012;110(5):1701–9.
Beck R, Sprong H, Lucinger S, Pozio E, Cacciò SM. A large survey of Croatian wild mammals for Giardia duodenalis reveals a low prevalence and limited zoonotic potential. Vector Borne Zoonotic Dis. 2010;11(8):1049–55.
Armson A, Yang RC, Thompson J, Johnson J, Reid S, Ryan UM. Giardia genotypes in pigs in Western Australia: prevalence and association with diarrhea. Exp Parasitol. 2009;121(4):381–3.
Budu-Amoako E, Greenwood SJ, Dixon BR, Barkema HW, Hurnik D, Estey C, et al. Occurrence of Giardia and Cryptosporidium in pigs on Prince Edward Island. Canada Vet Parasitol. 2012;184(1):18–24.
Fava NM, Soares RM, Scalia LA, Kalapothakis E, Pena IF, Vieira CU, et al. Performance of glutamate dehydrogenase and triose phosphate isomerase genes in the analysis of genotypic variability of isolates of Giardia duodenalis from livestocks. Biomed Res Int. 2013(4):417–22.
Hamnes IS, Gjerde BK, Forberg T, Robertson LJ. Occurrence of Cryptosporidium and Giardia in suckling piglets in Norway. Vet Parasitol. 2007;144(3-4):222–33.
Maddox-Hyttel C, Langkjaer RB, Enemark HL, Vigre H. Cryptosporidium and Giardia in different age groups of Danish cattle and pigs - occurrence and management associated risk factors. Vet Parasitol. 2006;141(1–2):48–59.
Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J Infect Dis. 2016;214(8):1256–9.
Slodkowicz-Kowalska A, Graczyk TK, Tamang L, Girouard AS, Majewska AC. Asymptomatic Enterocytozoon bieneusi microsporidiosis in captive mammals. Parasitol Res. 2007;100(3):505–9.
Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51(2):557–63.
Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, et al. Concurrent Infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. 2013;7(9):749–54.
Acknowledgements
We would like to thank Chao Gong, Jingchao Lan and Yue Zhang for collecting samples, and Xuefeng Cao, Yinan Tian, Liuhong Shen and Suizhong Cao for helping doing experiments, and Xuehan Liu, Haozhou Li, Na Xie and Xiaoping Ma for helping with dada analysis, and Ziyao Zhou for giving suggestions on experiment design.
Funding
The study was financially supported by the Chengdu Giant Panda Breeding Research Foundation (CPF2014-10; CPF2014-14; CPF2015-4) and the National Natural Science Foundation of China (number 31272620).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Representative sequences were submitted to the GenBank database under accession numbers: KU668880, KU668883–KU668893, KU668897, KU668898 and KX670577–KX670590.
Authors’ contributions
Experiments were conceived and designed by GP, ZZ and WL. HF collected the sample. Experiments were performed by WL, LD, KW, XH, YS and HS, and the data were analyzed by YH. The manuscript was written by WL. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
No animals were harmed during the sampling process. Permission was obtained from owners prior to collection of fecal specimens. This study followed guidelines in accordance with the Regulations for the Administration of Affairs concerning Experimental Animals, and was approved by the Animal Ethical Committee of Sichuan Agricultural University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, W., Deng, L., Wu, K. et al. Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasites Vectors 10, 10 (2017). https://doi.org/10.1186/s13071-016-1942-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1942-2