Abstract
Background
Rhodotorula toruloides is a robust producer of triacylglycerol owing to its fast growth rate and strong metabolic flux under conditions of high cell density fermentation. However, the molecular basis of fatty acid biosynthesis, desaturation and regulation remains elusive.
Results
We present the molecular characterization of four fatty acid desaturase (FAD) genes in R. toruloides. Biosynthesis of oleic acid (OA) and palmitoleic acid (POA) was conferred by a single-copy ∆9 Fad (Ole1) as targeted deletion of which abolished the biosynthesis of all unsaturated fatty acids. Conversion of OA to linoleic acid (LA) and α-linolenic acid (ALA) was predominantly catalyzed by the bifunctional ∆12/∆15 Fad2. FAD4 was found to encode a trifunctional ∆9/∆12/∆15 FAD, playing important roles in lipid and biomass production as well as stress resistance. Furthermore, an abundantly transcribed OLE1-related gene, OLE2 encoding a 149-aa protein, was shown to regulate Ole1 regioselectivity. Like other fungi, the transcription of FAD genes was controlled by nitrogen levels and fatty acids in the medium. A conserved DNA motif, (T/C)(G/A)TTGCAGA(T/C)CCCAG, was demonstrated to mediate the transcription of OLE1 by POA/OA. The applications of these FAD genes were illustrated by engineering high-level production of OA and γ-linolenic acid (GLA).
Conclusion
Our work has gained novel insights on the transcriptional regulation of FAD genes, evolution of FAD enzymes and their roles in UFA biosynthesis, membrane stress resistance and, cell mass and total fatty acid production. Our findings should illuminate fatty acid metabolic engineering in R. toruloides and beyond.
Similar content being viewed by others
Background
Unsaturated fatty acids (UFAs) are fatty acids containing one (mono-unsaturated fatty acid, MUFAs) or more double bonds (polyunsaturated fatty acids, PUFAs) in various positions and configurations in the carbon backbone. UFAs play important roles in membrane fluidity and serve as precursors for the biosynthesis of many bioactive molecules, such as lipid mediators, pheromones, eicosanoids and growth regulators [1].
Fatty acid desaturases (FADs) catalyze the sequential desaturation of fatty acids, leading to the production of MUFAs and PUFAs. FADs are classified into two groups, water-soluble acyl–acyl carrier protein (ACP) desaturases restricted in plant plastid [2, 3] and integral membrane type FADs, which share the highly conserved membrane-spanning motif, H(X)3-4H(X)7-41H(X)2-3HH(X)61-189H(X)2-3HH [4]. FADs can also be functionally categorized as front-end and methyl-end desaturase, which introduces a double bond towards the carboxyl terminus and methyl-end of the fatty acyl chain, respectively [3]. The front-end desaturases, such as Δ4, Δ5, Δ6 and Δ8 FADs, contain a specific N-terminal cytochrome b5-like domain, and are generally found in animals and lower eukaryotic microorganisms [5, 6].
The expression of FADs is usually regulated by fatty acids, nutrient and environmental cues. For example, Saccharomyces cerevisiae OLE1 expression is regulated at transcriptional and post-transcriptional (mRNA or protein stability) levels: the transcription is activated by unsaturated fatty acids through ubiquitin-mediated proteolytic processing of two membrane proteins, Mga2p and Spt23p, triggering their nuclear targeting to become transcriptional co-activators. Mga2p is believed to be a sensor for unsaturated fatty acid and it also regulates OLE1 transcripts stability by modulating exonuclease activity [7, 8]. These regulatory mechanisms appear to be quite conserved among different fungi [9].
Different fatty acids vary greatly in nutritional value and biological functions due to their unique fatty acid configuration in triacylglycerol, degree of desaturation and chain length. For example, γ-linolenic acid (GLA, 18:3Δ6,9,12) has anti-inflammation property and has applications in the treatment for atopic eczema, diabetes, heart disease, high blood pressure, arthritis, Alzheimer's disease, etc. [10, 11]. GLA is present in relatively low levels in oils extracted from a small number of plant seeds, such as those of evening primrose (Oenothera biennis) (8–10% of total fatty acid), blackcurrant (15–20%) and Borago officinalis (~ 20%) [12]. Although filamentous fungi such as Cunninghamella echinulata [13] and Mortierella isabellina [14] also produce GLA, they are not ideal hosts for industrial production due to slow growth, low lipid content and high viscosity during fermentation.
R. toruloides is an oleaginous yeast producing low levels of PUFAs, including linoleic acid (LA, 18:2Δ9,12) and α-linolenic acid (ALA, 18:3Δ9,12,15) [15]. Metabolic engineering offers an opportunity to drastically change its fatty acid composition and productivity [16,17,18,19,20]. As a highly robust oil producer, R. toruloides is a potentially powerful platform for fatty acid engineering and production [21,22,23,24]. R. toruloides remains a challenging host to work with due to its highly GC-rich genome (~ 62%); unusual regulation of gene expression and limited engineering tools [25,26,27]. To date, two Rhodotorula FAD genes have been reported, a stearoyl-CoA desaturase gene from R. toruloides IFFO 0880 [28] and a Δ12/Δ15 bifunctional desaturase gene from R. kratochvilovae YM25235 [29]. To further facilitate PUFA metabolic engineering in R. toruloides, we characterized four FADs identified, analyzing their gene/protein organizations, transcriptional regulations and effects of gene deletion and overexpression in the native host on fatty acid biosynthesis, stress responses and cell mass production. We illustrated multi-step metabolic engineering routes, via loss-of-function and gain-of-function approaches, for the efficient production of high-value fatty acids in R. toruloides.
Results and discussion
Identification of fatty acid desaturase genes in R. toruloides
To identify FAD genes, several well-studied enzymes were used as the queries for tBLASTn search (NCBI, USA) against the genomes of R. toruloides strain ATCC 10657 and 204091. Using S. cerevisiae Δ9 stearoyl-CoA desaturase (GenBank accession no. CAA96757, ScOle1p), Mortierella alpina Δ12 FAD (ADE06660, MaFAD2), M. alpina Δ6 FAD (AAL73949, MaFAD6) and Euglena gracilis Δ8 FAD (ADD51570) as queries, we identified 3 homologous genes, which were tentatively named OLE1, FAD2 and FAD4. However, homolog of M. alpina Δ5 FAD (ACM89303), Thraustochytrium sp. Δ4 FAD (AAM09688) and Saprolegnia diclina Δ17 FAD (AY373823) was not found.
Gene organization of FADs, such as coding sequence (CDS), 5′ and 3′UTR (untranslated region), was determined by incorporating the sequences of 5′ and 3′ RACE (rapid amplification of cDNA ends), RT-PCR and whole transcriptome. OLE1, FAD2 and FAD4 contain 7, 4 and 4 exons, encoding 545, 451 and 609 aa, respectively (Table 1 and Additional file 1: Fig. S1a). All splicing junctions abide strictly to the canonical GU-AG rule. Notably, OLE1 and FAD2 transcripts have long 3′UTRs, 296 nt and 261 nt, respectively, while FAD4 has a long 5′UTR of 349 nt (Table 1 and Additional file 1: Fig. S1a).
Ole1 was predicted to contain two transmembrane helices while Fad2 and Fad4 have three (Additional file 1: Fig. S1b). All three FADs contain the pfam00487 membrane domain that is highly conserved in different organisms [30], and other FAD-signature motifs, such as cd03505 (Δ9 FAD-like), cd03507 (Δ12 FAD-like) or cd03506 (Δ6 FAD-like) [31]. Surprisingly, only Ole1 contains the fungus-specific fused cytochrome b5 heme/steroid binding domain (pfam00173) at the carboxyl terminus (Additional file 1: Fig. S1b), suggesting Fad2 and Fad4 rely on the free form cytochrome b5 reductase to couple the fatty acid desaturation reaction. Like many reported membrane-bound FADs, all three FADs contain three conserved histidine boxes, H(X)3-4H, H(X)2-3HH and H/Q(X)2-3HH (Additional file 1: Fig. S1b and Additional file 1: Fig. S2), which form the di-iron complex that is essential for the desaturation reaction. The positions of the first two histidine boxes are highly conserved, separated by 31–32 aa. The 3rd histidine box is located 130 aa from the 2nd one in Ole1 while the spacing in Fad2 and Fad4 is 185 aa and 193 aa, respectively (Additional file 1: Fig. S2). Notably, the 3rd histidine box of Fad4 has an imperfect sequence, QxxHH, which is often observed in the front-end desaturases [32].
Phylogenetic analysis of eukaryotic FADs showed that R. toruloides homologs fell into three distinct groups (Additional file 1: Fig. S3). Consistent with previous work [3], it is difficult to distinguish mono-functional Δ12 FAD and Δ15 FAD from bifunctional Δ12/Δ15 FAD based on the amino acid sequences. Bifunctional enzymes with both Δ12 and Δ15 regioselectivity are believed to derive from Δ12 FAD [3]. In nature, R. toruloides strains are usually haploids with two mating types (A1 and A2) [33]. Sequence comparison revealed no amino acid sequence difference in strains of the same mating type (mating type A1 strains ATCC 10657, ATCC 204091 and IFFO0880 or mating type A2 strains ATCC 10788, MTCC457 and CECT 1137) [34] while 94.3 – 97.2% identities were observed between different mating types (Additional file 1: Table S1). However, the nucleotide sequence identities were much lower, ranging from 87.1 to 88.9%.
Regulation of FAD gene transcription
Fatty acid biosynthesis and lipid accumulation are often regulated by environmental and nutrient cues [35]. Indeed, qRT-PCR analysis showed that OLE1, FAD2 and FAD4 mRNA levels were significantly increased under 6-h nitrogen starvation, and the level of OLE1, FAD2 and FAD4 transcripts in nitrogen-free YNB medium was 1.6, 5.9- and 2.1-fold higher than in YNB medium, respectively (Fig. 1a). These suggest the involvement of common nitrogen-regulated transcriptional factors.

source-free YNB broth, which was individually supplemented with different fatty acids (10 g/L) and cultured at 28 °C for 8 h with 280 rpm agitation. Glucose (10 g/L) was used as the control, and Tergitol NP40 was supplemented at 1% (w/v) to facilitate fatty acid absorption in all treatments. For qPCR analysis, actin encoding gene (ACT1) was used as the reference and error bars represent the standard derivations of triplicates
qRT-PCR analysis of FAD genes. a Effect of nitrogen starvation on FAD gene transcription. R. toruloides seed culture established in YPD broth was washed in water, inoculated to yeast nitrogen base (YNB) or YNB-N− (without amino acid supplement and ammonium sulfate) and cultured at 28 °C. Cells for total RNA extraction were sampled at 12 and 24 h after inoculation. The relative mRNA levels were calculated by 2-ΔΔCt method and represented as the ratio between the two media. b Effects of exogenous fatty acid supplementation on transcription of FADs. The 2-ΔCt method was applied for the data analysis and represented as relative expression against the reference gene. The inlet shows the levels of each mRNA from cells cultured in different media after normalizing against that in the control medium (glucose as the carbon source). Seed culture was washed with water twice and inoculated in carbon
Studies on the gene transcriptional effects of exogenous fatty acids were concentrated on Δ9 FAD. Strong repressive effects of UFAs were reported in several yeasts [36,37,38], however, different effects were also reported, with minor repression or no effect in other yeasts [39, 40]. With regard to FAD2 and FAD4, it is interesting to note that UFAs resulted in different regulatory patterns [41,42,43]. Thus, R. toruloides FADs were investigated on their responses to different fatty acids as the sole carbon source. The three FAD genes showed significantly different transcription levels (Fig. 1b). OLE1 mRNA was the most abundant, the transcription of which could be strongly induced by palmitoleic acid (POA, 16:1∆9) and significantly depressed by most other fatty acids (Fig. 1b). FAD2 transcription was significantly induced by most fatty acids tested except 18:2. Like OLE1, POA was the strongest inducer for FAD2. This regulatory pattern was similar to the OA-inducible pattern of Yarrowia lipolytica ∆12 FAD [44]. FAD4 transcription, on the other hand, was significantly induced by POA, 18:2 and 18:3 although the overall mRNA level was the lowest among the three FAD transcripts. Taken together, the general regulatory network of FAD gene transcription is quite conserved among different fungi. Nevertheless, R. toruloides has evolved distinct regulatory controls, such as the strong induction of FAD gene expression by POA. It would be interesting to see how common this phenomenon is in other microbes although, as suggested previously [45], it could result from its unique evolutionary history and specific niche it resides. POA is a rare fatty acid present in the cells at very low levels in most systems. We speculate that supplying high levels of this may drastically change the membrane structure or fluidity, resulting in a stress response in the cells.
Molecular basis of transcriptional control of FAD genes
To investigate the transcriptional regulations, the upstream sequence of OLE1 (-843 to -1 from the 1st ATG codon) was cloned and analyzed by luciferase gene reporter assay. Time course study showed that the promoter was strongly induced by POA (peaked by 16.4-fold at 1 h) (Additional file 1: Fig. S4), which agrees well with the qRT-PCR results (Fig. 2b). The promoter was also induced by OA (peaked by 5.9-fold at 4 h). As POA is much more costly, OA was used as the inducer in later transcriptional studies.
Identification of putative transcriptional regulatory elements. a Distribution of putative fatty acid responsive elements in the upstream sequences of 3 FAD genes. Common DNA motifs were identified using the MEME suite at http://meme-suite.org. E-value is the estimate of the number of motifs expected to find by chance if the letters in the input sequences were shuffled. tss: transcription start site. b Sequence alignment of two candidate OLE1 Regulatory Element (ORE) in the OLE1 promoter. ORE1m is the artificially generated mutant of ORE1, containing 2-nt substitutions. c Luciferase gene reporter assay of hybrid GPD1 promoters. The promoter structures are illustrated on the left, where green, blue and red bars show the location of the inserted DNA motifs. d Effect of OLE1 terminator on gene expression. The Cauliflower mosaic virus 35S gene terminator in POLE1::RtLUC2::T35S was replaced with the 328 bp terminator of OLE1. The relative promoter activity (RPA) was calculated by normalizing the value against the reading of control construct (POLE1::RtLUC2::T35S) of the same culture conditions and sampling time point
To identify the common cis-acting elements involved in the transcriptional regulation by fatty acids, the upstream sequences of OLE1, FAD2 and FAD4 (Additional file 1) were analyzed using the MEME suite [46], leading to the identification of a 15-nt conserved DNA motif (Fig. 2a). In OLE1, two such motifs with 3-nt variations were found, at -638 and -434 from the translational start site. The motifs were tentatively named ORE1 and ORE2 (OLE1 Regulatory Element), respectively, Fig. 2b). To confirm its function, ORE1 and ORE2 was individually fused to the 5′ end of the basal GPD1-176 promoter (-176 to the 1st ATG codon) [25]. Neither ORE1 nor ORE2 significantly affected GPD1-176 activity when the reporter strains were cultured in YPD medium (Fig. 2c). In contrast, the ORE1-GPD1-176 promoter showed 1.5-fold higher activity than GPD1-176 when cultured in OA-supplemented medium, whereas ORE2 showed negligible effect. To determine which of the three substituted nucleotides was functionally important, an ORE1 mutant (ORE1m) was created by converting its first 2 nucleotides to the corresponding residues in ORE2 (Fig. 2b). Reporter assay revealed a complete loss of oleate-inducing effect after the sequence change (Fig. 2c). This suggests that ORE1, and possibly some related motifs, plays a significant role in regulating FAD gene transcription. The data also suggest the possibility to engineer a strong OA/POA-inducible gene expression system in R. toruloides by using the OLE1 promoter and ORE1 motif.
The transcripts of OLE1 and FAD2 have long 3′UTRs (Table 1 and Additional file 1: Fig. S1a). To investigate if the 3′UTR of OLE1 has any role in regulating OLE1 expression, the luciferase reporter construct was modified by replacing the terminator of Cauliflower mosaic virus (CaMV) 35S gene with that of OLE1, including the entire 296-bp 3′ UTR and 32-bp downstream sequence (Additional file 1). This resulted in a significant drop in the luciferase activity (Fig. 2d). This explains the discrepancy between the results of qRT-PCR (Fig. 1b) and promoter reporter assay (Additional file 1: Fig. S4). Thus, OA and POA modulate OLE1 transcription via the cis-acting elements located in both the upstream and downstream regions of the gene.
OLE1 is essential for cell viability and biosynthesis of oleic acid and palmitoleic acid
We reported previously that gene deletion frequency could reach more than 95% when using the KU70 knockout mutant [47]. However, no OLE1 deletion mutant was obtained after repeated attempts, regardless of OA supplementation to culture media. Subsequently, a true deletion mutant (ole1Δ) was generated in another strain, R. toruloides C3 (Additional file 1: Fig. S5a). Sequence analysis revealed that the single-copy OLE1 gene is highly conserved between C3 and ATCC 10657 strains, with only 7-nt substitutions that occurred in intron regions.
As expected, ole1Δ was unable to grow in medium with glucose or saturated fatty acids as the sole carbon source (Fig. 3a). In contrast, supplementation of any UFAs, such as 16:1, 18:1, 18:2 or 18:3, rescued the growth defect caused by the lack of OLE1 gene (Fig. 3a). Therefore, ole1Δ is an UFA-auxotrophic mutant, resembling its counterpart in S. cerevisiae [48]. ole1Δ was inactive in UFA biosynthesis (Fig. 3b). The small amount of 18:1 detected was probably derived from the inoculant cells that had been cultured in OA-supplemented medium (Fig. 3b). As expected, re-introduction of wild-type OLE1 gene into the ole1Δ genome completely restored the growth defects (Fig. 3a) and lipid biosynthesis (Fig. 3b). Furthermore, UFA, such as 16:1, 18:1, 18:2 and 18:3, partially restored the fatty acid profile of ole1Δ although fatty acid titer remained much lower due to the defect in cell growth (Fig. 3c). Supplement of 16:1 or 18:1, but not 16:0 or 18:0, was able to complement the growth and fatty acid biosynthesis defects of ole1Δ (Fig. 3a and c). These suggest that other FADs were functional in the absence of Ole1. Overexpressing OLE1 using the strong GPD1 promoter resulted in a 5.3- and 1.3-fold increase in 16:1 and 18:1 level, respectively (Fig. 3d). Collectively, our results suggest that, similar to its homologs in S. cerevisiae and Y. lipolytica [49], Ole1 is a Δ9 desaturase with a strong substrate preference to stearoyl-CoA over palmitoyl-CoA. Our data also support the previous studies that oleic acid plays a central role in fungal growth and metabolism [4, 48, 50]. To date, Δ9 FAD null mutants have been reported only in two ascomycetous yeasts, S. cerevisiae and Candida parapsilosis [4, 51]. To the best of our knowledge, this is the first report on the phenotypes of OLE1 null mutant in basidiomycetous fungi and oleaginous yeasts.
Functional characterization of OLE1. a Genetic and chemical complementation of ole1Δ. ole1C strain is an ole1Δ derivative containing a randomly inserted construct comprising 641-bp OLE1 promoter, full-length OLE1 cDNA and 328-bp OLE1 terminator derived from ATCC 10,657. Cells were cultured on YPD agar in the presence ( +) or absence (−) of various fatty acids at 1% w/v. b Unsaturated fatty acid profiles in R. toruloides wild-type (WT), ole1Δ and ole1C strains. Strains were cultured in YPD broth or YPDtO broth (YPD broth supplemented with 0.1% w/v OA and 0.5% w/v Tergitol NP40) until exponential phase. Water-washed (2 times) cells were inoculated to GJm3 medium and cultured for 5 days. c Fatty acid profiles of R. toruloides WT and ole1Δ mutant. Cells were cultured in GJm3 medium supplemented with different fatty acids (0.1%, w/v) for 4 days. CTL represents the cells cultured in GJm3 medium in the absence of any fatty acid. (d) Fatty acid profiles in wild-type strain (WT) and OLE1 overexpression mutant (OLE1GPD1). Both strains were cultured in GJm3 medium for 5 days. %TFA represents the weight percentage of total fatty acid. Error bars represent the standard derivations of triplicates. Student’s t-test was used for statistical analysis, where possibility less than 0.05% and 0.01% was marked as * and **, respectively
FAD2 encodes a Δ12/Δ15 bifunctional fatty acid desaturase
The FAD2 null mutant (fad2Δ, Table 2) was also generated (Additional file 1: Fig. S5b). Unlike ole1Δ, fad2Δ was able to grow normally without the presence of 18:2 in media (Fig. 6a, b), suggesting that Fad2 is dispensable for cell growth. The production of both 18:2 and 18:3 was abolished in fad2Δ while 18:1 was normal (Fig. 4a). These data suggest that Fad2 functions as both the Δ12 and Δ15 FAD to convert 18:1 to 18:2 and 18:3.
Characterization of FAD2. a GCMS chromatographs of FAMEs from WT and FAD2 deletion mutant (fad2Δ). + 18:2 indicates cells cultured in medium supplemented with 18:2. b Complementation of fatty acid production defect of fad2Δ by overexpression of known heterologous Δ12- and/or Δ15-fatty acid desaturase. WT wild-type strain, fad2MF2 fad2Δ expressing the codon-optimized gene encoding M. alpina Δ12 FAD (MaFAD2-2), fad2LF3 fad2Δ expressing the codon-optimized flax Δ15 FAD (LuFAD3-2), fad2ML fad2Δ expressing both MaFAD2-2 and LuFAD3-2. c Fatty acid profiles of WT, fad2Δ, fad2FAD2 and fad2FAD4. %TFA represents weight percentage of total fatty acids. Error bars represent standard derivations of triplicates
The enzymatic function of Fad2 was further investigated by complement with certain substrates and genes. Addition of 18:2 did not restore the 18:3 level in fad2Δ (Fig. 4a), which is consistent with the lack of Δ15 FAD in fad2Δ. As M. alpina Δ12 FAD (MaFAD2) and Linum usitatissimum Δ15 FAD (LuFAD3) have been functionally confirmed [52, 53], the encoding genes were synthesized after codon optimization (MaFAD2-2 and LuFAD3-2, respectively) and used in the test. Introduction of MaFAD2-2 (strain fad2MF2, Table 2) only restored the production of 18:2 (Fig. 4b). Introduction of LuFAD3-2 (strain fad2LF3, Table 2), had little effect on fatty acid profile (Fig. 4b). 18:3 was produced only when MaFAD2-2 and LuFAD3-2 were co-expressed (strain fad2ML, Table 2) (Fig. 4b). As expected, re-introduction of the endogenous FAD2 gene into fad2Δ (strain fad2FAD2, Table 2) restored the biosynthesis of UFAs (Fig. 4c). Collectively, Fad2 is a bifunctional FAD with Δ12 and Δ15 activities.
FAD4 encodes a minor multi-functional desaturase with low regioselectivity
The primary structure of Fad4 is more related to Δ4, Δ5, Δ6 and Δ8 FADs (Additional file 1: Fig. S3). However, there was no Δ6 (e.g., γ-linolenic acid, GLA) or Δ8 fatty acid detected in R. toruloides oil. Fad4 and Fad2 share 20.5% identity at amino acid level and 44.9% identity at cDNA level, suggesting that FAD4 may be derived from gene duplication or horizontal gene transfer from FAD2-related gene. Due to the presence of a di-proline motif at the N-terminus (P3-P4), Fad4 might be destabilized by membrane fatty acid desaturation [54]. To investigate its function, FAD4 null mutant (fad4Δ) was generated (Additional file 1: Fig. S5c). Deletion of FAD4 resulted in a significant drop in 18:1 (14.7%, p < 0.001) and 18:2 (6.1%, p < 0.001), suggesting that Fad4 has Δ9 and Δ12 FAD activities (Fig. 5a). However, it is puzzling to see the rise of 18:3 in fad4Δ (Fig. 5a). The increase of 18:0 level (Fig. 5a) might have resulted from pathway overflowing due to the downstream blockage.
Identification and characterization of FAD4. a Fatty acid profiles of WT, fad4Δ, fad4FAD2 and fad4FAD4. b Biomass, lipid titers and lipid contents of WT, fad4Δ, fad4FAD2 and fad4FAD4. c 18:2 production (%TFA) in WT and fad2Δ strains. d Fatty acid profiles of WT, fad24Δ, fad24FAD2 and fad24FAD4. e Unsaturated fatty acid yields (titer, g/L) in WT, ole1Δ and ole1FAD4 strains. f Fatty acid profiles in WT, ole1Δ and ole1FAD4 strains. The inlet shows the profile of 16:1. %TFA represents weight percentage of total fatty acids. Error bars represent standard derivations of triplicates
To further investigate Fad4 function, FAD2 and FAD4 were overexpressed in fad4Δ (strain fad4FAD2 and fad4FAD4, respectively, Table 2) using the strong GPD1 promoter. The increase of 18:3 level upon overexpression of either FAD2 or FAD4 strongly suggests both enzymes have the bifunctional ∆12/∆15 FAD activity (Fig. 5a). The drop of 18:2 level was likely the result of substrate consumption by the ∆15 FAD activity. Notably, the lack of FAD4 significantly enhanced lipid content and cell mass production (Fig. 5b), suggesting its role in suppressing lipid biosynthesis and cell growth.
These results prompted us to re-examine the fatty acid profile of fad2Δ. Indeed, trace amount of 18:2 (0.06 ± 0.01% TFA) was found, accounting for 0.7% of wild-type strain (Fig. 5c). Thus, we created a double mutant for FAD2 and FAD4 (fad24Δ, Table 2). As expected, 18:2 disappeared completely in fad24Δ (Fig. 5d). Overexpression of FAD2 in fad24Δ (strain fad24FAD2, Table 2) largely restored the fatty acid profile. Significantly, 18:2 and 18:3 levels were increased at the expense of their precursor 18:1 (Fig. 5d), reinforcing our conclusion for the Δ12/Δ15 FAD activity of Fad2. In contrast, FAD4 overexpression in fad24Δ background (strain fad24FAD4, Table 2) led to a slight increase of 18:1 (Fig. 5d), suggesting that Fad4 possessing a weak ∆9 FAD activity. To test this hypothesis, FAD4 was overexpressed in ole1Δ (strain ole1FAD4, Table 2). Although this strain remained defective in growth (Fig. 5e and Additional file 1: Fig. S6), 18:1, 18:2 and 18:3 levels were all increased (Fig. 5f). Notably, the increase in 18:2 and 18:3 levels were not accompanied by a drop of 18:1, a phenomenon observed in Δ12/Δ15 FAD overexpression. This further implied that ole1FAD4 strain contained a weak Ole1-like activity. Collectively, our results suggest that Fad4 is an unusual enzyme with Δ9/Δ12/Δ15 trifunctional FAD activity. To the best of our knowledge, only one similar case has been reported to date, a Δ6 FAD with Δ9 and Δ12 FAD activity [55].
Physiological roles of FAD
Being the major constituent, the number of double bonds in the fatty acids of phospholipids is critical for the physical property of cell membranes [56]. OLE1 deletion led to almost complete halt of cell division (cell budding) (Fig. 3a), although cell morphology was little changed (Additional file 1: Fig. S7). It was puzzling that ∆9 MUFA (18:1), but not its saturated precursor (18:0) or further desaturation products (18:2 or 18:3), was critical for cell viability. Studies in animal cells show that OA is not simply a structural element of membranes; it plays complex signaling roles also [57].
FAD2 deletion also led to slower cell growth under most conditions, which appeared to enhance the sensitivity to thermal stress (37 °C and 24 °C) and osmotic stress (glycerol or sorbitol) (Fig. 6a). As expected, the growth defect of fad2Δ was relieved by genetic complementation with a heterologous Δ12 FAD (strain fad2MF2) or Δ12 + Δ15 FAD (strain fad2ML, Fig. 6a).
Stress responses. a Effects of various stress stimuli on cell growth. b Effects of unsaturated fatty acids on cell growth. c Effects of 18:3 on cell growth of fad4Δ and fad4FAD4a. Cell cultured at exponential phase were water-washed twice and spotted in tenfold serial dilutions on YPD agar plates supplemented with the indicated UFAs (0.1%, w/v) or stress-inducing chemical, and incubated at different temperatures. Cells cultured at 30 °C on YPD agar in the absence of any supplement was used as the control. fad4FAD4, fad4Δ harboring FAD4 cDNA driven under GPD1 promoter; fad4FAD4a, fad4Δ harboring whole FAD4 allele; Supp., supplementation; Temp., temperature; CPL, β-caryophyllene. Concentrations used: glycerol, 2 M; sorbitol, 1 M; ethanol, 3% (w/v); CPL, 0.1% (w/v); NaCl, 0.8 M
Although Fad4 displayed weak activity in fatty acid desaturation, deletion of the gene significantly enhanced cell sensitivity to PUFA (18:3 in Fig. 6b). It is believed that ethanol alters plasma membrane transport [58] and increases membrane fluidity [59]. This is consistent with the previous observation that high unsaturation of fatty acids correlates with high cytotoxicity [60]. Surprisingly, overexpression of FAD4 cDNA under the strong GPD1 promoter only slightly relieved 18:3 sensitivity (Fig. 6b). On the other hand, re-introduction of the genomic FAD4 allele to fad4 mutant (fad4FAD4a) successfully complemented the growth defect (Fig. 6c), suggesting the significant role of introns in regulating FAD4 expression. This is consistent with our earlier report on other genes in this host [26]. Taken together, Fad2 and Fad4 both play important roles in protecting cells from membrane stress. These findings open a new avenue to enhance fatty acids and terpenoid productivity in R. toruloides.
OLE2 encodes a weak regulator of Ole1
During the annotation of OLE1, another DNA fragment sharing high homology to OLE1 was found in the genome. The 240-nt sequence shares 86% identity with OLE1. We tentatively named the gene OLE2 (Table 1). The conservation of this gene in different isolates, such as ATCC 10,788, NP11 and C3 strains [34, 61], suggest that it is functionally important. qRT-PCR analysis showed the sequence was abundantly transcribed and the transcripts level was regulated by fatty acids (Fig. 7a). In strain ATCC 10657, the sequence is located ~ 13 kb from OLE1. Notably, the predicted 149-aa Ole2 protein exhibits high identity to the region around the 3rd histidine box of Ole1, a region believed to be crucial for regioselectivity of FADs (Fig. 7b). Targeted deletion of OLE2 (Additional file 1: Fig. S5d) showed that 16:1 level dropped by 46.5% (p < 0.01) while 16:0 level increased by 16% (p < 0.05) (Fig. 7c). The levels of most other fatty acid species were not changed significantly. Deletion of OLE2 did not appear to affect cell growth significantly. Thus, OLE2 encodes a weak regulator of fatty acid desaturation, modulating the regioselectivity of Ole1. The effects of OLE2 deletion were consistent with the overexpression of OLE1 as both preferentially affected 16:1 level. Considering the strong role of 16:1 in inducing FAD gene transcription, it is possible that Ole2 can also regulate the transcription of OLE1, FAD2 and FAD4 indirectly.
Identification and analyses of OLE2. a qRT-PCR analysis of OLE1 and OLE2 expression. Seed cultures were water-washed twice and inoculated in YNB broth (without carbon source) supplemented with different fatty acids (10 g/L) and Tergitol NP40 (1%, w/v), and cultured at 28 °C, 280 rpm for 8 h. Glucose (10 g/L) was used as the control. The values presented are the relative expression against ACT1 mRNA (2-ΔCt method). Error bars represent the standard derivations of triplicates. b Sequence alignment of Ole1 (partial) and Ole2. c Fatty acid profiles of WT and ole2Δ. 16:1 was shown in the inlet plot
Metabolic engineering of fatty acids
As a proof of concept, we demonstrated how the FAD genes could be exploited for high-level production of OA and a novel fatty acid, γ-linolenic acid (GLA) (Fig. 8a). High OA oil has many applications, including food, cosmetics, textiles, adhesives and biofuels [62, 63]. Deletion of FAD2 led to little change in the volumetric productivity (titer) of OA, however, it significantly increased OA content, from 33.2 to 63.5% TFA (Fig. 8b). Further deletion of FAD4 resulted in a 1.3-fold improvement in OA titer, although OA content was slightly decreased (Fig. 8b). To further improve OA production, the endogenous OLE1 was overexpressed using the strong and lipogenic LDP1in promoter [26], resulted in a strain with two copies of OLE1 in the genome (mutant fad2OLE1 and fad24OLE1, Table 2 and Fig. 8a). This resulted in increased OA titer (15 ~ 17%, Fig. 8b). Surprisingly, OA content was reduced in fad2Δ, but not fad24Δ background (Fig. 8b). This difference may result from the interplay between cell growth mediated by the FAD4 gene and fatty acid imbalance conferred by OLE1 overexpression. Notably, increased expression of OLE1 was reported to be toxic to the cells in S. cerevisiae [48].
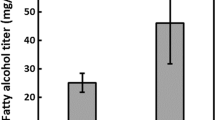
Metabolic engineering for oleic acid (OA) and γ-linolenic acid (GLA). a Metabolic engineering routes for production of OA and GLA. More information on the promoters and strains are shown in Table 2. b Production titer of OA of various strains. c Production titer of GLA of various strains. All strains were cultured in GJm3 medium for 5 days in shake flasks (30 °C and 250 rpm). The numbers shown on top the column show the weight percentage of total fatty acids (%TFA) of OA (in b) or GLA (in c). ALA is the α-linolenic acid produced in wild-type strain. Error bars represent standard derivations of triplicates. Symbols “ + ” and “−” represent the presence and absence of the gene, and “ + + ” represents the presence of two copy of the gene. d Comparison of OA and GLA yields by flask and bioreactor fermentation
M. alpina is a natural GLA producer, and its Δ12 and Δ6 FAD catalyze the final two steps of GLA biosynthesis [64]. Overexpression of M. alpina MaFAD2-2 (synthetic Δ12 FAD) along with MaFAD6-2 (synthetic Δ6 FAD) (Fig. 8a) in fad2Δ and fad24Δ (mutant fad2MF26 and fad2MF26, Table 2) successfully turned R. toruloides into a GLA producer, resulting in a titer of 0.37 g/L (28.4%TFA) and 0.53 g/L (27.4%TFA), respectively (Fig. 8c). A preliminary 2-L-scale fed-batch fermentation showed that the maximal OA and GLA titer reached 3.5 and 2.6 g/L, representing 60.1% and 27.3% TFA, respectively (Fig. 8d).
Recently, an engineered Y. lipolytica strain has been reported to produce GLA to 4.6% of total fatty acids. The low yield could have resulted from the toxicity of GLA as lowering the culturing temperature increased GLA yield about 61% [65]. Notably, our GLA content has far exceeded the dominant commercial product, evening primrose oil [12]. We expect further improvement in OA and GLA yields when PUFA degradation and fatty acid selectivity in TAG synthesis can be manipulated. Thus, R. toruloides can be a strong platform for PUFA metabolic engineering and production.
Conclusion
R. toruloides genome encodes a single-copy highly conserved ∆9 FAD, which is essential for cell viability and biosynthesis of MUFAs and PUFAs. The mutant ole1Δ provided a rare genetic insight into the role of Δ9 FAD on cell growth, fatty acid desaturation and lipid accumulation. As a yeast that is phylogenetically distant to the popular yeast hosts, such as S. cerevisiae and Y. lipolytica, it was not surprising to see R. toruloides has evolved significantly in the control of fatty acid biosynthesis and FAD gene expression, which were exemplified by the regulation of gene transcription via the GC-rich ORE1 motif, preferential induction by palmitoleic acid and involvement of two similar multi-functional FADs for PUFA biosynthesis. Fad4 is particularly interesting, not only for its relaxed regioselectivity of fatty acid desaturation, but also its roles in stress tolerance and maintaining healthy biomass and lipid production. Our data should illuminate PUFA engineering beyond this host.
Materials and methods
Strains, media, and culture conditions
Strains used are listed in Table 2. R. toruloides strain ATCC 10657 was obtained from ATCC (USA), and strain C3 was isolated from a fish sample in Singapore. Both are haploid (mating type A1) and share high genome sequence homology to R. toruloides ATCC 204091 (GenBank No. AEVR02000000) [34, 66]. R. toruloides strain Δku70e, a KU70 null mutant with high frequency of homologous recombination [47], is referred as the wild-type strain in this study. Yeast strains were maintained at 28–30 °C in YPD broth (1% yeast extract, 2% peptone, 2% glucose, w/v) or on potato-dextrose agar (PDA, Sigma-Aldrich, USA). YPDtO is YPD broth supplemented with 0.1% (w/v) oleic acid and 0.5% (w/v) Tergitol NP40) and was used for propagation of ole1Δ.
GJm3 is a lipid accumulation medium modified from the previous report [15]. It contains (per liter) 70 g glucose, 2.5 g yeast extract, 0.4 g KH2PO4, 1.5 g MgSO4·7H2O, 40 mg CaCl2·2H2O, 5.5 mg FeSO4·7H2O, 5.2 mg citric acid·H2O, 1 mg ZnSO4·7H2O, 0.76 mg MnSO4·H2O and pH was adjusted to 6.0 with sulfuric acid.
Yeast nitrogen base (without amino acid or ammonium sulfate) containing glucose (20 g/L) was used as nitrogen starvation medium (YNB-N−) while nitrogen rich medium (YNB-N+) was YNB-N− supplemented with 5 g/L ammonium sulfate. Cells were cultured until exponential phase; washed twice with water; inoculated to either YNB-N− or YNB-N+ and cultured at 28 °C with 280 rpm agitation. In fatty acid supplementation experiments, R. toruloides cells were cultured in YNB broth containing 5 g/L ammonium sulfate, a fatty acid of interest or glucose (10 g/L) and Tergitol™ NP40 (1%, w/v).
DNA constructs
DNA constructs used are listed in Table 2. Oligonucleotide sequences are listed in Additional file 1: Table S2. DNA constructs were verified by restriction mapping and DNA sequencing using the BigDye method (ABI). Details for DNA vector construction are shown in Additional file 1: Fig. S8.
Extraction of genomic DNA and total RNA
Genomic DNA and total RNA extraction were performed as reported previously [25]. Nucleic acids were quantified using NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA) and the quality was checked by agarose gel electrophoresis.
Rapid amplification of cDNA ends (RACE)
5′ RACE and 3′ RACE were done using BD SMARTer™ RACE cDNA Amplification Kit (BD CLONTECH Laboratories, Palo Alto, CA, USA) according to the manufacturer’s instructions. Oligo pair OLE1U1/OLE1L1, FAD2U1/FAD2L1 and FAD4U1/FAD4L1 (Additional file 1: Table S2) was used as the specific primer for 5′ and 3′ RACE of OLE1, FAD2 and FAD4, respectively.
Gene annotation and phylogenetic analysis
As the genome sequences of R. toruloides strain ATCC 10657, IFFO 0880 and ATCC 204091 [34, 66] are highly similar, the annotated genome database of R. glutinis ATCC 204091 was used as the reference. FAD genes were identified using tBLASTn at NCBI against the reference genome database using protein sequences of various types of well-characterized FADs as the queries. Full-length cDNA sequences were obtained by RT-PCR after the 5′ and 3′ ends were determined by RACEs. The FAD orthologs of other Pucciniomycotina species were predicted by BLASTp program. Sequence alignment and phylogenetic analysis were performed with the MEGA 6 program (www.megasoftware.net) using the Neighbor-Joining algorithm [67]. The membrane configurations of the proteins were predicted at the transmembrane prediction server TMHMM-2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The consensus sequences were analyzed through the MEME suite server (http://meme-suite.org/) [46].
Genetic manipulation
Agrobacterium tumefaciens-mediated transformation (ATMT), targeted gene deletion and fungal colony PCR were performed as described previously [25, 47]. Oleic acid was supplemented to the media for co-culture and selection in order to obtain OLE1 the deletion mutant while LA was supplemented to culture media to facilitate the generation of FAD2 and FAD4 mutants.
Gene expression cassettes were usually site-specifically integrated to the CAR2 locus (encoding phytoene synthase/lycopene cyclase) to eliminate positional effects. Knock-in mutants were selected based on the albino phenotype [26, 47, 68]. At least 3 biological replicates were used in the assays.
Southern blot analysis
Genomic DNA (5 µg) was digested with a restriction enzyme and separated by electrophoresis in a 0.8% agarose gel. Southern blotting was performed as described using digoxigenin-labeled DNA as the probes [69]. The restriction enzymes and DNA probes used are shown in Additional file 1: Fig. S5a-d.
Analyses of gene expression
Total RNA preparation, cDNA synthesis, real-time PCR analysis and luciferase gene reporter assay were performed as reported previously [26]. Briefly, R. toruloides strains harboring different reporter cassettes were cultured in YPD broth until exponential phase. Cells were cultured for 8 h in fresh YPD broth, which may be supplemented with a fatty acid of interest at 0.1% (w/v).
Quantification methods
Quantification of cell biomass (dry cell weight), residual glucose and lipids were performed as previously reported [70]. Fatty acid profile was determined by gas chromatography–mass spectrometry (GCMS) after esterified to fatty acid methyl esters (FAMEs) as described previously [70]. The specific fatty acids were quantified by normalization against the internal standard (ISTD, 15:0) and the corresponding response factor against ISTD as calculated through a pre-run of the standard FAME mixture (Supelco 37 Component FAME Mix, Sigma, USA).
GenBank accession numbers
Based on the codon preference of highly expressed genes in R. toruloides, MaFAD2-2, MaFAD6-2, LuFAD3-2 were synthesized by GenScript (USA) according to the protein sequence of M. alpina FAD2 (ADE06660), M. alpina FAD6 (AAL73949), L. usitatissimum omega-3 desaturase (AFN53677), respectively. The nucleotide sequences have been deposited to GenBank under the accession number MF152712 through MF152717.
Availability of data and materials
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files or from the corresponding author on request.
Abbreviations
- ATCC:
-
American Type Culture Collection
- CDS:
-
Coding sequence
- ER:
-
Endoplasmic reticulum
- 16:1/POA:
-
Palmitoleic acid (16:1Δ9)
- 18:1/OA:
-
Oleic acid (18:1Δ9)
- 18:2/LA:
-
Linoleic acid (18:2Δ9,12)
- 18:3/ALA:
-
α-Linolenic acid (18:3Δ9,12,15)
- GLA:
-
γ-Linolenic acid (18:3Δ6,9,12)
- FAD:
-
Fatty acid desaturase
- Ole1:
-
Δ9 Stearoyl-CoA desaturase
- Fad2:
-
Δ12/Δ15 Bifunctional fatty acid desaturase
- Fad4:
-
Δ9/Δ12/Δ15 Trifunctional fatty acid desaturase
- OLE2 :
-
OLE1-Related gene
- RACE:
-
Rapid amplification of cDNA ends
- TFA:
-
Total fatty acids
- UTR:
-
Untranslated region
References
Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu Rev Nutr. 2005;25:317–40.
Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol. 1998;49(1):611–41.
Sperling P, Ternes P, Zank TK, Heinz E. The evolution of desaturases. Prostaglandins Leukot Essent Fatty Acids. 2003;68(2):73–95.
Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265(33):20144–9.
Los DA, Murata N. Structure and expression of fatty acid desaturases. Biochim Biophys Acta. 1998;1394(1):3–15.
Meesapyodsuk D, Qiu X. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 2012;47(3):227–37.
Kandasamy P, Vemula M, Oh CS, Chellappa R, Martin CE. The endoplasmic reticulum membrane protein, Mga2p, a transcription Activator of the OLE1 gene, regulates the stability of the OLE1 mRNA through exosome-mediated mechanisms. J Biol Chem. 2004;279(35):36586–92.
Chellappa R, Kandasamy P, Oh CS, Jiang Y, Vemula M, Martin CE. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of S. cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem. 2001;276(47):43548–56.
Oh C-S, Martin CE. Candida albicans Spt23p controls the expression of the Ole1p Δ9 fatty acid desaturase and regulates unsaturated fatty acid biosynthesis. J Biol Chem. 2006;281(11):7030–9.
Kapoor R, Huang YS. Gamma linolenic acid: an antiinflammatory omega-6 fatty acid. Curr Pharm Biotechnol. 2006;7(6):531–4.
Horrobin D.F. Clinical applications of n-6 essential fatty acids: atopic eczema and inflammation, diabetic neuropathy and retinopathy, breast pain and viral infections. In: Sinclair A, Gibson R editors. Essential fatty acids and eicosanoids. Champaign: American Oil Chemists’ Society; 1992. p. 367–372.
Huang YS, Ziboh VA. Chapter 1. Gamma linolenic acid: an introduction. In: Huang Y-S, Ziboh VA editors. Gamma linolenic acid: recent advances in biotechnology and clinical applications. Champaign: American Oil Chemists’ Society; 2001, p. 1–5.
Li S, Yue Q, Zhou S, Yan J, Zhang X, Ma F. Trehalose contributes to gamma-linolenic acid accumulation in Cunninghamella echinulata based on de novo transcriptomic and lipidomic analyses. Front Microbiol. 2018;9(1296):1296.
Papanikolaou S, Sarantou S, Komaitis M, Aggelis G. Repression of reserve lipid turnover in Cunninghamella echinulata and Mortierella isabellina cultivated in multiple-limited media. J Appl Microbiol. 2004;97(4):867–75.
Jin G, Zhang Y, Shen H, Yang X, Xie H, Zhao ZK. Fatty acid ethyl esters production in aqueous phase by the oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. 2013;150:266–70.
Qiao K, Wasylenko TM, Zhou K, Xu P, Stephanopoulos G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35:173.
Yu T, Zhou YJ, Huang M, Liu Q, Pereira R, David F, Nielsen J. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174(6):1549-1558.e1514.
Shi S, Zhao H. Metabolic engineering of oleaginous yeasts for production of fuels and chemicals. Front Microbiol. 2017;8:2185.
Kim HM, Chae TU, Choi SY, Kim WJ, Lee SY. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nat Chem Biol. 2019;15(7):721–9.
Ledesma-Amaro R, Dulermo R, Niehus X, Nicaud JM. Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metab Eng. 2016;38:38–46.
Wen Z, Zhang S, Odoh CK, Jin M, Zhao ZK. Rhodosporidium toruloides—a potential red yeast chassis for lipids and beyond. FEMS Yeast Res. 2020. https://doi.org/10.1093/femsyr/foaa038.
Tiukova IA, Prigent S, Nielsen J, Sandgren M, Kerkhoven EJ. Genome-scale model of Rhodotorula toruloides metabolism. Biotechnol Bioeng. 2019;116(12):3396–408.
Coradetti ST, Pinel D, Geiselman GM, Ito M, Mondo SJ, Reilly MC, Cheng YF, Bauer S, Grigoriev IV, Gladden JM, et al. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. Elife. 2018;7:e32110.
Park YK, Nicaud JM, Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36(3):304–17.
Liu Y, Koh CM, Sun L, Hlaing MM, Du M, Peng N, Ji L. Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol. 2013;97(2):719–29.
Liu Y, Yap SA, Koh CM, Ji L. Developing a set of strong intronic promoters for robust metabolic engineering in oleaginous Rhodotorula (Rhodosporidium) yeast species. Microb Cell Fact. 2016;15(1):200.
Schultz JC, Cao M, Zhao H. Development of a CRISPR/Cas9 system for high efficiency multiplexed gene deletion in Rhodosporidium toruloides. Biotechnol Bioeng. 2019;116(8):2103–9.
Zhang S, Ito M, Skerker JM, Arkin AP, Rao CV. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl Microbiol Biotechnol. 2016. https://doi.org/10.1007/s00253-016-7815-y.
Cui J, He S, Ji X, Lin L, Wei Y, Zhang Q. Identification and characterization of a novel bifunctional Delta(12)/Delta(15)-fatty acid desaturase gene from Rhodosporidium kratochvilovae. Biotechnol Lett. 2016;38(7):1155–64.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222-230.
Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:348–52.
Pereira SL, Leonard AE, Mukerji P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids. 2003;68(2):97–106.
Coelho MA, Rosa A, Rodrigues N, Fonseca Á, Gonçalves P. Identification of mating type genes in the bipolar basidiomycetous yeast Rhodosporidium toruloides: first insight into the MAT locus structure of the Sporidiobolales. Eukaryot Cell. 2008;7(6):1053–61.
Hu J, Ji L. Draft genome sequences of Rhodosporidium toruloides strains ATCC 10788 and ATCC 10657 with compatible mating types. Genome Announc. 2016;4(2):e00098-e116.
Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol. 2002;51:1–51.
McDonough VM, Stukey JE, Martin CE. Specificity of unsaturated fatty acid-regulated expression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1992;267(9):5931–6.
Yu AQ, Shi TL, Zhang B, Xing LJ, Li MC. Transcriptional regulation of desaturase genes in Pichia pastoris GS115. Lipids. 2012;47(11):1099–108.
Meesters PA, Eggink G. Isolation and characterization of a delta-9 fatty acid desaturase gene from the oleaginous yeast Cryptococcus curvatus CBS 570. Yeast. 1996;12(8):723–30.
Lu SF, Tolstorukov II, Anamnart S, Kaneko Y, Harashima S. Cloning, sequencing, and functional analysis of H-OLE1 gene encoding delta9-fatty acid desaturase in Hansenula polymorpha. Appl Microbiol Biotechnol. 2000;54(4):499–509.
Kajiwara S. Molecular cloning and characterization of the Δ9 fatty acid desaturase gene and its promoter region from Saccharomyces kluyveri. FEMS Yeast Res. 2002;2(3):333–9.
Watanabe K, Oura T, Sakai H, Kajiwara S. Yeast Delta 12 fatty acid desaturase: gene cloning, expression, and function. Biosci Biotechnol Biochem. 2004;68(3):721–7.
Oura T, Kajiwara S. Saccharomyces kluyveri FAD3 encodes an omega3 fatty acid desaturase. Microbiology. 2004;150(Pt 6):1983–90.
Sangwallek J, Kaneko Y, Tsukamoto T, Marui M, Sugiyama M, Ono H, Bamba T, Fukusaki E, Harashima S. Cloning and functional analysis of HpFAD2 and HpFAD3 genes encoding Delta12- and Delta15-fatty acid desaturases in Hansenula polymorpha. Gene. 2014;533(1):110–8.
Tezaki S, Iwama R, Kobayashi S, Shiwa Y, Yoshikawa H, Ohta A, Horiuchi H, Fukuda R. Delta12-fatty acid desaturase is involved in growth at low temperature in yeast Yarrowia lipolytica. Biochem Biophys Res Commun. 2017;488(1):165–70.
Santomartino R, Riego-Ruiz L, Bianchi MM. Three, two, one yeast fatty acid desaturases: regulation and function. World J Microbiol Biotechnol. 2017;33(5):89.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–8.
Koh CM, Liu Y, Du M, Ji L. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014;14(1):50.
Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264(28):16537–44.
Tsakraklides V, Kamineni A, Consiglio AL, MacEwen K, Friedlander J, Blitzblau HG, Hamilton MA, Crabtree DV, Su A, Afshar J, et al. High-oleate yeast oil without polyunsaturated fatty acids. Biotechnol Biofuels. 2018;11:131.
Martin CE, Oh CS, Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta. 2007;1771(3):271–85.
Nguyen LN, Nosanchuk JD. Lipid droplet formation protects against gluco/lipotoxicity in Candida parapsilosis: an essential role of fatty acid desaturase Ole1. Cell Cycle. 2011;10(18):3159–67.
Wang Y, Zhang S, Potter M, Sun W, Li L, Yang X, Jiao X, Zhao ZK. Overexpression of delta12-fatty acid desaturase in the oleaginous yeast Rhodosporidium toruloides for production of linoleic acid-rich lipids. Appl Biochem Biotechnol. 2016;180(8):1497–507.
Indo Y, Tatemizo A, Abe Y, Suzuki I, Matsumoto K, Hosoi Y, Kinoshita M, Mikami K, Murata N, Iritani A, et al. Functional expression of a humanized gene for an omega-3 fatty acid desaturase from scarlet flax in transfected bovine adipocytes and bovine embryos cloned from the cells. Biochim Biophys Acta. 2009;1791(3):183–90.
Murakami A, Nagao K, Juni N, Hara Y, Umeda M. An N-terminal di-proline motif is essential for fatty acid–dependent degradation of Δ9-desaturase in Drosophila. J Biol Chem. 2017;292(49):19976–86.
Damude HG, Zhang H, Farrall L, Ripp KG, Tomb JF, Hollerbach D, Yadav NS. Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants. Proc Natl Acad Sci U S A. 2006;103(25):9446–51.
Sakamoto T, Murata N. Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr Opin Microbiol. 2002;5(2):208–10.
Lim JH, Gerhart-Hines Z, Dominy JE, Lee Y, Kim S, Tabata M, Xiang YK, Puigserver P. Oleic acid stimulates complete oxidation of fatty acids through protein kinase A-dependent activation of SIRT1-PGC1alpha complex. J Biol Chem. 2013;288(10):7117–26.
Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 2001;498(1):98–103.
Mendanha SA, Moura SS, Anjos JL, Valadares MC, Alonso A. Toxicity of terpenes on fibroblast cells compared to their hemolytic potential and increase in erythrocyte membrane fluidity. Toxicol In Vitro. 2013;27(1):323–9.
Garbarino J, Sturley SL. Saturated with fat: new perspectives on lipotoxicity. Curr Opin Clin Nutr Metab Care. 2009;12(2):110–6.
Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H, et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun. 2012;3:1112.
Cosgrove JP, Hayden JG, Robinson PL. Process for making high-purity oleic acid. 1993, U.S. Patent No. 5,194,640.
Liu Q, Singh S, Green A. High-oleic and high-stearic cottonseed oils: nutritionally improved cooking oils developed using gene silencing. J Am Coll Nutr. 2002;21(3 Suppl):205S-211S.
Huang YS, Chaudhary S, Thurmond JM, Bobik EG Jr, Yuan L, Chan GM, Kirchner SJ, Mukerji P, Knutzon DS. Cloning of delta12- and delta6-desaturases from Mortierella alpina and recombinant production of gamma-linolenic acid in Saccharomyces cerevisiae. Lipids. 1999;34(7):649–59.
Sun M-L, Madzak C, Liu H-H, Song P, Ren L-J, Huang H, Ji X-J. Engineering Yarrowia lipolytica for efficient γ-linolenic acid production. Biochem Eng J. 2017;117:172–80.
Paul D, Magbanua Z, Arick M 2nd, French T, Bridges SM, Burgess SC, Lawrence ML. Genome sequence of the oleaginous yeast Rhodotorula glutinis ATCC 204091. Genome Announc. 2014. https://doi.org/10.1128/genomeA.00046-14.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Liu Y, Koh CM, Ngoh ST, Ji L. Engineering an efficient and tight D-amino acid-inducible gene expression system in Rhodosporidium/Rhodotorula species. Microb Cell Fact. 2015;14(1):170.
Liu Y, Koh CM, Sun L, Ji L. Tartronate semialdehyde reductase defines a novel rate-limiting step in assimilation and bioconversion of glycerol in Ustilago maydis. PLoS ONE. 2011;6(1):e16438.
Liu Y, Koh CMJ, Yap SA, Du M, Hlaing MM, Ji L. Identification of novel genes in the carotenogenic and oleaginous yeast Rhodotorula toruloides through genome-wide insertional mutagenesis. BMC Microbiol. 2018;18(1):14.
Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y). 1991;9(10):963–7.
Araki K, Okada Y, Araki M, Yamamura K. Comparative analysis of right element mutant lox sites on recombination efficiency in embryonic stem cells. BMC Biotechnol. 2010;10:29.
Wickens M, Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3’ end formation. Science. 1984;226(4678):1045–51.
Acknowledgements
We thank Mr Jian Feng for the isolation and taxonomical characterization of R. toruloides C3 strain.
Funding
This work was financially supported by Singapore National Research Foundation CRP-8-2011-02, Singapore Economic Development Board, Temasek Trust and Temasek Foundation Innovate.
Author information
Authors and Affiliations
Contributions
LJ and YL conceived and designed the experiments. YL and CMJK performed sequence analysis, gene deletion, real-time PCR, Southern blotting, cell cultures, metabolite analysis and physiological studies. SAY contributed to plasmid construction, yeast transformation and fatty acid profiling. CL generated the ole1∆ mutant. YL, CMJK, SAY and CL interpreted the data. YL drafted and LJ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. Temasek Life Sciences Laboratory has interests in developing Rhodotorula toruloides as an industrial biotechnology platform.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Additional tables and figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, Y., Koh, C.M.J., Yap, S.A. et al. Understanding and exploiting the fatty acid desaturation system in Rhodotorula toruloides. Biotechnol Biofuels 14, 73 (2021). https://doi.org/10.1186/s13068-021-01924-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-021-01924-y