Abstract
Background
An economically viable production of biofuels and biochemicals from lignocellulose requires microorganisms that can readily convert both the cellulosic and hemicellulosic fractions into product. The yeast Candida intermedia displays a high capacity for uptake and conversion of several lignocellulosic sugars including the abundant pentose d-xylose, an underutilized carbon source since most industrially relevant microorganisms cannot naturally ferment it. Thus, C. intermedia constitutes an important source of knowledge and genetic information that could be transferred to industrial microorganisms such as Saccharomyces cerevisiae to improve their capacity to ferment lignocellulose-derived xylose.
Results
To understand the genetic determinants that underlie the metabolic properties of C. intermedia, we sequenced the genomes of both the in-house-isolated strain CBS 141442 and the reference strain PYCC 4715. De novo genome assembly and subsequent analysis revealed C. intermedia to be a haploid species belonging to the CTG clade of ascomycetous yeasts. The two strains have highly similar genome sizes and number of protein-encoding genes, but they differ on the chromosomal level due to numerous translocations of large and small genomic segments. The transcriptional profiles for CBS 141442 grown in medium with either high or low concentrations of glucose and xylose were determined through RNA-sequencing analysis, revealing distinct clusters of co-regulated genes in response to different specific growth rates, carbon sources and osmotic stress. Analysis of the genomic and transcriptomic data also identified multiple xylose reductases, one of which displayed dual NADH/NADPH co-factor specificity that likely plays an important role for co-factor recycling during xylose fermentation.
Conclusions
In the present study, we performed the first genomic and transcriptomic analysis of C. intermedia and identified several novel genes for conversion of xylose. Together the results provide insights into the mechanisms underlying saccharide utilization in C. intermedia and reveal potential target genes to aid in xylose fermentation in S. cerevisiae.
Similar content being viewed by others
Background
In a sustainable society that relies on a circular bio-based economy, lignocellulosic biomasses such as energy crops, agricultural and forestry residues will be important starting materials for the production of fuels, chemicals and materials. Lignocellulosic biomass that consists of 35–50% cellulose, 20–35% hemicelluloses and 15–20% lignin must be pre-treated and hydrolysed to obtain the monomeric sugars that can be converted into desired product(s) through microbial fermentation (review in [1]). The hemicellulosic pentoses d-xylose and l-arabinose are currently underutilized resources, as many of the industrially relevant microorganisms (e.g. Saccharomyces cerevisiae) lack the ability to ferment these sugars. For economically viable fermentation-based production processes, discovery and development of microorganisms that can readily ferment all lignocellulosic sugars is of foremost importance [2].
Xylose fermentation has attracted a lot of research attention, as xylose is the second, after glucose, most abundant monomeric sugar in lignocellulosic biomass [2, 3]. A number of native xylose-fermenting yeasts (e.g. Scheffersomyces stipitis, Spathaspora passalidarum, Sugiyamaella lignohabitans, and Candida tenuis) have been identified and characterized to various degrees in terms of metabolism, physiology, genomics and proteomics. Their usefulness for lignocellulosic conversion processes has been explored, but as cell factories they are limited by their (i) dependence on micro-aerobic conditions for fermentative metabolism, (ii) low tolerance to lignocellulosic inhibitors and (iii) poor overall fermentation titers [4, 5]. As an alternative, xylose-fermenting yeasts can serve as a source of knowledge and genes for xylose uptake and assimilation which may be transferrable to other microorganisms including S. cerevisiae that are better suited as industrial workhorses.
Numerous S. cerevisiae strains capable of xylose fermentation have been engineered through heterologous expression of either the xylose oxidoreductive pathway (xylose reductases (XR) and xylitol dehydrogenases (XDH)) donated from various xylose-utilizing yeasts, or the xylose isomerase (XI) pathway of either bacterial or fungal origin [3, 6]. Despite the many attempts to optimize the xylose metabolic pathway fluxes by Adaptive Laboratory Evolution (ALE) and metabolic engineering [7,8,9,10] and as reviewed in [11], several physiological issues remain that prevent xylose from being fermented as efficiently as glucose in this species. The primary reason for the low xylose-fermenting capacity of S. cerevisiae is that it lacks an efficient and specific xylose transport system, which hampers the metabolite’s uptake and the cell’s subsequent metabolic flux. Instead, xylose enters the cell through hexose transporters (in particular Hxt1, Hxt4, and Hxt7), which all have 100-fold greater affinities for glucose over xylose [12]. Moreover, transporter discrimination in combination with glucose repression mechanisms make S. cerevisiae ferment glucose prior to xylose in mixed sugar conditions [13]. This is problematic in fermentations of lignocellulose hydrolysates containing inhibitory compounds, as cells are more sensitive to these substances during xylose-conversion than during glucose-conversion, which leads to stalled fermentations in the xylose phase [14]. Simultaneous saccharification and co-fermentation (SSCF) setups where glucose/xylose co-fermentation is key for an efficient process are also severely limited by suboptimal utilization of xylose in recombinant strains of S. cerevisiae [15]. Lastly, S. cerevisiae strains relying on the co-factor-dependent XR/XDH system often stall during xylose fermentation as incomplete co-factor recycling leads to accumulation of xylitol and ultimately results in a lower overall product yield [16]. The incomplete co-factor recycling results from the majority of XRs having a preference for NADPH over NADH while XDHs use NAD+, which ultimately leads to insufficient amounts of NAD+ being regenerated [11, 17].
In a previous ALE experiment using repetitive batch cultures with increasing concentrations of the liquid fraction of a lignocellulosic hydrolysate, we sought to improve the xylose-fermentative capacity of an industrial strain of S. cerevisiae equipped with the XR/XDH genes from S. stipitis. To our surprise, we learned that the cell population had been contaminated with a xylose-utilizing yeast species that managed to compete with S. cerevisiae in the xylose-rich, highly inhibiting environment that characterizes a typical lignocellulosic hydrolysate. Several clones were isolated throughout the experiment, and the contaminants were identified to the species level by PCR amplification and sequencing of the internal transcribed spacer (ITS) regions and the D1–D2 region of the large-subunit RNA gene [18, 19]. The sequences were 98–100% (ITS) and 99–100% (D1–D2) identical with a large number (> 20) of Candida intermedia strains. As strains with higher than 99% identity in the D2 domain are considered the same or sister species [19], we concluded that the isolates were of C. intermedia origin.
C. intermedia is an interesting species for industrial conversion of lignocellulosic biomass, as it possesses many of the traits that xylose-fermenting S. cerevisiae strains lack. Its specific growth rate on xylose outcompetes most other yeast species [20], and two different C. intermedia xylose-transporters have previously been identified and characterized; the glucose/xylose facilitator 1 Gxf1, which facilitates high-capacity, low-affinity diffusion of glucose and xylose, and the glucose/xylose symporter 1 Gxs1 which is a high-affinity glucose/xylose-proton symporter [20,21,22,23,24,25]. C. intermedia also expresses an XR with preference for both NADH and NAPDH [26], which can alleviate co-factor imbalance during xylose fermentation. Moreover, we have shown in a previous study that a clone isolated from the end population of the ALE experiment, CBS 141442, is capable of glucose and xylose co-fermentation in wheat straw hydrolysates [14].
In the present study, the aim was to elucidate the genetic features of C. intermedia, which form the basis for its industrially interesting traits. We report here on the complete genome sequence of C. intermedia, its transcriptional response to xylose and glucose and the identification of several novel genes with potential to contribute to xylose conversion in lignocellulose-based production of biofuels and biochemicals.
Results and discussion
De novo genome sequencing of C. intermedia
To enable analysis of the in-house isolated C. intermedia strain CBS 141442 on a genomic and molecular level, we set out to determine its genome sequence. For comparative reasons we also sequenced the C. intermedia strain PYCC 4715, a strain previously characterized in terms of xylose uptake and assimilation [20]. The genome of CBS 141442 was sequenced to 61X coverage and assembled into nine chromosomal contigs and a mitochondrial contig. Essentially, each chromosomal contig resembles one chromosome, except the largest chromosome that consists of three contigs with the middle one being flanked by ribosomal repeats, resulting in seven chromosomes in total. The genome of PYCC 4715 was sequenced to 32× coverage, and the 27 contigs were reference-guided with the CBS 141442 chromosomes as a backbone to create full-length chromosomes. The genome sizes were summed up to 13.2 Mbp (CBS 141442) and 13.1 Mbp (PYCC 41715), both with a mean G+C content of 43.5%. A total of 5936 and 6073 protein coding genes were identified for CBS 141442 and PYCC 4715, respectively, and genome completeness according to CEGMA was above 95% for both strains (Table 1) [27]. Overall, we can conclude that the two complete C. intermedia genomes are highly similar to each other in terms of size and protein coding genes, and the numbers correlate well with other sequenced ascomycetous yeast species [28].
Genetic distance and genome synteny
To determine the genetic distance between the two strains on a genome level, the average nucleotide identity (ANI) was calculated using both BLAST and MUMmer as search engines [29]. The sequence identity between the two assemblies was 97% for both runs, and cut-off scores of > 95% confirm that they belong to the same species [30]. We also determined the chromosomal map conservation between the strains. For visualization of the results, the two sets of chromosomes were placed in a circle with the ribbon tracks displaying synteny blocks for the whole genome alignment (Fig. 1a). Although the overall order of genes is highly conserved, every chromosome displays 1 or 2 translocations of large genome blocks, resulting in that all CBS 141442 chromosomes are composed of a mosaic pattern with 2–3 conserved syntenic segments of individual PYCC 4715 chromosomes, and vice versa. In addition, numerous smaller syntenic blocks are translocated throughout the genomes in a non-reciprocal manner. As the chromosomes are numbered in order of size with the largest labelled 1, the large rearrangements result in that the CBS 141442’s chromosome 4, 5 and 6 are labelled as chromosome 5, 6 and 4 in PYCC 4715. Reciprocal read mapping was used as an alternative approach to assess the translocations, which confirmed the results of the full genome alignment (Fig. 1b, c). Finally, two randomly chosen chromosomal fusion points were confirmed by PCR (indicated by * in Fig. 1b, c). These results collectively indicate that the translocations observed in the different strains are real and not due to errors in genome assembly.
Genome differences due to numerous chromosomal translocations. a Circos plot of the syntenic block translocations between chromosomes in the two C. intermedia strains. The chromosomes are arranged in a circle, and each aligned segment was drawn as a ribbon from the chromosomes of CBS 141442 to PYCC 4715, where each CBS 141442 chromosome is marked with a unique colour to clearly visualize the genomic rearrangements. b, c Reciprocal read mapping, where b displays the CBS 141442 genome reads coloured according to which chromosome they belong to in the CBS 141442 assembly, and mapped to the PYCC 4715 chromosomes, and c marked reads from PYCC 4715 mapped to the CBS 141442 assembly. *Breakpoints confirmed with PCR
For most non-conventional yeast species, knowledge of sequence identity and genome synteny of different strains is sparse. Typically, only one or a very small number of strains per species have been sequenced, and most isolated strains are classified using ITS/D1–D2 sequences alone. For model organisms such as S. cerevisiae where multiple strains have been sequenced, the relevance of genome diversity in natural and domestic populations for adaptation to new and changing environments has been demonstrated [28]. We can only speculate about the reason for the chromosomal translocations observed in the two C. intermedia genomes. The translocations might shed light on a common phenomenon among C. intermedia and ascomycetous yeast as a strategy of survival and/or the continuous process of speciation [31]. Alternatively, these translocations could be a result of CBS 141442 being isolated from a highly challenging condition of repetitive batch cultures with toxic wheat straw hydrolysates [14], as such evolutionary engineering schemes are prone to generate genome alternations [32].
Phylogenetic relationship of C. intermedia with other yeasts
To determine the evolutionary relationship of the two C. intermedia strains with other yeasts of industrial and clinical relevance, we selected 15 genome sequenced ascomycetous yeast strains with industrial or clinical importance and performed a phylogenetic analysis. The resulting tree shows that the two C. intermedia strains cluster together with species within the CTG clade (Fig. 2a), in good correlation with previous work based on ITS/D1–D2 sequence alignments alone [33]. The webserver Bagheera [34] was used to confirm CUG codon usage and the haploid identity of C. intermedia CBS 141442 was determined using flow cytometry analysis (Fig. 2b).
Phylogenetic placement and ploidy determination of C. intermedia. a Phylogenetic tree of C. intermedia CBS 141442 and PYCC 4715 and 15 other ascomycetous yeasts. The molecular phylogenetic analysis was based on ten protein-encoding genes using the maximum likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The numbers at each branch indicate bootstrap values, and the CTG branch is highlighted. b Ploidy determination of C. intermedia CBS 141442 by flow cytometry. Three S. cerevisiae strains were used as reference; haploid BY4741, diploid CEN.PK and tetraploid 122 MDS
Many members of the CTG clade are commensal with humans or other host organisms, and can emerge as opportunistic pathogens (Candida albicans, Candida tropicalis [35, 36]). Other species, such as S. stipitis and S. passalidarum, have adapted to living in the guts of wood-consuming beetles and are capable of fermenting xylose, yet others are associated with marine environments (Metschnikowia bicuspidata and Debaryomyces hansenii) or plants and soil (Clavispora lusitaniae) [37,38,39,40,41,42]. The biological niche of C. intermedia has not been thoroughly investigated, but strains deposited in different databases have been isolated from a range of environmental sources including fruits and flowers, dairy products and humans (listed in Additional file 1). From the information collected, we can conclude that C. intermedia can be found in diverse habitats around the world, including also a number of xylose-rich substrates such as decaying forest residues and biomass hydrolysates, where C. intermedia’s high capacity of xylose assimilation could be an advantageous trait.
Transcriptional profiles of C. intermedia grown in xylose and glucose containing media
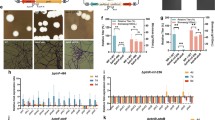
To identify the genes responsible for utilization of xylose in C. intermedia, we performed a transcriptome analysis using RNA-sequencing (RNA-seq) technology. The CBS 141442 strain was cultivated in bioreactors in medium containing xylose or glucose (as reference), both at a concentration of 20 g L−1 to promote rapid cell growth and at 200 g L−1 to assess the transcriptional reponse to high sugar concentrations encountered during industrial fermentations. The strain displayed similar specific growth rates in glucose and xylose at 20 g L−1 (0.37 and 0.34 h−1, respectively) (Fig. 3a, c), which correlates well with what has been shown previously for C. intermedia PYCC 4715 and FL023 [20, 43]. The specific growth rate was only slightly affected in 200 g L−1 of glucose (0.33 h−1) (Fig. 3b), confirming our previous experiments showing that C. intermedia is a highly osmotolerant yeast (unpublished results). However, the yeast was clearly challenged in 200 g L−1 of xylose as judged by the drop in specific growth to 0.17 h−1 (Fig. 3d), suggesting that high concentrations of xylose exert an osmotic stress on the cells. This is in line with previous observations for C. intermedia, as well as for other yeasts, where cells grown in xylose are comparatively more sensitive to stress factors in the environment than glucose-grown cells, possibly due to lower intracellular ATP levels [14, 44,45,46,47,48,49]. Moreover, C. intermedia switched from a respiratory to fermentative metabolism when oxygen became limited in the cultures (due to fixed stirring and sparging settings in the experimental design), evidenced by the formation of ethanol and the by-products glycerol and xylitol (Fig. 3a–d). This is indicative of Crabtree-negative yeasts, where the onset of fermentation is regulated by a decrease in oxygen levels [50].
C. intermedia growth, sugar consumption and transcriptional profiles in media with different sugar compositions. a–d Growth (OD600) and extracellular sugar and metabolite concentrations over time. µmax represents the maximum growth rate (h−1) of the cultures. Vertical dashed lines represent the sampling time points for RNA-seq. e MDS plot of gene expression of 12 RNA-seq libraries. Samples cluster by carbon source and concentration. f Heatmap plot of differently expressed genes using the hierarchical clustering method, displaying seven main clusters. Each row of the heatmap represents the z-score transformed log 2 values of one differentially expressed gene across all samples (blue, low expression; red, high expression). Colours above the heatmap indicate carbon source and concentration (G20—glucose 20 g L−1; G200—glucose 200 g L−1; X20—xylose 20 g L−1; X20—xylose 200 g L−1). The dendrogram displays how the samples cluster based on their differential expression patterns
The transition from respiratory to fermentative growth in Crabtree-negative yeasts is characterized by a significant change in transcription [51]. Thus, to enable comparison of the overall transcriptional profiles for the different conditions in a straightforward way, samples for RNA-seq were collected in mid-exponential, respiratory phase at maximum growth rate when no metabolites were accumulating (dotted lines Fig. 3a–d), and purified mRNA was sequenced as outlined in the materials and methods section. The resulting RNA-seq datasets were used in an unsupervised hierarchical clustering of the 5761 expressed genes across the four growth conditions. The multi-dimensional scaling (MDS) plot shows that the triplicates for each condition cluster together, and that the gene expression profiles from the cells grown in 20 and 200 g L−1 of glucose and 20 g L−1 of xylose are more similar to each other than to cells grown in 200 g L−1 of xylose (Fig. 3e). This difference is consistent with the observed slower specific growth rate in 200 g L−1 of xylose as discussed above. The trend is also demonstrated very clearly in the heat map, which revealed 7 distinct, condition-dependent clusters of differentially expressed genes and enriched gene ontology (GO) annotations (Fig. 3f, Additional file 2). In clusters 1, 2 and 6, the three fast-growing cultures displayed the opposite expression patterns from the culture with 200 g L−1 of xylose. Cluster 1 is composed of genes with an increased expression in cells with higher growth rates, and the annotations revealed an enrichment of genes associated with growth, including genes for organization of the cytoskeleton and the plasma membrane as well as RNA processing and translation. On the other hand, clusters 2 and 6 comprise genes that had higher expression in cells grown in 200 g L−1 of xylose and include a significant number of genes involved in regulation of transcription, protein phosphorylation and response to stress. Cluster 3 and 4 are markedly upregulated in 200 g L−1 of glucose and are composed of genes for translation and rRNA processing. Cluster 7 follows a carbon source-dependent transcription pattern and comprise genes upregulated in xylose conditions and downregulated in glucose-grown cells. Here, we found an enrichment of genes associated with carbohydrate metabolism and oxidation–reduction reactions, similar to what was previously shown for S. stipitis when grown in xylose [52]. To summarize, the major differences in transcriptional output seem to be correlated with the specific growth rate, osmotic stress and carbon source provided.
Metabolic map of xylose and glucose utilization pathways
The genomic and transcriptomic data collected allowed identification of genes encoding putative transporters and enzymes for xylose and glucose metabolism in C. intermedia. Accordingly, we scanned the CBS 141442 genome for genes encoding Major Facilitator Superfamily (MFS) sugar transporters and the RNA-seq dataset for the corresponding transcriptional profiles [53], and identified several orthologs to xylose transporters from other yeasts (Additional file 3) [21, 25, 54, 55]. Of particular interest were two novel genes encoding putative homologs to the previously characterized glucose/xylose transporters Gxf1 and Gxs1 (named Gxf1_2 and Gxs1_2, respectively). The four genes displayed different expression patterns which may indicate diverse physiological roles: whereas relatively high levels of GXF1 and GXF1_2 transcripts were observed in all conditions tested, GXS1 expression appeared induced in xylose conditions and GXS1_2 displayed a stable but low expression across all conditions assessed (Fig. 4). Further experimental characterization is needed to determine the transporters’ substrate specificities and their contribution to the total sugar uptake capacity of C. intermedia.
Expression profiles of genes encoding putative transporters and enzymes in the glucose and xylose metabolic pathways. Arrows represent enzyme reactions catalysed by the associated proteins. Normalized expression of enzyme-encoding genes is represented in all carbon sources and concentrations tested using variance-stabilized counts (VST). Enzymatic reactions substrates and products as follow: XYL—xylose; XOL—xylitol; XUL—xylulose; XUL5P—xylulose 5-phosphate; RL5P—ribulose 5-phosphate; R5P—ribose 5-phosphate; S7P—sedoheptulose 7-phosphate; GA3P—glyceraldehyde 3-phosphate; E4P—erythrose 4-phosphate; D6PGC—6-phosphogluconate; G6PGL—glucono 1,5-lactone 6-phosphate; G6P—glucose 6-phosphate; GLC—glucose; F6P—fructose 6-phosphate; FDP—fructose 1,6-biphosphate; DHAP—dihydroxyacetone phosphate; 13DPG—1,3-diphosphoglycerate; 3PG—3-phosphoglycerate; 2PG—2-phosphoglycerate; PEP—phosphoenolpyruvate; PYR—pyruvate. Accession numbers of all proteins are available in Additional file 4
To identify the putative genes responsible for xylose and glucose catabolism in C. intermedia, we searched the CBS 141442 genome for genes encoding proteins with high homology to proteins involved in central metabolism in the S. stipitis CBS 6054 strain [56]. As expected, the genes ascribed a role in central carbon metabolism were strongly expressed in all four conditions, indicative of respiratory growth. The expression of genes encoding a putative XR (XYL1, EC 1.1.1.307), XDH (XYL2, EC 1.1.1.9) and xylulokinase (XK, XKS1, EC 2.7.1.17) was higher during growth on xylose than during growth on glucose, pointing towards that these genes are important for xylose utilization under the set conditions. Moreover, we found two additional putative XR genes, XYL1_2 and XYL1_3, which share 62 and 75% identity to XYL1, respectively. XYL1_2 and XYL1_3 are minimally expressed in xylose (as well as in glucose-grown cells) (Fig. 4), suggesting minor roles for these enzymes during xylose growth in aerobic conditions.
Characterization of the C. intermedia xylose reductases
The finding of multiple genes encoding putative XRs in the C. intermedia CBS 141442 genome is in line with the previous report that suggests C. intermedia to have two different enzymes with XR activity, one with preference for both NADH and NADPH and one with strict NADPH-dependence [26]. XRs with specificity for NADH are of particular interest as they can alleviate cofactor imbalance during xylose fermentation. To assess the cofactor preferences and substrate specificities of the three putative XRs identified, the genes were individually expressed in S. cerevisiae and characterized in vitro. The S. stipitis XYL1_K270R gene encoding an XR with dual cofactor specificity was also included as reference.
All of the CiXYL1 gene products had XR activity using NADPH as cofactor and xylose as substrate, which confirmed that the enzymes were expressed and active in the S. cerevisiae host (Table 2). No, or minimal, reductase activity was observed using glucose as substrate indicating a specificity towards xylose (data not shown). When NADH was supplied as a cofactor, no XR activity was detected for CiXR1 and CiXR3, indicating that these enzymes have a high preference for NADPH over NADH. In contrast, both CiXR2 and SsXR_K270R could utilize NADH and NADPH with similar preferences. Taken together, we can conclude that C. intermedia possesses genes for three functional XRs and that CiXR2 is responsible for the previously reported NADH-dependent XR activity in this species [26].
Amino acids contributing to the co-factor preference of xylose reductases
While the majority of characterized yeast XRs exclusively use NADPH as co-factor, the XR from C. tenuis displays dual co-factor specificity although with a 10-fold preference for NADPH over NADH. Solved structures of the apo- and holo-forms of CtXR show the co-factor binding pocket and how the enzyme can accommodate co-factors both with and without 2′-phosphate groups [57]. A conserved, key motif of KSN-X(3)-R found within the pocket aids in defining co-factor specificity, and numerous mutagenesis studies have revealed that changes in this motif can result in a loss of specificity of NADPH over NADH [58,59,60]. A multiple sequence alignment with XR yeast homologs revealed conservation of the KSN-X(3)-R motif in CiXR1 and CiXR3, consistent with their high preference for NADPH (Fig. 5a, Table 2).
Comparison of characterized yeast XRs. a Excerpt of a multiple sequence alignment of yeast XRs showing key residues in the cofactor binding site. The KSN-X3-R protein motif implicated in NADPH specificity is highlighted, and mutations introduced into the genes are noted with arrows beside the species name. Identical residues are shown as white text on a red background and similar residues in red text on a white background. The kinetic parameters of each XR are shown where available alongside the parameters determined in this work. The species from which the XR originate are abbreviated as follows: Ct, C. tenuis; Mg, M. guilliermondii; Sp, S. passalidarum; Ss, S. stipitis; Ci, C. intermedia. b The cofactor binding site of C. tenuis XR in complex with NADP+ (PDB ID: 1k8c) and c with NAD+ (PDB ID: 1mi3). The hydrogen bonds stabilizing the NADP+ in the C. tenuis XR structure is shown with black dashes. d A homology model of the C. intermedia XR2 created with PHYRE [82] using the XR from C. tenuis as a template (PDB ID: 1k8c) to visualize the differences in the active site which may contribute to the lack NADP+ specificity
In the structure where CtXR is bound to NADP+, the residues of the KSN-X(3)-R motif assist in co-factor binding by making hydrogen bonds with the 2′-phosphate moiety with a notable hydrogen bond contributed by the main chain amide of Asn276 (Fig. 5b). When CtXR is instead bound to NAD+, which lacks the 2′-phosphate moiety of NADP+, only minimal rearrangements are observed in the side chains of the KSN-X(3)-R motif, suggesting an active site fine-tuned for NADP+ (Fig. 5c).
In contrast to CtXR, CiXR2 showed similar affinity for both co-factors (Km of 52 µM for NADH and 47 µM for NADPH) (Fig. 5a). The residues in the binding-pocket motif in CiXR2 are KGD-X(3)-H (rather than the conserved KSN-X(3)-R), and these differences in the active site motif likely contribute to the enzyme’s lack of co-factor preference. A homology model of CiXR2, using CtXR as template, suggests that the N276D and S275G substitutions are key contributors to the differences in co-factor affinities observed for the two enzymes (Fig. 5d). The introduction of the acidic aspartate residue likely leads to electrostatic repulsion with the NADP+ 2′-phosphate, while the introduction of a neighbouring glycine would give an increased degree of flexibility for the region. This could lead to an alternate position of the Asp276 residue and a loss of the hydrogen bond made by the backbone amide with the 2′-phosphate.
The three substitutions in the CiXR2 motif have all been observed previously in XR variants from other species, where the cofactor specificity has been altered in favour of NADH (Fig. 5a, [58,59,60]). Improving the NADH/NADPH ratio can be achieved either by increasing the affinity for NADH or by decreasing the affinity for NADPH. Here, enzyme variants with improved affinity for NADH are more useful than those where the affinity for NAPDH is lost, as poor co-factor affinity negatively affects the enzyme’s xylose conversion-activity. CiXR2 belongs to the former category, as although it lacks specificity for NADPH, the xylose conversion-activity (Vmax) is maintained at levels comparable to other XRs [58,59,60]. This holds true also for the S. passalidarum XR2 (SpXR2), which just like CiXR2 contains an aspartate in the position equivalent to CtXR Asn276 and displays a NADH/NADPH ratio of 1.21 (Fig. 5a). When expressed in S. cerevisiae, the SpXR2 enzyme improve ethanol yields and productivity from xylose under low oxygen conditions compared to cells expressing SsXR [60]. Moreover, XR1 from M. guilliermondii also contains Gly275 and the Asp276, and likely displays dual co-factor specificity with high affinity for NADH. Thus, both CiXR2 and MgXR1 are new, interesting candidates to express in S. cerevisiae for elucidating the relation between the catalytic characteristics of the enzymes and the efficiency of xylose fermentation of lignocellulosic biomass.
Most yeast species capable of utilizing xylose as a carbon source present a single gene coding for XR, but some species including S. passalidarum and M. guilliermondii possess two XR genes. To the best of our knowledge, C. intermedia is the first yeast identified that has three genes encoding functional XRs, and their different expression patterns and co-factor specificities suggest that they have different physiological roles. With the genome sequenced, the possibility to develop a genome editing toolbox for this yeast arises, which would enable phenotypic characterization of XR deletion and/or overexpression strains as well as targeted genome-editing approaches for developing strains for industrial use. In fact, overexpression of CiXR2 has the potential to improve the xylose-fermentative capacity of C. intermedia by lowering the xylitol accumulation seen under oxygen-limited conditions [27].
Conclusions
The non-conventional yeast C. intermedia can metabolize several important lignocellulosic sugars, including the abundant monosaccharide xylose that S. cerevisiae and many other industrial microorganisms cannot naturally ferment. The aim of the present work was to understand the genetics underlying C. intermedia’s capacity to ferment lignocellulosic-derived xylose and to generate knowledge that can be used to improve lignocellulose-based fermentation processes. We describe for the first time the genome analysis of the C. intermedia strains CBS 141442 and PYCC 4715, which revealed two strains with high overall genomic similarity yet noticeably different due to multiple chromosomal translocations. The phylogenetic analysis firmly established the species within the CTG clade of ascomycetes yeasts, adding to the list of xylose-fermenting yeasts with importance in biotechnology and lignocellulosic-based production of biofuels and biochemicals. Moreover, the transcriptomic analysis of strain CBS 141442 grown in high and low concentrations of glucose and xylose provides a first insight into the xylose and glucose metabolism of C. intermedia, and several novel genes for xylose conversion were identified. In particular, a gene encoding an XR with equal affinity for NADH and NADPH was found, and the subsequent characterization put forward this as a valuable candidate gene to express in S. cerevisiae to relieve its co-factor imbalance during xylose fermentation. Collectively, the results presented expand our knowledge on the metabolism of different carbon sources in yeast and enable new comparative genomic studies.
Materials and methods
Yeast strains and media
The C. intermedia strain CBS 141442 was isolated in-house from a co-culture with S. cerevisiae [27]. The C. intermedia PYCC 4715 strain, originally isolated from sewage in Oeiras, Portugal, was obtained from the Portuguese Yeast culture collection. The S. cerevisiae strain BY4741 Δgre3 was used for heterologous expression of the xylose reductases, and BY4741 (haploid), CEN.PK 122MDS (diploid) and G26 (tetraploid) were used for ploidy determination.
Cells were cultured in rich medium (YPD) (20 g L−1 yeast extract, 10 g L−1 peptone and 20 g L−1d-glucose), in minimal mineral medium (MM) (7.5 g L−1 (NH4)2SO4, 3.5 g L−1 KH2PO4, 0.75 g L−1 MgSO4·7H2O, 2 mL L−1 trace metal solution, and 1 mL L−1 vitamin solution) [61] or on Yeast Nitrogen Base (YNB-Ura) medium plates supplemented with glucose (20 g L−1), (NH4)2SO4 (5 g L−1) and Complete Supplement Mixture without uracil (CSM-Ura) (1.9 g L−1), and agar (20 g L−1).
Species determination using PCR
PCR amplification and sequencing of the Internal Transcribed Spacer (ITS) regions and the D1-D2 region of the large-subunit RNA gene was performed using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′), ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′), respectively [18, 62]. The PCR products were sent for sequencing, and the results were blasted against the NCBI nucleotide database.
DNA sequencing and raw data analysis
DNA isolation, library preparation, single-molecule real-time (SMRT) sequencing (Pacific Biosciences), genome assembly and annotation were performed as previously described [27]. To assess the completeness of each genome assembly, we used the CEGMA pipeline that scans assemblies for 248 highly conserved eukaryotic genes [63].
Bioreactor cultivation and RNA sampling
For bioreactor cultivation, starter cultures were grown overnight in an orbital shaker, 200 rpm, at 30 °C. Cells were harvested and washed with sterile distilled water and the pellets were weighed. Sterile water was added to the pellets to aid resuspension in a 1:1 ratio. The inoculum volume was calculated to achieve 0.25 g L−1 dry weight, considering a dilution factor of 2, a cell density of ≈ 1 g mL−1 and a dry weight corresponding to 25% of the wet weight.
The cells were then inoculated in 400 mL of MM containing glucose (20 or 200 g L−1) or xylose (20 or 200 g L−1) in controlled stirred 1-L bioreactor vessels (DASGIP fermenter system with 8 parallel SR0700ODLS vessels, Eppendorf, Hamburg, Germany) at 30 °C. The medium pH was maintained at 5.5 with 2 M potassium hydroxide. All reactors were equipped with pH and dissolved oxygen probes (Mettler Toledo probes, Mettler Toledo, Greifensee, Switzerland) and off-gas analysers. The CO2 (v/v, %) and O2 (v/v, %) in the off-gas were monitored continuously using BlueSens gas analysers (BlueSens Gas Technology GmbH, Herten, Germany) to calculate the volumetric off-gas rates, namely the CO2 production rate (mmol L−1 h−1) and the O2 consumption rate (mmol L−1 h−1). The aeration rate was fixed at 1 Vessel Volume per Minute, with stirring at 300 rpm.
Growth was followed by measurement of OD at 600 nm in a Spectrophotometer (Genesys 20, ThermoFisher—Waltham, MA, USA). Cell samples (10 mL) were collected when the culture dissolved oxygen was 35–40% (v/v). The cells were diluted in 35 mL of very cold distilled water in a falcon tube and place it in an ice bath. Cells were then harvested by centrifugation at 5000 rpm for 5 min, at 2 °C. The pellets were resuspended in ice-cold distilled water and centrifuged again at 10,000 rpm for 5 min, at 0 °C. The pellets were frozen in liquid nitrogen and then stored at − 80 °C until extraction.
RNA sequencing and raw data analysis
The frozen pellets were thawed in 500 μL of TRIzol (Ambion—Foster City, CA, USA) and thoroughly resuspended, and then transferred to 2-mL tubes with Lysing Matrix C (MP Biomedical—Santa Ana, CA, USA). The tubes were incubated in a FastPrep FP120 (Savant—Carlsbad, CA, USA) for five cycles, at intensity 5.5 for 30 s. Tubes were cooled on ice for a minute between cycles. Another 500 μL of TRIzol were added to each tube and vortexed thoroughly. The tubes were incubated at room temperature for 5 min and then centrifuged 10 min at 12,000 rpm, 4 °C. The supernatants were collected, chloroform was added (200 μL of chloroform per mL of supernatant) and vortexed vigorously for 30 s. The samples were incubated at room temperature for 5 min, and then centrifuged 15 min at 12,000 rpm, 4 °C. The top clear aqueous phase was collected and transferred to a new RNase-free tube. An equal amount of absolute ethanol was slowly added, mixing it as needed. Each sample was loaded into a RNeasy column (RNeasy Mini Kit, Qiagen—Hilden, Germany), further steps followed the protocol of the manufacturer. The RNA was treated with DNase in-column twice at room temperature for 30 min. The RNA was eluted with RNase-free water and the samples were stored at − 80 °C until use.
RNA samples were analysed in a TapeStation (Agilent—Santa Clara, CA, USA), and only samples with RNA integrity number above 8 were used for library preparation. Sequencing libraries were prepared from 1 μg total RNA using the TruSeq stranded mRNA library preparation kit (Cat# RS-122-2101/2102, Illumina Inc.—San Diego, CA, USA) including polyA selection. The library preparation was performed according to the manufacturers’ protocol (#15031047). The libraries were then sequenced using the HiSeq 2500 system (Illumina Inc.—San Diego, CA, USA), with paired-end 125 bp read length, and v4 sequencing chemistry. A sequencing library for the phage PhiX was included as 1% spike-in in the sequencing run. The sequencing generated 180 M of read-pairs, yielding a coverage of 5–20 M reads per condition.
The obtained read data were quality controlled using the software FastQC version 0.11.5 [64]. Following this, reads were mapped to the reference genome with the software Star version 2.5.2b [65] after using the annotation information to generate the genome index. Read counts were extracted from the BAM files by counting all reads mapping to exonic regions and merging exon counts to gene level using featurecounts [66]. Gene counts were imported into R and all subsequent analysis were done using R [67]. First the counts were normalized with weighted trimmed mean of M-values using the calcNormFactor function from the package edgeR [68]. Second, the Limma package [69] were used to transform and make data suitable for linear modelling. Finally, significant effects of growth conditions were identified using a multifactor linear model. The estimated p-values were corrected for multiple testing with the Benjamini–Hochberg procedure, and genes were considered significant if the adjusted p-values were lower than 0.05.
Genome synteny and reciprocal read mapping
For genome synteny analysis, the two genomes where aligned using nucmer from the MUMmer package version 3.2 [70], applying the CBS 141442 assembly as reference and PYCC 4715 as query. Each aligned segment was drawn as a link from the reference to the query and each chromosome was plotted relative to its size.
For reciprocal read mapping, the CBS 141442 genome reads were mapped using BLASR v.1.3.1.127046 to the CBS 141442 assembly, with the option-bestn 1 to only report 1 alignment [71]. The reads that mapped with a mapping quality above 1 were marked with the chromosome identity, and these marked reads were then mapped with BLASR to the PYCC 4715 assembly. The procedure was repeated by mapping the marked reads from PYCC 4715 to the CBS 141442 assembly. Here, the raw read counts were used and thus the number of reads is not always correlated with chromosome size (as for the mitochondria where many reads mapped).
Sequence alignments, phylogenetic analysis and structural figures
BLAST search hits of the NCBI’s nucleotide and protein databases were used to supplement the sequence data generated in this study. For species phylogenetic analysis, nucleotide sequences for 10 protein coding genes (ACT1, FBP1, PGK1, GPD1, HXK2, SNF1, HIS3, LEU2, PMA1 and TAL1) from the two C. intermedia strains and 15 other yeast species with S. pombe as outgroup were aligned, trimmed and concatenated. From this dataset, containing 10,652 characters, a maximum likelihood (ML) phylogenetic tree with bootstrap value 1000 was constructed using MEGA X [72, 73].
The multiple sequence alignment of the XR motifs was completed with Clustal Omega [74, 75] and was visualized with ESPRIPT [76]. The structural figures were made in Pymol (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC) using the coordinates for the C. tenuis XR in complex with NADP+ (b, PDB ID: 1k8c) and NAD+ (c, PDB ID: 1mi3). The C. intermedia XR2 homology model was created with PHYRE2 [77] using the XR from C. tenuis as a template (PDB ID: 1k8c).
Heatmap
The raw counts were filtered for lowly expressed genes. Genes with CPM > 3.84 in at least 12% (5/43) of samples were retained. Raw counts were converted to variance-stabilized-counts (VST) using function ‘varianceStabilizingTransformation()’ from R package ‘DESeq2’ [78]. Sample-to-sample euclidean distances were computed on the VST counts. Multi-dimensional scaling (MDS) was run on the distance matrix using R function ‘cmdscale()’. Scatterplots from MDS axes were created in R. VST counts were scaled genewise to z-scores. Z-scores were hierarchically clustered on genes and samples to generate the heatmap. The heatmap was created using R package ‘pheatmap’.
Flow cytometry
The ploidy of C. intermedia CBS 141442 was investigated by flow cytometry of cells stained with Sytox green (Thermo Fischer Scientific). Exponentially growing cells were harvested and fixated in cold 70% ethanol. The cells were washed in 10 mM EDTA, pH 8.0, followed by treatment with RNase A (Thermo Fischer Scientific), 0.1 mg mL−1 at 37 °C for 2 h. Sytox green was added to a final concentration of 1 µM and the cells were analysed with a Guava easyCyte 8HT flow cytometry System (Merck Millipore, Darmstadt, Germany). S. cerevisiae strains of different ploidy were used as references.
HPLC analysis of sugar consumption and metabolite formation
The glucose and xylose consumption and production of metabolites (glycerol, xylitol and ethanol) were analysed using HPLC (LC-4000, Jasco Inc—Easton, MD, USA) with a Rezex ROA H+ (8%) column (Phenomenex—Torrance, CA, USA). The oven temperature was set to 80 °C and the eluent was 5 mM H2SO4 at a flow rate of 0.8 mL min−1. A Jasco RI-4030 refractive index detector (Jasco Inc.—Easton, MD, USA) was used to quantify the sugars and the extracellular metabolites.
Cloning of xylose reductases
The C. intermedia XR genes were codon optimized, with special attention given to the CTG codons, flanked with sequences for the restriction sites for BamHI and XhoI and synthesized at Eurofins Genomics (Ebersberg, Germany). The codon-optimized S. stipitis XR gene XYL1_K270R gene was obtained by PCR amplification from a plasmid developed previously in the group [79], where the forward primer included the restriction for ClaI and the reverse primer included the restriction site for the XhoI enzyme. The C. intermedia and S. stipitis genes were digested by BamHI/XhoI and ClaI/XhoI, respectively, and cloned into the plasmid p426GPD linearized with the corresponding enzymes. All constructs were sequenced and then transformed into the S. cerevisiae strain BY4741 Δgre3, using the modified lithium acetate/PEG/ssDNA methodology [80]. Positive transformants were selected for on YNB-Ura plates and confirmed with colony PCR.
Enzymatic activity assay
Transformants were grown in shake flasks under aerobic conditions in CSM-Ura at 30 °C, 200 rpm. The cells were collected in the mid-exponential growth phase by centrifugation (5 min at 5100 rpm) and washed twice with cold water. The cell pellets were resuspended in 1 mL breaking buffer (0.1 M triethanolamine buffer (pH 7.0); 1 mM phenylmethylsulfonyl fluoride (PMSF); 0.5 mM dithiothreitol; 0.5 mM EDTA) and transferred to 2-mL tubes with Lysing Matrix C (MP Biomedical—Santa Ana, CA, USA). The tubes were incubated in a FastPrep FP120 for five cycles, at a speed of 5.5 for 30 s, with ice incubations for a minute between cycles. The supernatants were collected, and the protein concentrations were determined with Bradford reagent following the supplier instructions, using BSA as standard.
Enzyme activity was determined by following the oxidation of NAD(P)H in the microplate reader (FLUOstar Omega—BMG LabTech, Ortenberg, Germany) operating at 340 nm and 30 °C. Biological duplicates were performed. The activity was measured as reported before [81], with minor changes. The reaction was started by the addition of aldose (final concentration 300 mM) and followed for 5 min. Protein extracts were diluted to a final concentration of 400-600 μg mL−1 of total protein. One unit was defined as the generation of 1 μmol NAD(P) per minute. The specific enzyme activities were given in units (U) per mg protein [58]. Determination of Km for NADH and NADPH was performed measuring the activity in a range of different co-factor concentrations (5–400 μM) while keeping the substrate concentration constant (300 mM).
Availability of data and materials
The genome and protein datasets generated and/or analysed during the current study are available in the in the European Nucleotide Archive (ENA) with the accession numbers LT635756 to LT635763 and LT635764 to LT635771, respectively. RNA-Seq accession number is E-MTAB-6670.
Abbreviations
- ALE:
-
Adaptive laboratory evolution
- ITS:
-
Internal transcribed spacer
- MFS:
-
Major facilitator superfamily
- NAD(P)H:
-
Nicotinamide adenine dinucleotide (phosphate)
- XDH:
-
Xylitol dehydrogenase
- XR:
-
Xylose reductase
References
Mood SH, Golfeshan AH, Tabatabaei M, Jouzani GS, Najafi G, Gholami M, Ardjmand M. Lignocellulosic biomass to bioethanol, a comprehensive review with a focus on pretreatment. Renew Sust Energ Rev. 2013;27:77–93.
Avanthi A, Kumar S, Sherpa KC, Banerjee R. Bioconversion of hemicelluloses of lignocellulosic biomass to ethanol: an attempt to utilize pentose sugars. Biofuels. 2017;8(4):431–44.
Kwak S, Jin YS. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb Cell Fact. 2017;16:82.
Du C, Li YM, Zhao XY, Pei XZ, Yuan WJ, Bai FW, Jiang Y. The production of ethanol from lignocellulosic biomass by Kluyveromyces marxianus CICC 1727-5 and Spathaspora passalidarum ATCC MYA-4345. Appl Microbiol Biotechnol. 2019;103(6):2845–55.
Nakanishi SC, Soares LB, Biazi LE, Nascimento VM, Costa AC, Rocha GJM, Ienczak JL. Fermentation strategy for second generation ethanol production from sugarcane bagasse hydrolyzate by Spathaspora passalidarum and Scheffersomyces stipitis. Biotechnol Bioeng. 2017;114(10):2211–21.
Moyses DN, Reis VCB, de Almeida JRM, de Moraes LMP, Torres FAG. Xylose Fermentation by Saccharomyces cerevisiae: challenges and Prospects. Int J Mol Sci. 2016;17(3):207.
Morikawa Y, Takasawa S, Masunaga I, Takayama K. Ethanol productions from D-xylose and cellobiose by Kluyveromyces cellobiovorus. Biotechnol Bioeng. 1985;27(4):509–13.
Hou X. Anaerobic xylose fermentation by Spathaspora passalidarum. Appl Microbiol Biotechnol. 2012;94(1):205–14.
Slininger PJ, Shea-Andersh MA, Thompson SR, Dien BS, Kurtzman CP, Balan V, da Costa Sousa L, Uppugundla N, Dale BE, Cotta MA. Evolved strains of Scheffersomyces stipitis achieving high ethanol productivity on acid- and base-pretreated biomass hydrolyzate at high solids loading. Biotechnol Biofuels. 2015;8:60.
Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, Labutti KM, Sun H, Clum A, Pangilinan JL, Lindquist EA, et al. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci USA. 2011;108(32):13212–7.
Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007;108:147–77.
Apel AR, Ouellet M, Szmidt-Middleton H, Keasling JD, Mukhopadhyay A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci Rep. 2016;6:1.
Hou J, Qiu CX, Shen Y, Li HX, Bao XM. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 2017;17:4.
Moreno AD, Carbone A, Pavone R, Olsson L, Geijer C. Evolutionary engineered Candida intermedia exhibits improved xylose utilization and robustness to lignocellulose-derived inhibitors and ethanol. Appl Microbiol Biotechnol. 2018;103(3):1405–16.
Nielsen F, Tomas-Pejo E, Olsson L, Wallberg O. Short-term adaptation during propagation improves the performance of xylose-fermenting Saccharomyces cerevisiae in simultaneous saccharification and co-fermentation. Biotechnol Biofuels. 2015;8:219.
Lee YG, Jin YS, Cha YL, Seo JH. Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour Technol. 2017;228:355–61.
Skoog K, Hahn-Hagerdal B. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl Environ Microbiol. 1990;56(11):3389–94.
Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35(5):1216–23.
Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73(4):331–71.
Gardonyi M, Osterberg M, Rodrigues C, Spencer-Martins I, Hahn-Hagerdal B. High capacity xylose transport in Candida intermedia PYCC 4715. FEMS Yeast Res. 2003;3(1):45–52.
Young E, Poucher A, Comer A, Bailey A, Alper H. Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl Environ Microbiol. 2011;77(10):3311–9.
Saloheimo A, Rauta J, Stasyk OV, Sibirny AA, Penttila M, Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol. 2007;74(5):1041–52.
Du J, Li S, Zhao H. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol BioSyst. 2010;6(11):2150–6.
Runquist D, Hahn-Hagerdal B, Radstrom P. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2010;3:5.
Leandro MJ, Goncalves P, Spencer-Martins I. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem J. 2006;395(3):543–9.
Mayr P, Bruggler K, Kulbe KD, Nidetzky B. D-Xylose metabolism by Candida intermedia: isolation and characterisation of two forms of aldose reductase with different coenzyme specificities. J Chromatogr B Biomed Sci Appl. 2000;737(1–2):195–202.
Moreno AD, Tellgren-Roth C, Soler L, Dainat J, Olsson L, Geijer C. Complete genome sequences of the xylose-fermenting candida intermedia strains CBS 141442 and PYCC 4715. Genome Announc. 2017;5:14.
Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59:135–53.
Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126–31.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91.
Rajeh A, Lv J, Lin ZG. Heterogeneous rates of genome rearrangement contributed to the disparity of species richness in Ascomycota. BMC genomics. 2018;19:282.
Voordeckers K, Kominek J, Das A, Espinosa-Cantu A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang YD, et al. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 2015;11:11.
Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, LaFe K, Yarfitz SL, Limaye AP, Cookson BT. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J Clin Microbiol. 2000;38(6):2302–10.
Muhlhausen S, Kollmar M. Predicting the fungal CUG codon translation with Bagheera. BMC Genom. 2014;15:411.
Sankari SL, Gayathri K, Balachander N, Malathi L. Candida in potentially malignant oral disorders. J Pharm Bioallied Sc. 2015;7(5):S162–4.
Wingard JR. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20(1):115–25.
Tanahashi M, Kubota K, Matsushita N, Togashi K. Discovery of mycangia and the associated xylose-fermenting yeasts in stag beetles (Coleoptera: Lucanidae). Naturwissenschaften. 2010;97(3):311–7.
Urbina H, Schuster J, Blackwell M. The gut of Guatemalan passalid beetles: a habitat colonized by cellobiose- and xylose-fermenting yeasts. Fungal Ecol. 2013;6(5):339–55.
Long TM, Su YK, Headman J, Higbee A, Willis LB, Jeffries TW. Cofermentation of glucose, xylose, and cellobiose by the beetle-associated yeast Spathaspora passalidarum. Appl Environ Microbiol. 2012;78(16):5492–500.
Medina-Cordova N, Lopez-Aguilar R, Ascencio F, Castellanos T, Campa-Cordova AI, Angulo C. Biocontrol activity of the marine yeast Debaryomyces hansenii against phytopathogenic fungi and its ability to inhibit mycotoxins production in maize grain (Zea mays L.). Biol Control. 2016;97:70–9.
Chi ZM, Liu GL, Zhao SF, Li J, Peng Y. Marine yeasts as biocontrol agents and producers of bio-products. Appl Microbiol Biotechnol. 2010;86(5):1227–41.
Canto A, Herrera CM, Rodriguez R. Nectar-living yeasts of a tropical host plant community: diversity and effects on community-wide floral nectar traits. Peerj. 2017;5:e3517.
Wu J, Hu J, Zhao S, He M, Hu G, Ge X, Peng N. Single-cell protein and xylitol production by a novel yeast strain Candida intermedia FL023 from lignocellulosic hydrolysates and xylose. Appl Biochem Biotechnol. 2018;185(1):163–78.
Moreno AD, Tomas-Pejo E, Ibarra D, Ballesteros M, Olsson L. In situ laccase treatment enhances the fermentability of steam-exploded wheat straw in SSCF processes at high dry matter consistencies. Bioresour Technol. 2013;143:337–43.
Ikeuchi T, Azuma M, Kato J, Ooshima H. Screening of microorganisms for xylitol production and fermentation behavior in high concentrations of xylose. Biomass Bioenerg. 1999;16(5):333–9.
Sonderegger M, Jeppsson M, Hahn-Hagerdal B, Sauer U. Molecular basis for anaerobic growth of Saccharomyces cerevisiae on xylose, investigated by global gene expression and metabolic flux analysis. Appl Environ Microbiol. 2004;70(4):2307–17.
Toivari MH, Aristidou A, Ruohonen L, Penttila M. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng. 2001;3(3):236–49.
Skoog K. The influence of oxygen in pentose fermentation by yeasts. Lund: Lunds Universitet; 1992.
Shi NQ, Cruz J, Sherman F, Jeffries TW. SHAM-sensitive alternative respiration in the xylose-metabolizing yeast Pichia stipitis. Yeast. 2002;19(14):1203–20.
Pfeiffer T, Morley A. An evolutionary perspective on the Crabtree effect. Front Mol Biosci. 2014;1:17.
Bellasio M, Peymann A, Steiger MG, Valli M, Sipiczki M, Sauer M, Graf AB, Marx H, Mattanovich D. Complete genome sequence and transcriptome regulation of the pentose utilizing yeast Sugiyamaella lignohabitans. FEMS Yeast Res. 2016;16:4.
Yuan T, Ren Y, Meng K, Feng Y, Yang P, Wang S, Shi P, Wang L, Xie D, Yao B. RNA-Seq of the xylose-fermenting yeast Scheffersomyces stipitis cultivated in glucose or xylose. Appl Microbiol Biotechnol. 2011;92(6):1237–49.
Saier MH Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1(2):257–79.
Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148(Pt 9):2783–8.
Katahira S, Ito M, Takema H, Fujita Y, Tanino T, Tanaka T, Fukuda H, Kondo A. Improvement of ethanol productivity during xylose and glucose co-fermentation by xylose-assimilating S. cerevisiae via expression of glucose transporter Sut1. Enzyme Microb Technol. 2008;43(2):115–9.
Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–64.
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK. Structure of xylose reductase bound to NAD(+) and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases. Biochem J. 2003;373:319–26.
Petschacher B, Leitgeb S, Kavanagh KL, Wilson DK, Nidetzky B. The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem J. 2005;385:75–83.
Cadete RM, de Las Heras AM, Sandstrom AG, Ferreira C, Girio F, Gorwa-Grauslund MF, Rosa CA, Fonseca C. Exploring xylose metabolism in Spathaspora species: XYL1.2 from Spathaspora passalidarum as the key for efficient anaerobic xylose fermentation in metabolic engineered Saccharomyces cerevisiae. Biotechnol Biofuels. 2016;9:167.
Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2009;2:9.
Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis D, Gelfand D, Sninsky J, White T, editors. PCR—protocols and applications—a laboratory manual. San Diego: Academic Press; 1990.
Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–7.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30(7):923–30.
Team RC: R: A language and environment for statistical computing. In: Computing RFfS. R Foundation for Statistical Computing; 2013.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12.
Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinf. 2012;13:238.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9.
Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–35.
Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–45.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:1.
Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–8.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Westman JO, Bonander N, Taherzadeh MJ, Franzen CJ. Improved sugar co-utilisation by encapsulation of a recombinant Saccharomyces cerevisiae strain in alginate-chitosan capsules. Biotechnol Biofuels. 2014;7:102.
Gietz RD. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2014;1163:33–44.
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66(8):3381–6.
Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–71.
Acknowledgements
We acknowledge the support from Christian Tellgren-Roth at the Science for Life Laboratory (SciLifeLab), and Roy Francis and Thomas Källman at the National Bioinformatics Infrastructure Sweden (NBIS) for assistance with massively parallel sequencing, bioinformatics analysis and access to the UPPMAX computational infrastructure. We also thank Johan Westman and Eduard Kerkhoven at the Department of Biology and Biological Engineering at Chalmers University of Technology for help in experimental data analysis.
Funding
Open access funding provided by Chalmers University of Technology. This work was financed by the Swedish Energy Agency (project no. 35372-1 and 38779-1 and 38779-2) and the Knut and Alice Wallenberg Foundation through the Wallenberg Wood Science Center.
Author information
Authors and Affiliations
Contributions
CG and FFO designed the study, performed the phylogenetic analyses, analysed the data and wrote the manuscript. GC performed the genome sequence analysis and flow cytometry, FFO performed the RNA sequence analysis and cloning, expression and activity analysis of the XRs. ADM and FFO completed the bioreactor cultivations. SS made the genome synteny, the reciprocal read mapping and the ANI analysis. SM completed the sequence alignment and the structural figures of the XRs. LO supervised the project and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
List of the locations and substrates where different C. intermedia strains have been isolated.
Additional file 2.
GO enrichment analysis by high-throughput functional annotation for gene product properties.
Additional file 3.
Phylogenetic tree of putative sugar transporters from C. intermedia CBS 141442 and other pentose-assimilating yeasts and accession numbers for C. intermedia putative MFS sugar transporters and known xylose-transporters from other yeast species. References for reported xylose uptake activity are included.
Additional file 4.
C. intermedia CBS 141442 central carbon metabolism associated proteins and corresponding accession numbers.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Geijer, C., Faria-Oliveira, F., Moreno, A.D. et al. Genomic and transcriptomic analysis of Candida intermedia reveals the genetic determinants for its xylose-converting capacity. Biotechnol Biofuels 13, 48 (2020). https://doi.org/10.1186/s13068-020-1663-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-020-1663-9