Abstract
This review article aims to investigate the genotypic profiles of Plasmodium falciparum and Plasmodium vivax isolates collected across a wide geographic region and their association with resistance to anti-malarial drugs used in Indonesia. A systematic review was conducted between 1991 and date. Search engines, such as PubMed, Science Direct, and Google Scholar, were used for articles published in English and Indonesian to search the literature. Of the 471 initially identified studies, 61 were selected for 4316 P. falciparum and 1950 P. vivax individual infections. The studies included 23 molecular studies and 38 therapeutic efficacy studies. K76T was the most common pfcrt mutation. K76N (2.1%) was associated with the haplotype CVMNN. By following dihydroartemisinin–piperaquine (DHA–PPQ) therapy, the mutant pfmdr1 alleles 86Y and 1034C were selected. Low prevalence of haplotype N86Y/Y184/D1246Y pfmdr1 reduces susceptibility to AS–AQ. SNP mutation pvmdr1 Y976F reached 96.1% in Papua and East Nusa Tenggara. Polymorphism analysis in the pfdhfr gene revealed 94/111 (84.7%) double mutants S108N/C59R or S108T/A16V in Central Java. The predominant pfdhfr haplotypes (based on alleles 16, 51, 59,108, 164) found in Indonesia were ANCNI, ANCSI, ANRNI, and ANRNL. Some isolates carried A437G (35.3%) or A437G/K540E SNPs (26.5%) in pfdhps. Two novel pfdhps mutant alleles, I588F/G and K540T, were associated with six pfdhps haplotypes. The highest prevalence of pvdhfr quadruple mutation (F57L/S58R/T61M/S117T) (61.8%) was detected in Papua. In pvdhps, the only polymorphism before and after 2008 was 383G mutation with 19% prevalence. There were no mutations in the pfk13 gene reported with validated and candidate or associated k13 mutation. An increased copy number of pfpm2, associated with piperaquine resistance, was found only in cases of reinfection. Meanwhile, mutation of pvk12 and pvpm4 I165V is unlikely associated with ART and PPQ drug resistance. DHA–PPQ is still effective in treating uncomplicated falciparum and vivax malaria. Serious consideration should be given to interrupt local malaria transmission and dynamic patterns of resistance to anti-malarial drugs to modify chemotherapeutic policy treatment strategies. The presence of several changes in pfk13 in the parasite population is of concern and highlights the importance of further evaluation of parasitic ART susceptibility in Indonesia.
Graphical Abstract

Similar content being viewed by others
Background
Malaria is an infectious disease that still poses a public health problem in 87 countries worldwide. In 2020, 1 year after the COVID-19 pandemic and service disruptions, the number of malaria cases rose to 241 million, an additional 14 million cases compared with 2019. In 2020, malaria deaths increased by 12% compared with 2019, to 627,000, an estimated additional 69,000 deaths. Nine countries in the World Health Organization (WHO) Southeast Asia Region were endemic for malaria in 2020, with approximately 5 million cases accounting for 2% of the global malaria caseload. Despite significant progress in reducing malaria incidence, Indonesia remains one of Southeast Asia’s nine malaria-endemic countries. It is responsible for 21% of reported cases and 6% of deaths in the area [1,2,3].
The rapid emergence and spread of parasite strains resistant to anti-malarial drug mainstay, such as chloroquine (CQ), sulfadoxine–pyrimethamine (SP), and, more recently, artemisinin (ART)-based combination therapy (ACT) poses a constant challenge to the malaria control and elimination programmes. It proposes to eliminate malaria in all provinces in Indonesia by 2030 based on regional target, with Jawa-Bali in 2023, Sumatra, Sulawesi, West Nusa Tenggara in 2025, Kalimantan and North Maluku in 2027, Maluku and East Nusa Tenggara in 2028 and Papua, West Papua in 2029 [3, 4]. Indonesia's malaria control and elimination programmes have successfully eliminated malaria in 67% of the 514 regencies and municipalities. However, malaria is still persistently highly endemic in Papua, West Papua, and East Nusa Tenggara provinces. It represents the vast majority of the country’s cases [5, 6]. Since 2004, Indonesia adopted ACT as the first-line anti-malarial drug regimen to replace the failing CQ and SP. However, to prevent ACT resistance, the government has tightened controls on ACT deployment. Only those with a proven laboratory diagnosis, either by microscopy or rapid diagnostic test, will receive DHA–PPQ [7].
In addition, anti-malarial drugs are divided into different families, some of which are still in use or removed due to resistance [8,9,10,11,12,13,14,15]. The history of using anti-malarial drugs, as described in Table 1, provides a detailed description of the development of anti-malarial drugs in several regions in Indonesia. Quinolines are the oldest type of anti-malarial medication. Quinine (QN) was the first medicine in this class to be proven safe and effective, and it is still used as a second-line treatment for severe malaria to this day for both Plasmodium sp. [16]. Since the first reports of CQ resistance in East Kalimantan and Indonesian Papua Provinces in 1975, CQ resistance has been documented in all parts of the archipelago through in vivo and in vitro drug tests [8,9,10,11,12,13,14,15]. except in one remote area in Indonesia, CQ therapy is more amenable to treatment and control [15].
Reports of decreasing CQ efficacy have highlighted the necessity for alternative P. falciparum and P. vivax treatment options [15, 17]. Furthermore, molecular studies have provided evidence that polymorphisms in the P. falciparum chloroquine resistance transporter (pfcrt) and P. falciparum multidrug resistance 1 (pfmdr1) genes modulate higher levels of CQ, mefloquine (MQ), halofantrine (HL), and QN resistance [18,19,20,21,22]. Due to increased cases of CQ treatment failure in various parts of Indonesia [23,24,25], a combination of antifolates, pyrimethamine (PYR), and sulfadoxine (SX) have become the first-line drug [26, 27]. This combination uses inhibitors of dihydrofolate reductase (DHFR) and dihydropteroate synthase (DHPS) [28]. The molecular basis of resistance to PYR and SX has been more clearly defined. From 1996 until 2015, several Indonesian studies in Indonesia have revealed many molecular markers in several codons responsible for antifolates resistance. SNPs mutation in pfdhfr such as A16V, C59R, S108R/N/T, I164L, and N51I were associated with PYR resistance. Meanwhile, SNPs mutation in pfdhps such as A437G, K540E, A581T, I588G, and I588F were linked to SX resistance [29,30,31,32,33,34,35,36]. The failure of SP was first reported in 1979 [26, 27, 37,38,39]. However, resistance to this medication combination was discovered in Indonesia and has spread throughout the archipelago [13, 40,41,42,43].
The global deployment of ACT to treat asexual blood-stage infections was recommended by the WHO and has successfully decreased the global prevalence of malaria [44, 45]. The Ministry of Health (MoH) Republic of Indonesia adopted ACT in its treatment strategy in 2004. To avoid the drug resistance problem, the National Malaria Control Programme, Republic of Indonesia, has taken steps to ensure effective drug administration and regulation [7, 16, 46]. ACT is an anti-malarial drug regimen that combines ART derivatives with other anti-malarial drugs, including lumefantrine (LUM), amodiaquine (AQ), MQ, piperaquine (PPQ), and SP [47]. Initially, the MoH used artesunate–amodiaquine (AS–AQ) as the first-line drug. However, several therapeutic efficacy studies reported high failure of these combinations in Central Java, Papua, and Sumatra [48, 49]. AS–AQ has lower efficacy than AL for treating uncomplicated malaria in children [50]. In line with a study from Hasugian et al. comparing two artemisinin-based combinations, patients treated with AS–AQ have a higher risk of failure than those treated with DHA–PPQ. The authors concluded that DHA–PPQ was a better tolerated and more efficient treatment for the multidrug-resistant P. vivax in Papua [49]. An artemisinin-based combination, dihydroartemisinin–piperaquine (DHA–PPQ), was recommended as a first-line treatment for falciparum and vivax malaria in 2008 [7]. Price et al. evaluated DHA–PPQ for P. vivax infection between 2004 and 2005. The study showed that the median time to recurrence was 43 days (range 22–45 days), and DHA–PPQ was an effective treatment of P. vivax in Papua [49]. In addition, for P. vivax therapy, Primaquine (PQ) will be given for 14 days with the dosage of 0.25 mg/BW/day. If it relapses, an additional dosage will be necessary (0.5 mg/BW/day) [7]. Later, in 2012, DHA–PPQ was adopted as the only ACT used for uncomplicated malaria cases throughout Indonesia. ART and its derivatives rapidly clear parasite load in the blood, within a few hours after oral administration, and yield a decrease in gametocyte carriage. The parasites are normally cleared after three days and the partner drug, which has a longer plasma half-life, is responsible for eliminating any surviving parasites [51, 52]. Therefore, successful treatment with ACT may depend on the parasite’s response to the partner drug, transmission severity, and parasite load [52, 53]. The combination of DHA–PPQ has contributed to new results of the piperaquine resistance marker, which is the copy number of plasmepsin 2–3. Patients with multicopy-plasmepsin2 parasites were 20 times more likely to experience treatment failure [53,54,55]. Although ART has been a component of ACT, the drug has been used as monotherapy in many Greater Mekong Subregion communities. As evidenced by a delay in parasite clearance, the situation has rapidly selected for P. falciparum and P. vivax resistance to ART in the region [56,57,58,59,60]. To monitor the efficacy of the DHA–PPQ, the MoH regularly conducted therapeutic efficacy studies (TES) [61].
Despite a long history since anti-malarial drug resistance emerged in the 1970s with several studies revealing the molecular basis of drug resistance in P. falciparum and P. vivax isolates, no systematic review reported the genotypic profile of molecular maker drug resistance based on TES study or epidemiological survey of malaria in Indonesia has yet been undertaken. Genotyping has been proposed to identify early dynamics changes in the parasite population and genetic diversity [42, 62, 63].
This study reviewed all publications on anti-malarial drug resistance in Indonesia since 1991. In addition, the frequency distribution of SNPs of relevant gene(s) related to resistance to anti-malarial drug mainstay, CQ, SP, and ACT was included. Importantly, molecular drug resistance studies indicated that the time and spatial distribution of malaria cases reflect an epidemiological process. Although broad in scope, this review highlights the need to understand the dynamics pattern of anti-malarial drug resistance in different periods. First, the authors describe the history of anti-malarial drug use in several areas in Indonesia. Second, it identifies information about the distribution of molecular markers associated with drug resistance in different periods. Third, dynamics map of SNPs prevalence mutant allele for each molecular marker associated with drug resistance based on spatial and temporal in three time periods 1995–2003, 2004–2012, and 2013–present of P. falciparum and P. vivax isolates include putative mutations in Indonesia. Finally, this review provides the following data that should be beneficial to prevent local malaria transmission and treatment strategy development as the Indonesian government recommends modifying chemotherapeutic treatment plans that effectively prevent further development of the resistance and mitigate or eliminate malaria transmission in the country. It can help guide the country’s anti-malarial policy for using ACT.
Methods
Study identification
The review was carried out following a predefined protocol and described as per Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) recommendations [42]. A computerized search was carried out with references that were screened using literature descriptors for PubMed as follows: (over 20 years, ending March 2022) combining the terms [(falciparum OR vivax resistance, Indonesia)] AND with different combinations of ACT (chloroquine) OR (quinine OR kina, which is the local Indonesia term of tree for quinine, Cinchona spp.) which has been known to humans since ancient times was first discovered by the Peru Indians as a cure for malaria, because this plant contains quinoline alkaloids (quinoline) on the bark, which OR quinidine OR amodiaquine OR mefloquine OR lumefantrine OR halofantrine OR dihydroartemisinin OR dihydroartemisinin–piperaquine OR artemisinin OR artemether OR artesunate OR pyrimethamine sulphadoxine OR trimethoprim OR pyrimethamine OR sulphadoxine OR antifolates OR primaquine OR artemether OR artemether lumefantrine AND according to single-nucleotide polymorphisms known to be associated with treatment failure (mutation OR polymorphism OR SNP OR mutant allele OR mutant or allele change OR allelic changing). PubMed, Science Direct, and Google Scholar databases implemented the complete search strategies.
Study selection
Four authors (FVR, PBSA, FKD, and CR) independently reviewed abstracts and the full text of the references identified to select the articles for inclusion suitability. They extracted the data, with disagreements addressed and agreement with other authors (MI, DS, and PC). The authors considered studies of various designs that identified molecular profiles of anti-malarial drug resistance across a wide geographic region of Indonesia in P. falciparum and P. vivax isolates. However, examiners were not blinded to authors, institutions, or journal names.
Inclusion criteria
Studies would be included in the analysis if all of the following distinguishing features could be obtained from the publication:
-
1.
Published in English and Indonesian.
-
2.
Original articles and short reports, but no review articles.
-
3.
Patients with uncomplicated P. falciparum or P. vivax infections in Indonesia since 1991.
-
4.
The standards of drug resistance were defined following the WHO guidelines.
-
5.
A study comprising an analysis of anti-malarial drug resistance molecular marker.
Data extraction
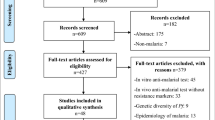
Data were extracted using a standard form created particularly for this review. The sections entered in the form were as follows: study identification (study title, author, journal, year of collecting sample, year of publication, country, language, and financial institution), study characteristics (design, number of patients, number of sites, and study period), and population characteristics under study (drug type, hemoglobin, gametocytes, days of follow-up, gender, therapeutic response, P. falciparum count, molecular marker, SNPs, SNPs frequency). Therefore, this systematic review focuses on reporting studies in evidence, their findings, and qualitative synthesis. Figure 1 shows the search strategy used. The list of 61 articles included in this review is shown in Additional File 1.
Results
The literature search yielded 471 studies, 124 of which were duplicated after the inclusion criteria were checked. The title, abstract, and full text were then examined, and an additional 286 studies were eliminated, as shown in the flowchart (Fig. 1). Finally, 61 studies were included for 4316 P. falciparum and 1950 P. vivax isolates. Of the 4316 individual isolates infected with P. falciparum, 1458 (29.05%) were treated with CQ, 271 (4.3%) with SP, treatment of CQ plus SP with a single dose of primaquine (PQ) 28 (0.73%) for gametocytaemia, 32 (0.83%) received artemether (AM) and PQ, 138 (0.85%) received artesunate (AS) plus SP, 53 (1.37%) received AS–AQ–PQ, 31 (0.8%) received CQ plus PQ, 195 (5.05%) received artemether-lumefantrine (AL), 235 (6.1%) received AS–AQ, 24 (0.62%) received (AS–AQ) plus PQ, 1642 (42.6%) received DHA–PPQ, and 209 (5.41%) received DHA–PPQ plus PQ.
In this review, PQ was administered as a single dose (0.75 mg/kg) on day 3 or PQ single dose of 45 mg on days 0 and 2 [12, 13, 43,44,45,46, 64,65,66,67,68,69,70]. PQ was administered as a gametocytocide. Of the 1950 individual isolates infected with P. vivax, 793 (40.5%) were treated with CQ, 11 (0.65%) were treated with ART-SP, 84 (5%) received CQ plus PYR, 83 (5%) received AQ, 212 (12.6%) received SP, 78 (4.6%) received CQ plus PQ, 19 (1.1%) received HL, 167 (10%) were treated with AS–AQ plus PQ for 14 days (0.25 mg base/kg BW), 164 (9.7%) received DHA–PPQ plus PQ for 14 days (0.25 mg base/kg BW), and 266 (10.9%) were treated with DHA–PPQ [71,72,73,74,75,76,77,78,79,80,81,82,83,84]. In P. falciparum observation, patients were followed up for 3 days in one study (n = 119), 28 days in six studies (n = 420), 35 days in one study (n = 114), and 42 days in five studies (n = 993). In terms of P. vivax isolates, patients were followed up for 8 days in one study (n = 46), 21 days in two studies (n = 315), 28 days in six studies (n = 407), 42 days in one study (n = 164).
This systematic review included studies conducted in western Indonesia, including North Sumatra, South Sumatra, Nias Island (North Sumatra), Lampung, Central Java, East Kalimantan, South Kalimantan, North Sulawesi, West Sulawesi, Lombok, Sumbawa, and eastern parts, such as East Nusa Tenggara, North Maluku, Alor, Kupang, Flores Island in East Nusa Tenggara, West Papua, and Papua. The dynamics of molecular markers analysed included SNPs in the pfcrt, pfmdr1, pfdhfr, pfdhps, and their homologs in P. vivax, as shown in Fig. 2. The vast majority of the isolates carried the pfcrt (C72S; V73I; M74I; N75D/K; K76T/N; H97L/Y; T152A; S163R; A220S; N326D; T333A/S; I356L/T) and pvcrt-o (AAG insertion), pfmdr1 (N86Y; Y184F; N1042D; S1034C; D1246Y) and pvmdr1 (Y976F; F1076L), pfdhfr (A16V; C59R; S108R; S108N; S108T; I164L) and pvdhfr (15S; 49R; N50K; F57L; S58R; T61M; 111L; S117N; S117T; 173F), and pfdhps (S436A; A437G; K540E; A581T; A581G; I588G; I588F; A613S/T) and pvdhps (383G) in various endemic areas.
Spatiotemporal map of molecular marker anti-malarial drug resistance (A) 1995–2003, (B) 2004–2012, (C) 2012 to date. Original map was obtained from Natural Earth (https://www.naturalearthdata.com) and modifed according to data from the references
From 1995 to 2003, epidemiology studies in molecular markers of anti-malarial drug resistance P. falciparum have mostly spread in Papua. Subsequently, from 2004 to 2012, after the ACT recommendation, the molecular marker P. falciparum has continuously spread in Sumatra, East Nusa Tenggara, Kalimantan, Sulawesi, and Papua. In terms of co-endemic from 2004 to 2012, molecular markers of anti-malarial drug resistance for P. vivax have been detected predominantly compared to other observation periods.
Distribution of mutant alleles associated with resistance to aminoquinoline
In this study, anti-malarial drug administration to patients included the 4-aminoquinolines, including CQ and AQ, which act as schizontocidal and gametocytocidal. The analysis of 18.3% (789/4316) individual isolates of pfcrt from western and eastern parts of Indonesia demonstrated that polymorphism in the pfcrt gene, K76T, has spread to all sample collection sites in North Sumatra, Nias Island in North Sumatra, South Sumatra, Lampung, North Sulawesi, North Maluku, East Nusa Tenggara, and Flores Island in East Nusa Tenggara during 1995. A highly variant in pfcrt SNPs mutation was detected in Papua isolates (Table 2; Fig. 3). Several SNPs formed four pfcrt haplotypes, such as C72S/V73/M74/N75/K76T haplotype as the dominant allele being present in 336 of 387 (86.8% of evaluable isolates), C72/V73/M74/N75/K76 being present in 66 of 387 (17.05%), and C72/V73/M74I/N75E/K76T being present in 42 of 387 isolates (10.85%). Parasites carrying the C72/V73/M74/N75/K76N haplotype were the rarest in all study areas, comprising 1 out of 387 in one individual in Lombok (0.25%).
Dynamics map of pfcrt and pvcrt in three time periods (A) 1995–2003, (B) 2004–2012 (C) 2013 to date. Map source from Natural Earth (https://www.naturalearthdata.com) and modifed according to data from the references
The treatment failure rate for CQ in P. vivax infection appeared in 14% of infections among residents of Nias in North Sumatra [75]. Thereafter in 1995, CQ therapeutic failure rate for P. vivax and P. falciparum was 15% (n = 34) and 30% (n = 37), respectively. Based on Baird et al., among 50% of the individual treated, there was in vivo resistance to CQ, and the weekly 300 mg prophylaxis base tablet of CQ was not effective against P. vivax [85]. In West Kalimantan during 1996, Fryauff et al. reported another analysis of the cumulative incidence of therapeutic failure among P. falciparum cases, in 1997, representing at day 28 was 7%. In addition, all 20 P. vivax parasitaemias were sensitive to chloroquine, and the blood remained clear, except for one case in which an asymptomatic parasitaemia appeared on day 28 [11]. The sample analysis obtained in 1996 by Fryauff et al. revealed parasitaemias cleared initially within four days of beginning supervised chloroquine therapy (25 mg base/kg over a 48-h period), but asexual parasites reappeared within 28 days in 52% (27 of 52) P. vivax and 25% (3 of 12) P. falciparum cases [43]. In the study between November 1996 and July 1999 in Papua of a 28-day observation, in vivo test revealed clinical resistance to CQ in 79 (74%) of the 107 individuals’ samples [12]. Twenty-eight-day cumulative incidence of confirmed resistance to chloroquine was 56% of infections evaluated. Chloroquine should not be considered adequate for treating acute vivax malaria acquired in this region [86].
The prevalence of the pfcrt K76T, C72S, A220S, N326D, T333A/S, and I356T reached 93.75% (15 of 16) from 2004 to 2012. Afterward, the mutant prevalence pfcrt K76T during the study period 2013–2017 was predominant overall (42 of 42, 100%) in South Sumatra, North Sulawesi, and North Maluku but varied among sites (12 of 25, 48%) to (12 of 13, 92.3%). Other SNPs pfcrt mutations such as C72S, A220S, N326D, T333A/S, and I356T were found in 12 of 13, or 92.3% of the cases (Table 2).
Of the 19% of isolates (821/4316) analysed for pfmdr1, 35.4% (17/48) isolates carried the pfmdr1 N86Y in 2002, 91.9% (102/111) in 2003, 37.7% (40/106) in 2004, and 62.7% (94/150) in 2005, as shown in Table 2. From 2013 to 2017, this allele was highly prevalent, occurring in more than 80% of isolates in Papua, West Sulawesi, North Sulawesi, and South Sumatra (Fig. 4). The mutant SNP Y184F occurred (21 of 35, 60%) in 2004 and, after that, until 2012, varied in Papua (4 of 8, 50%) to (13 of 16, 81.25%). The other pfmdr1 allele N1042D was also present 5/111 (4.5%) in 2003, 2/106 (1.9%) in 2004, and 16/254 (10.3%) in 2005. After 2008, the pfmdr1 S1034C 131/131 (100%) and N86Y 131/131 (100%) were found only in Lombok, Sumbawa, Alor, Kupang, and Lampung without any mutation in codon 1042. Polymorphisms at codons D1246Y of the pfmdr1 gene were observed only in isolates from Papua from 1995 to 2003. Subsequently, from 2004 to 2012, pfmdr1 SNP mutation changed to wild-type alleles D1246 accompanied by N86Y, Y184F, and N1042D (Fig. 4).
Dynamics map of pfmdr1 and pvmdr1 in three time periods (A) 1995–2003, (B) 2004–2012, (C) 2013 to date. Map source from Natural Earth (https://www.naturalearthdata.com) and modifed according to data from the references
Several authors have reported that N86Y/Y184/D1246Y and N86/Y184F/D1246 haplotypes in pfmdr1 may be associated with reduced AQ sensitivity and decreased sensitivity to AL, respectively [10, 77]. The N86Y/Y184/D1246Y haplotype was detected in (24 / 203, 11.8%) before 2008, reflecting the moderate prevalence during AS–AQ deployment in 2004–2008, suggestive reduce AQ susceptibility. Subsequent observation in the following year showed a decreased number of this haplotype (3/203, 1.5%). Thereafter, other haplotypes were detected N86/Y184/D1246Y (47 of 203, 23.15%) and N86Y/Y184F/D1246Y (28 of 203, 13.79%).
Regarding P. vivax, as shown in Table 3, the highest distribution of SNPs in pvmdr1 in Indonesia was observed in Maluku Province. The SNP variation in pvmdr1 before 2008 included F1076L 3/4 (75%) and Y976F 617/643 (96%). However, since 2008, polymorphism has only been found in codon Y976F 123/128 (96.1%) in both areas; Papua and East Nusa Tenggara. Another mutation in pvcrt-o with AAG insertion (2.2%) could be found only in Papua persistently before and after DHA–PPQ treatment. Data related to the pvmdr1 gene copy number were unavailable in any isolates examined (Fig. 4).
Distribution of mutant alleles associated with resistance to antifolate
Antifolate drug administration to patients in this review study used SP combination therapy. Antifolate drugs are used as anti-malarials through their inhibition of the folate metabolism of the parasite, both in the synthesis and use of folate cofactors. The key enzyme targets are dihydropteroate synthase (DHPS), inhibited by sulfa drugs, and dihydrofolate reductase (DHFR), inhibited by pyrimethamine and cycloguanil.
Table 2 show that the SNPs related to pfdhfr included double mutant C59R//S108N and A16V/ S108T 94/111 (84.7%) in Central Java during 2003. Other variations including A16V 11/46 (22%), N51 17/17 (100%), C59R 256/467 (55%), S108N/T 318/401 (79.3%), and I164L 8/27 (30%) were observed in P. falciparum isolates before 2008. SNP A16V was not observed in North Sulawesi and North Maluku in the following 2008 but increased in the prevalence of S108N 167/172 (91.7%), C59R 76/98 (62.5%), and I164L 24/81 (29.5%). According to study from Basuki et al. [37], pfdhfr haplotypes (based on alleles 16, 51, 59, 108, 164) found in Indonesia were A16/N51/C59/S108N/I164 (ANCNI), A16/N51/C59/S108/I164 (ANCSI), A16/N51/C59R/S108N/I164L (ANRNL), and A16/N51/C59R/S108N/I164 (ANRNI). In additional observation from 2013 to 2015, four pfdhfr haplotypes A16/C50/N51/C59R/108 (N/T), A16/C50/N51/C59R/108 (S/T), A16/C50/N51/C59R/108(S/N), and A16/C50/N51/C59R/S108 were found in North Sulawesi and North Maluku (Fig. 5). The double mutant C59R/S108N/T was dominant in North Sumatra, Lampung, Central Java, East Kalimantan, North Sulawesi, West Sulawesi, and Papua. The triple mutant C59R/S108N/I164L was commonly detected in South Kalimantan. In North Sulawesi, there was any change in mutation between 2004 to 2012, and 2013 to date from pfdhfr triplet mutation A16V/C59R/S108N and A16V/C59R/S108T confer to pfdhfr duplet mutation A16/C50/N51/59R/108N/T, A16/C50/N51/59R/108S/T, and A16/C50/N51/59R/108S/N. In Lampung, Central Java, and East Nusa Tenggara from 1995 to 2003, SNP mutation was detected in P. vivax from single mutant, duplet, and triplet. SNP mutation pvdhfr quintuple 49R/57L/58R/61M/117T was only found in Papua. Meanwhile, in East Nusa Tenggara, there were no differences in SNPs mutation compared between 2004 to 2012, and 2013 to date (Fig. 5).
Dynamics map of pfdhfr and pvdhfr in three time periods (A) 1996–2003, (B) 2004–2012, (C) 2013 to date. Map source from Natural Earth (https://www.natur alearthdata.com) and modifed according to data from the references
In addition, SNPs in pfdhps included A581T 9/27 (33.3%), I588G 12/27 (44.4%), and I588F 6/27 (22.2%). Before 2008, P. falciparum isolates only carried A437G and K540E (Table 2). The pfdhps haplotype S436/A437/K540/A581G/A613 was found in North Maluku, North Sulawesi, and East Nusa Tenggara in 2015. Six different mutant alleles S436/A437G/K540/A581/A613 (SGKAA) (0.03%) in Sumatra, South Kalimantan, Sulawesi, and West Nusa Tenggara; S436/A437G/K540/A581G/A613 (SGKGA) (0.03%) in East Java, South Kalimantan, East Kalimantan, and Sulawesi; S436/A437G/K540E/A581/A613 (SGEAA) (0.02%) in East Java, South Kalimantan, East Kalimantan, Central Kalimantan, and West Nusa Tenggara; S436/A437G/K540E/A581/A613 (SGEAA) (588F) (0.04%) in South Kalimantan, Sulawesi, West Nusa Tenggara, and Papua; S436/A437G/K540T/A581G/A613 (SGTGA) (0.1%) in East Java, South Kalimantan, East Kalimantan, and Central Kalimantan; and S436/A437G/K540E/A581G/A613 (SGEGA) (0.006%) in Jambi, East Java and Central Kalimantan were identified (Table 4) [51]. The pfdhps double mutant A437G/K540E was found predominantly in Papua from 1995 to 2003. The other quadruple mutation accompanied by I588F had been identified from 2004 to 2012 isolates from East Kalimantan and Java. From 2013 to date, haplotypes S436/A437/K540/A581G/A613 were identified in North Sulawesi, North Maluku, and East Nusa Tenggara. Furthermore, similar variant haplotypes were detected in Sumatra, Java, Kalimantan, West Nusa Tenggara, and Sulawesi (Fig. 6). However, slightly different haplotype variations were observed in the parasite isolates from various research areas in Papua and East Nusa Tenggara. The substitution of I588G/F was detected as A437G/K540E/A581T (GET) only in South Kalimantan.
Dynamics map of pfdhps and pvdhps in three time periods (A) 1996–2003, (B) 2004–2012, (C) 2013 to date. Map source from Natural Earth (https://www.natur alearthdata.com) and modifed according to data from the references
The novel mutations of pfdhps genes K540T, I588G, and I588F were detected as the specifically combined haplotypes (A16/N51/C59R/S108N/I164 + S436/A437G/K540T/A581G/A613) (ANRNI/SGTGA), (A16/N51/C59R/S108N/I164L + S436/A437G/K540T/A581G/A613) (ANRNL/SGTGA), (A16/N51/C59R/S108N/I164 + S436/A437G/K540E/A581/A613 (ANRNI/SGEAA) (588F)), and (A437G/K540E/A581T (GET) (588G)) (Fig. 5).
According to data from Basuki et al. and this review study, the combination of eleven pfdhr and eight pfdhps haplotypes, a totally of 29 different combined pfdhfr/pfdhps genotypes were determined; 3 combined haplotypes in 1995–2003, 34 combined haplotypes in 2004–2012 isolates and 10 combined haplotypes in 2013–present isolates. Polymorphisms in the pfdhfr/pfdhps combined haplotypes were observed to be dominant in West Papua during 1995–2003, Lampung, North Sumatra, Central Java, Middle Kalimantan, South Kalimantan, East Nusa Tenggara, Papua in 2004–2012 and North Sulawesi, North Maluku, East Nusa Tenggara in 2013–present (Table 4). Three quadruple mutants (59R/108R + 437G/540E); (59R/108N + 437G/540E), and (59R/108T + 437G/540E) were observed only in West Papua isolates during 1996–2003, and many different combined haplotypes were found in up to 2015 samples (Fig. 7).
Dynamics map of pfdhfr/pfdhps haplotypes in three time periods (A) 1996–2003, (B) 2004–2012 (C) 2013 to date. Map source from Natural Earth (https://www.naturalearthdata.com) and modifed according to data from the references
Increased mutations in a combination of haplotypes enhance parasite resistance levels against SP. The parasites with pfdhfr/pfdhps quintuple mutant, a genotype marker of SP resistance (A16V/C59R/S108N/T + A437G/K540E), were found in Lampung, North Sumatra, Central Java, East Kalimantan, West Sulawesi, and North Sulawesi. Previously reported results from Kalimantan (A16/N51/C59R/S108N/I164 + S436/A437G/K540E/A581/A613 (588F)) (ANRNI + SGEAA); (A16/N51/C59R/S108N/I164 + S436/A437G/K540T/A581G/A613) (ANRNI + SGTGA); (A16/N51/C59R/S108N/I164L + S436/A437G/K540/A581G/A613) (ANRNL + SGKGA); (A16/N51/C59R/S108N/I164L + S436/A437G/K540E/A581/A613) (ANRNL + SGEAA) and (A16/N51/C59R/S108N/I164 + S436/A437G/K540E/A581G/A613) ANRNI + SGEGA were detected from 2004 to 2012 (Fig. 7) [37]. In East Nusa Tenggara, pfdhfr/pfdhps quadruple mutants were found persistently during both observation periods, 2004 to 2012 and 2013. The parasites containing quintuple mutants from pfdhfr/pfdhps have been shown not to respond adequately to SP treatment [87, 88] and were detected mostly in Central Java and Papua from 2004 to 2012. The presence of a single pfdhfr mutation (C59R) with a single pfdhps mutation (K540E) accurately predicted the presence of the quintuple mutant [88]. The distribution of pfdhfr and pfdhps combined haplotypes in Indonesia is presented in Table 4.
Until 2008, SNPs of the pvdhfr in P. vivax included the double mutant (duplet) S58R/S117N 90/182 (49.5%), triplet (S58R/T61M/S117N) 6/137 (4.4%), (F57L/S58R/ S117T) 1/22 (4.5%), and quadruplet (15S/F57L/S117T/I173F) 1/22 (4.5%), (F57L/S111L/S117T/I173F) 1/22 (4.5%) (Table 3) [17, 18]. Quadruple mutations (F57L/ S58R/ T61M/S117T) were predominant in Central Java, Papua, Sumba, Purworejo, and Lampung. [89] Finally, pvdhps has a less frequent variation of haplotypes with codon involvement in position at 383G before and after 2008, with a prevalence of 19%.
Distribution of mutant alleles associated with resistance to artemisinin and its drug partner in Indonesia
Initially, AS–AQ was used in Indonesia from 2004 to 2008 as ACT's first-line treatment, until many resistance to this ACT was reported [48,49,50]. Since 2008, the ACT regimen has changed to DHA–PPQ. The ACT consists of a potent ART component classified as sesquiterpene lactone [51]. In any 1469 pfK13 isolates examined, no mutation associated with ART resistance was found. Other SNPs mutation pfK13 genes such as G453W (20%), V454C (20%), E455K (20%), and T474A (2.6%) were detected during 2015–2016. By concerning the ART partner drug, an increase in the copy number of the pfpm2 (3 of 6, 50%) gene was found among 808 isolates that survived the DHA–PPQ treatment in Papua [71, 90], but was not associated with PPQ resistance. This result demonstrates that DHA used in ACT is still highly productive in suppressing parasite density. Another study described a slight decline in PPQ susceptibility, although it did not appear to have reached clinically significant levels [91]. During observation periods, no 8 SNPs mutation in pvK12 associated with ART resistance, such as M448, T517, F519, I568, S578, D605, and D691, and L708, has been found in Papua and Jambi. P. vivax TES in Keerom and Merangin, Jambi province, revealed 100% ACPR of total analysed cases [61].
Discussion
This review analysed the genotypic patterns of P. falciparum and P. vivax isolates across wide geographic regions in Indonesia since 1991, when the MoH changed the first-line anti-malarial drugs from CQ to SP, then continued in 2004 with the use of ACT. It is the first longitudinal genotypic profile documenting the molecular marker of anti-malarial drug resistance of co-endemic P. falciparum and P. vivax analysis over 30 years in Indonesia [5, 92]. Therefore, this study review is expected to give comprehensive information through parasite genetic diversity patterns in the context of epidemiologic investigations.
The K76T of the pfcrt, a determining SNP that distinguishes resistance to CQ, was found in most P. falciparum isolates collected between 1991 and 2004 [12, 93, 94]. Mutations in the pfmdr1 gene can occur at several codon positions such as 86 (asparagine to tyrosine), 184 (tyrosine to phenylalanine), 1034 (serine to cysteine), 1042 (asparagine to aspartic acid), and 1246 (aspartic acid to tyrosine) [95]. SNP mutation at codon D1246Y was only observed in Papua. Studies on the pfmdr1 gene have identified N86Y and Y184F being more frequent in Asian and African parasites, whereas S1034C, N1042D, and D1246Y are more common in South American parasites [88]97. However, another study could find it in Asia [98]. This spread of South American alleles could explain the possibility of importing parasite isolate from its location, and a hard selective sweep may induce a higher level of mutant allele prevalence [99]. The isolates carried SNPs in pfmdr1 associated with CQ. resistance, such as N86Y 644/968 (67%), N1042D 191/558 (34.2%), and S1034C 131/334 (39.2%) [100]. The 184F allele is slowly disappearing in Southeast Asia, including Sumatra, Indonesia [101], except in western Cambodia and eastern Thailand [101]. Overall, the genotypic profiles of the P. falciparum isolate reflect a continual selective pressure by CQ and other similar drugs, such as QN, AQ, and PPQ, on the isolates in Indonesia. However, CQ is no longer used for P. falciparum treatment. That low prevalence of pfmdr1’s 184F mutation was due to QN, AQ, and PPQ exposures [102,103,104]. In addition, it is significant to recognize the interesting evidence that ART selects for the pfmdr1 N86/Y184F haplotype in vivo [94] and in vitro [104] experiments. The selective impact of ACT supports the pfmdr1 haplotype N86/Y184F, originally explained in an African study [105] and Papua [104, 106]. The pfmdr1 N86Y allele was once widespread in Southeast Asia, but it has slightly declined in frequency as CQ and AQ have been withdrawn [54]. The change to N86 resulted in a three- to four-fold increase in the IC50 values for LUM, MQ, and DHA. Codons N86Y, Y184F, and D1246Y are uniquely associated with sensitivity to LUM and AQ in sub-Saharan Africa [107]. Another study in Tanzania observed a high prevalence of pfmdr1 N86Y, Y184, and 1246Y in patients who failed treatment with AQ. Longitudinal cohort studies in Africa showed that the SNPs mutation pfmdr1 at codons N86Y, Y184F, and D1246Y is associated with AL or AS–AQ drug pressure [108, 109]. Meanwhile, Uganda detected a high prevalence of pfmdr1 N86, Y184F, and D1246 alleles after treatment with AL [89]. A dramatic fall in the prevalence of N86Y was also detected in Nias Island in North Sumatra, from 100% in 2003 [110] to 31.4% in 2005 [105, 111]. However, it is also related to high pfcrt K76T mutant prevalence [11]. All P. falciparum isolates from Central Java possessed a mutant allele K76T of the pfcrt gene paired with the N86Y or D1042 allele of the pfmdr1 gene [15]. SNP mutation of pfmdr1 D1246Y allele might reduce the chloroquine 50% inhibitory concentration (IC50) [99]. It is in line with the surveys conducted in the same area, which observed resistance of P. falciparum to CQ, QN, and MQ by either in vivo or in vitro drug resistance tests [75].
CQ resistance in P. vivax has spread in all the countries since 1989 [88]. According to previous in-vivo and drug analysis studies in Papua, Sumatra and Sulawesi observed high grade and frequent CQ-resistant in P. vivax isolates [75, 112,113,114,115]. A study in northeastern Papua [116] and Tjitra et al.[116] in eastern Indonesia showed that the CQ failure rate reached more than 50%. In contrast, a later study by Asih et al. in Sentani Papua in 2007 [114] observed an estimation of a failure rate of 17%. This significant difference might be because naturally acquired immunity by persistent infection among indigenous residents in Sentani contributed to a sharp decrease in failure rate. Another mechanism, such as bottle neck, was greater in P. falciparum than in P. vivax. It was shown by the adaptation of one minor subpopulation (K2) among 4 subpopulations accounting for 100% of infection in late 2016–2017 [92]. This bottleneck led to the decreased allelic richness, the near fixation of a few alleles, and missing pre-existing alleles [63].
Observations on P. vivax isolates identified several SNPs in pvmdr1, such as F1076L 3/4 (75%) and Y976F 617/643 (96%). Brega et al.[117] identified the P. vivax orthologue of the pfmdr1 gene (pvmdr1), which was shown to have a role in the drug resistance of P. falciparum. The Y976F alteration was responsible for a 1.7-fold higher IC50 to CQ in Thai isolates [72]. In a study between 2003 and 2006 by Suwanarusk et al.[72], Y976F mutation was significantly prevalent in Indonesian isolates that almost reached fixation (96%, 24/25). Two polymorphisms, pvmdr1 Y976F mutation and insertion in the 1st exon (amino acid substitutions 10) of pvcrt-o were associated with in vitro CQ susceptibility and a significant increase in CQ IC50. Another study reported the identification of the pvmdr1 976 and 1076 mutation in a small number of Thai and Indonesian isolates without in vitro and clinical correlates [117]. The pvmdr1 Y976F mutation, combined with quadruple mutant, refers to pvdhfr sequences such as F57L/S58R/T61M/S117T, F57L/S111L/S117T/173F, and 15S/F57L/S117T/173F correlated with treatment failure following AQ plus SP [118].
In this review, pfdhfr and pfdhps SNPs mutation included several mutant alleles, such as A16V, N51I, C59R, S108N/T, I164L, and S436F, A437G, K540E, A581T, A581G, and I588F/G, respectively. The proportion of pfdhfr codon position at 108 in various locations in Indonesia has been determined, including double mutant C59R/S108N and A16V/S108T (84.7%) in Purworejo-Central Java [14], single mutant S108N (71.2%) in Alor-East Nusa Tenggara [114], and double mutant S108N/S108T (81.3%) in Lampung [74]. The emergence of pfdhfr SNP mutation A16V is also particularly interesting in resistance to cycloguanil, although this drug has never been used in Indonesia. The highest prevalence of this SNP mutation in Central Java might be due to the involvement of other drugs with similar action to cycloguanil, such as trimethoprim, which is commonly used in combination with sulfamethoxazole for the treatment of bacterial diseases. Another explanation is the presence of the parasite isolates imported from an area where the drug has been applied [119]. However, pfdhfr SNP I164L mutation was discovered only in South Kalimantan between 2012 and 2014, and pfdhfr N51 was detected in North Sulawesi and North Maluku. The pfdhfr mutant allele at codons 50 and 51 was absent in the samples examined in Indonesia. Exclusively, two new pfdhfr SNP mutations in low-parasitemia Bolivian isolates were detected as a point mutation at codons 50 and 164, showing the continuous establishment of these polymorphisms in a restricted area [120]. Pfdhfr mutant allele C50R is detected in association with N51I and S108N in South America and confers midlevel resistance. The pfdhfr SNPs mutation S108N, N51I, and pfdhps SNP mutation A437G were as the “primary anti-folate resistance mutations,” meanwhile the pfdhfr SNPs mutation C50R, I164L, and pfdhps SNP mutation K540E, and A581G were as the “secondary anti-folate resistance mutations [121].
In Madagascar and Thailand, two haplotypes have been described as pfdhfr triple mutants N51I/S108N/I164L and C59R/S108N/I164L might induce six–ten fold higher IC50 to PYR [122, 123]. The pfdhfr double mutants N51I/S108N or C59R/S108N induced PYR resistance 2–16-fold higher than pfdhfr single mutant S108N [124]. In contrast with another study, the pfdhfr single mutant S108N conferred a 100-fold increase in resistance to PYR [120, 125]. The pfdhfr triple mutants A16/N51/C59R/S108N/I164L and pfdhps double mutants S436/A437G/K540E/A581/A613 were observed in all study sites except in North Maluku and North Sulawesi.
Mutations in S436A/F, A437G, K540E/T, A581G, I588F, and A613S/T in the pfdhps gene have been linked to SX resistance [37, 69]. In P. falciparum, the resistance rate will increase when the A437G mutation is combined with the different mutant allele K540E [29]. Another study also reported that a combination between I588F and K540E mutations could increase the SX resistance [55]. Isolates P. falciparum from Purworejo, Central Java, carried multiple mutations in the pfdhps A437G (35.3%) and K540E (26.5%) genes, which might suggest the wider use of the second-line SP anti-malarial following withdrawal of CQ [14]. Another pfdhps study [126] reported that point mutations at A437G and K540E are responsible for SX resistance. It was considered that the point mutation at 437 is the first event and reduced response to SX. A molecular study in Malaysia [127] reported that 87% of isolates had a triple mutation in pfdhfr, and all isolates had point mutation at pfdhps codon A437G, accompanied by 81% point mutation at codon A581G, indicating decreased responsiveness of SX. Mutations at A581G and A613S in the background of A437G were associated with high clinical resistance to SP in Thailand and India [128]. A previous African study demonstrated that resistance to SP in vivo was related to three mutant alleles, such as S108N, N51I, and C59R, in the dhfr gene with or without mutant alleles A437G and K540E dhps gene [129].
The P. vivax isolates carried double mutant (S58R + S117N) 11/22 (50%) and quadruplet mutant (F57L, S58R, T61M, and S117T) 2/16 (12.5%), (15S/F57L/S117T/I173F) 1/22 (4.5%), (F57L/S111L/S117T/I173F) 1/22 (4.5%) [14] have been linked to resistance to PYR [107]. In P. vivax, nonsynonymous SNPs that alter amino acid positions 49, 57, 58, 61, 117, and 173, corresponding to similarly homologous positions in P. falciparum, have been shown to confer resistance to PYR [130]. The analogous SNP mutation pfdhfr S108N with the pvdhfr SNP mutation S117N conferred approximately 4000- and approximately 1600-fold increased resistance to PYR and cycloguanil, respectively, compared to the wild-type pvdhfr [131]. Pvdhfr SNP mutation S117N could increase the IC50 of PYR by more than 80 times [85]. Additionally, the single mutated allele S117N was detected at a high frequency in Turkey and Azerbaijan sample isolates, areas where antifolate drug pressure or resistance is not obvious as the first-line treatment in these areas. It occurs because the areas are still use a combination of CQ-PQ [132]. According to this important role of S117N, it is assumed that the S117N mutation is the first step in the drug resistance selection process [132] and has been strongly associated with SP resistance in areas with extensive use of SP [31, 130]. The double mutant pvdhfr S58R/S117N was 10- to 25-fold less resistant than the S117N [131]. In the pvdhfr gene, 20 non-synonymous mutations have already been identified [133, 134]. It is contrary to the availability of data from Indonesia, which is still limited. Previous data from Lampung showed a triple mutation in this area [135], and a quadruple mutant F57L/S58R/T61M/S117T was found in Papua. This quadruple mutation confers higher resistance to SP than the mutant allele encoding a double mutation (S58R/S117N or N50K/S117N) [78]. In 13 of the 16 isolates from Southeast Asia, residues 58 and 117 are implicated in PYR resistance [136, 137]. Triple mutations were found exclusively in Thai parasites. In this study, parasites harbouring triple mutations at F57L/S58R/ S117N are associated with high levels of SP resistance and cleared significantly more slowly in P. vivax than those with double mutations of S57R/S117N [138].
This review found no parasite clearance delay or mutation in the pfk13 gene associated with ART resistance in any isolates examined after ACT treatment [111, 119, 139, 140]. The pfK13 propeller domain polymorphisms have been associated with decreased sensitivity to ART in Southeast Asia and have arisen separately in Cambodia and Myanmar [59, 141,142,143]. The eight non-synonymous mutations observed in Southeast Asia and China, including the F446I, N458Y, N537D, R539T, I543T, P553L, P574L, and C580Y are related to P. falciparum resistance to AS monotherapy or ACT on day 3 [144]. Analyses of the P. falciparum genotypes in eastern Indonesia identified another SNPs in the pfK13 gene, G497V, in 0.9% of the 106 samples from Sumba [144]. In western Indonesia, the other SNPs of the pfK13, including G453W, V454C, and E455K, were detected in 20% of the isolates. Previous molecular investigations of clinical isolates of P. falciparum collected from DHA–PPQ clinical efficacy trials during 2015 and 2016 revealed no kelch13 polymorphism associated with ART resistance [90]. Nevertheless, late treatment failures related to resistance to partner PPQ were increasingly detected in Papua. Analyses of the copy number of the pfplasmepsin 2–3 gene revealed several recurrent isolates that carried an increased copy number of the pfplasmepsin 2–3 gene but still failed to identify any association with PPQ resistance. The results suggested that PPQ resistance had slowly emerged among the field isolates and might indicate the preparation of alternative partner drugs to replace PPQ [55]. In another study, pfpm2 CNVs did not result in PPQ resistance in vitro [145]. In Cambodia, the pfplasmepsin gene cluster showed 2–3 amplification as an important molecular determinant of PPQ resistance in P. falciparum [54].
Until now, no cases of P. vivax resistance to ACT have been reported in South Pacific and Southeast Asia. However, several studies have monitored the P. vivax ortholog of pfk13, pvk12 for polymorphisms that might lead to ART resistance [144]. While several polymorphisms have been found in pvkelch12, with low frequencies and limited polymorphisms, such as V552I, K151Q, and M124I (7 of 734, 1%) [146,147,148] Study in Indonesia also found no polymorphism associated with ART resistance [61]. These results suggested a lack of strong selection pressure from ART on pvk12 [144, 147]. Drug pressure with ART in the GMS was not related to signatures of selection for mutations in the Pvk12, and additional observations, including analysis of associated clinical data from these regions, could further clarify current findings [148].
The orthologous gene for Pfpm 2/3 in P. vivax is Pvpm4, identified and mapped for P. vivax located on chromosome 13 with a sequence length of 1353 bp [11]. Recently, genetic variation in pvpm4 I165V has been reported in Malaysia, Thailand, and Indonesia. Unfortunately, this mutation is unlikely to be associated with PPQ drug resistance since its frequency was not associated with the level of PPQ drug pressure. Meanwhile, pvpm4 amplification was not observed in 141 P. vivax field isolates from Thailand and Cambodia [148]. Nowadays, the genetic diversity of P. vivax cases from Timika, Papua Indonesia, was detected to be higher than in P. falciparum. This result demonstrated the greater refractoriness of P. vivax to control measures and the risk of distinct parasite subpopulations persisting in the community undetected by passive surveillance [12]
ART resistance is considered to have emerged due to high rates of private-sector self-medication, presumptive fever treatment, misdiagnosis, and the unregulated use of anti-malarial agents, including low-quality ACT medicines [142, 149]. Genetic markers of pfk13, pfcrt, and pfmdr1 were diverse, corresponding to variations in populations' transmission levels, treatment-seeking patterns, access to medical care, and use of antimalarials [150]. In Indonesia, the procurement and deployment of DHA–PPQ are strictly regulated by the MoH. The drugs are provided only in carefully selected government-run health institutions and private facilities capable of confirming the malaria diagnosis through microscopy or rapid diagnostic tests. As a result, DHA–PPQ is still highly productive in treating uncomplicated malaria cases. However, in some areas, the second line of ACT should anticipate the increasing cases of DHA–PPQ late treatment failure [74].
Resistance can be caused by using drugs that do not meet the norms, which will encourage the emergence of Plasmodium, which is treatment-resistant. If it happens, the trend of increasing parasitic drug resistance in malaria-endemic areas will increase, which is a cause of malaria’s high morbidity and mortality. It indicates that there are still obstacles to the implementation of the provision. Therefore, serious consideration should be given to identifying the demographics of resistance to anti-malarial drugs to help modify chemotherapeutic treatment plans that effectively prevent further development of the resistance and mitigate or eliminate malaria transmission in the districts. The unique characteristics of SNPs in each pfmdr1, pfdhfr, pfdhps, and their orthologue in P. vivax play a major role in driving anti-malaria treatment failure. It can help guide the country’s anti-malarial policy for using ACT. The presence of several changes in pfk13 in the parasite population is of concern and highlights the importance of further evaluation of parasitic ART susceptibility in Indonesia. Although additional efficacy studies are needed, DHA–PPQ appears to be an effective treatment for P. falciparum and P. vivax infection.
Conclusion
Summarily, polymorphism genes related to resistance to CQ, SP, and recently ACT, were examined in P. falciparum and P. vivax field isolates from Indonesia. The findings implied that the prevalence of altered genotypes remained dominant more than 20 years after CQ was removed from this region. The frequency distribution of molecular markers among the P. falciparum and P. vivax isolates indicated that the currently recommended anti-malarial drug DHA–PPQ is still effective in treating uncomplicated falciparum and vivax malaria. The unique characteristics of SNP haplotypes in each pfcrt, pfmdr1, pfdhfr, pfdhps, and its orthologue in P. vivax played a key role in driving anti-malarial treatment failure.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
WHO. World Malaria Report 2021. Geneva, World Health Organization, 2021.
MoH. Informasi Malaria Indonesia. Jakarta; 2021.
malERA Refresh Consultative Panel on Insecticide and Drug Resistance. malERA: An updated research agenda for insecticide and drug resistance in malaria elimination and eradication. PLoS Med. 2017;14:e1002450.
WHO. Zero malaria starts with me”: history of malaria elimination in Indonesia helps to shape a malaria-free future. Geneva, World Health Organization, 2020.
Elyazar IR, Hay SI, Baird JK. Malaria distribution, prevalence, drug resistance and control in Indonesia. Adv Parasitol. 2011;74:41–175.
MoH. High Malaria Endemic Areas. Jakarta; 2021.
Ministry of Health. Buku Saku Tatalaksana Kasus Malaria. Jakarta: Ministry of Health of the Republic of Indonesia; 2020.
Clyde DF, McCarthy VC, Miller RM, Hornick RB. Chloroquine-resistant falciparum malaria from Irian Jaya (Indonesian New Guinea). J Trop Med Hyg. 1976;79:38–41.
Syafruddin D, Krisin, Asih P, Sekartuti, Dewi RM, Coutrier F, et al. Seasonal prevalence of malaria in West Sumba district, Indonesia. Malar J. 2009;8:8.
Pribadi W. In vitro sensitivity of Plasmodium falciparum to chloroquine and other antimalarials in east Timor and east Kalimantan, Indonesia. Southeast Asian J Trop. 1992;23:143–8.
Fryauff DJ, Baird JK, Candradikusuma D, Masbar S, Sutamihardja MA, Leksana B, et al. Survey of in vivo sensitivity to chloroquine by Plasmodium falciparum and P. vivax in Lombok, Indonesia. Am J Trop Med Hyg. 1997;56:241–4.
Maguire J, Susanti A, Krisin, Sismadi P, Fryauff D, Baird J. e T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Ann Trop Med Parasitol. 2001;95:559–72.
Nagesha HS, Din-Syafruddin, Casey GJ, et al. Mutations in the pfmdr1, dhfr and dhps genes of Plasmodium falciparum are associated with in-vivo drug resistance in West Papua, Indonesia. Trans R Soc Trop Med Hyg. 2001;95:43–9.
Baird JK, Wiady I, Sutanihardja A, Suradi, Purnomo, Basri H, et al. Short report: therapeutic efficacy of chloroquine combined with primaquine against Plasmodium falciparum in northeastern Papua, Indonesia. Am J Trop Med Hyg. 2002;66:659–60.
Syafruddin D, Asih PB, Aggarwal SL, Shankar AH. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Purworejo district, Central Java Province. Indonesia Am J Trop Med Hyg. 2003;69:614–20.
Ministry of Health, Republic of Indonesia. Keputusan menteri kesehatan republik indonesia nomor hk.01.07/menkes/556/2019 tentang pedoman nasional pelayanan kedokteran tata laksana malaria. 2019. https://yankes.kemkes.go.id/unduhan/fileunduhan_1610416186_13796.pdf
Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:982–91.
Djimdé A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–1.
Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–3.
Adam R, Mukhtar MM, Abubakar UF, Damudi HA, Muhammad A, Ibrahim SS. Polymorphism analysis of pfmdr1 and pfcrt from Plasmodium falciparum isolates in Northwestern Nigeria revealed the major markers associated with antimalarial resistance. Diseases. 2021;9:6.
Happi CT, Gbotosho GO, Folarin OA, Bolaji ON, Sowunmi A, Kyle DE, et al. Association between mutations in Plasmodium falciparum chloroquine resistance transporter and P. falciparum multidrug resistance 1 genes and in vivo amodiaquine resistance in P. falciparum malaria-infected children in Nigeria. Am J Trop Med Hyg. 2006;75:155–61.
Wurtz N, Fall B, Pascual A, Fall M, Baret E, Camara C, et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother. 2014;58:7032–40.
Ebisawa I, Fukuyama T. Chloroquine resistance of Plasmodium falciparum in West Irian and East Kalimantan. Ann Trop Med Parasitol. 1975;69:275–82.
Huaman MC, Yoshinaga K, Suryanatha A, Suarsana N, Kanbara H. Short report: polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok. Indonesia Am J Trop Med Hyg. 2004;71:40–2.
Saleh I, Handayani D, Anwar C. Polymorphisms in the pfcrt and pfmdr1 genes in Plasmodium falciparum isolates from South Sumatera. Indonesia Med J Indones. 2014;23:3–8.
Hutapea AM. Treatment of falciparum malaria with combined sulfadoxine and pyrimethamine in Jayapura. Indonesia Bul Penelit Kesehat. 1979;7:9–13.
Tjitra E, Gunawan S, Laihad F, Marwoto H, Sulaksono S. Evaluation of antimalarial drugs in Indonesia 1981–1995. Bul Penelit Kesehat. 1997;25:27–58.
WHO Scientific Group on the Chemotherapy of Malaria & World Health Organization. Practical chemotherapy of malaria: report of a WHO scientific group. 1990. World Health Organization. https://apps.who.int/iris/handle/10665/39778
Basuki S, Fitriah, Riyanto S, Budiono, Dachlan YP, Uemura H. Two novel mutations of pfdhps K540T and I588F, affecting sulphadoxine-pyrimethamine-resistant response in uncomplicated falciparum malaria at Banjar district, South Kalimantan Province, Indonesia. Malar J. 2014;13:135.
Syafruddin D, Asih PB, Wahid I, Dewi RM, Tuti S, Laowo I, et al. Malaria prevalence in Nias District, North Sumatra Province. Indonesia Malar J. 2007;6:116.
Asih PB, Rogers WO, Susanti AI, Rahmat A, Rozi IE, Kusumaningtyas MA, et al. Seasonal distribution of anti-malarial drug resistance alleles on the island of Sumba. Indonesia Malar J. 2009;8:222.
Fitriah F, Sulistyawati S, Riyanto S, Budiono, Basuki S, Dachlan YP, et al. Polymorphism of Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes among pregnant women with falciparum malaria in Banjar District, South Kalimantan Province, Indonesia. J Trop Life Sci. 2012;2:92–8.
Syafruddin D, Asih PB, Siregar JE, Tjitra E. Molecular basis of antimalarial drug resistance in Indonesia. Adv Exp Med Biol. 2003;531:103–15.
Mukh S, Darlina, Siti N. Deteksi spesies parasit malaria berbasis 18s rrna dan uji resistensinya terhadap obat untuk gen dhps sebagai pendukung pengembangan vaksin malaria iradiasi. Prosiding Seminar PSTA dan UNS: PSTA BATAN 2016:7–13. http://repo-nkm.batan.go.id/2752/1/DETEKSI%20SPESIES.pdf
Poespoprodjo JR, Fobia W, Kenangalem E, Lampah DA, Sugiarto P, Tjitra E, et al. Treatment policy change to dihydroartemisinin–piperaquine contributes to the reduction of adverse maternal and pregnancy outcomes. Malar J. 2015;14:272.
Hastings MD, Maguire JD, Bangs MJ, Zimmerman PA, Reeder JC, Baird JK, et al. Novel Plasmodium vivax dhfr alleles from the Indonesian Archipelago and Papua New Guinea: association with pyrimethamine resistance determined by a Saccharomyces cerevisiae expression system. Antimicrob Agents Chemother. 2005;49:733–40.
Basuki S, Fitriah, Risamasu PM, Kasmijati, Ariami P, Riyanto S, et al. Origins and spread of novel genetic variants of sulfadoxine–pyrimethamine resistance in Plasmodium falciparum isolates in Indonesia. Malar J. 2018;17:475.
Rumans LW, Dennis DT, Atmosoedjono S. Fansidar resistant falciparum malaria in Indonesia. Lancet. 1979;2:580–1.
Hoffman SL, Campbell J, Rustama D, Dimpudus AJ, Surumpaet B, Rusch J, et al. Pyrimethamine-sulfadoxine still effective against Plasmodium falciparum in Jayapura, Irian Jaya: RI-type resistance in 2 of 18 patients. Trans R Soc Trop Med Hyg. 1987;81:276–7.
Maguire JD, Lacy MD, Sururi, Sismadi P, Krisin, Wiady I, et al. Chloroquine or sulfadoxine–pyrimethamine for the treatment of uncomplicated, Plasmodium falciparum malaria during an epidemic in Central Java, Indonesia. Ann Trop Med Parasitol. 2002;96:655–68.
Hoffman SL, Dimpudus AJ, Campbell JR, Marwoto HA, Sukri N, Rustama D, et al. RII and RIII type resistance of Plasmodium falciparum to a combination of mefloquine and sulfadoxine/pyrimethamine in Indonesia. Lancet. 1985;2:1039–40.
Baird JK, Basri H, Jones TR, Purnomo, Bangs MJ, Ritonga A. Resistance to antimalarials by Plasmodium falciparum in Arso Pir, Irian Jaya, Indonesia. Am J Trop Med Hyg. 2002;44:640–4.
Fryauff DJ, Leksana B, Masbar S, Wiady I, Sismadi P, Susanti AI, et al. The drug sensitivity and transmission dynamics of human malaria on Nias Island, North Sumatra, Indonesia. Ann Trop Med Parasitol. 2002;96:447–62.
WHO. World health statistics 2018: monitoring health for the SDGs, sustainable development goals. Geneva: World Health Organization, 2018. https://apps.who.int/iris/handle/10665/272596.
WHO. Artemisinin resistance and artemisinin-based combination therapy efficacy: a status report. Geneva: World Health Organization, 2018. https://apps.who.int/iris/handle/10665/274362.
Ministry of Health, Republic of Indonesia. Epidemiologi Malaria di Indonesia. 2011. https://pusdatin.kemkes.go.id/resources/download/pusdatin/buletin/buletin-malaria.pdf
WHO. Guidelines for malaria. Geneva: World Health Organization, 2021. https://www.who.int/publications/i/item/guidelines-for-malaria
Djatmiko W. Uji efikasi terapi kombinasi artesunate + amodiaquine pada malaria falciparum tanpa komplikasi di kabupaten banjarnegara propinsi jawa tengah [Masters thesis]. Semarang: Universitas Diponegoro; 2005. http://eprints.undip.ac.id/12561/
Hasugian AR, Purba HL, Kenangalem E, Wuwung RM, Ebsworth EP, Maristela R, et al. Dihydroartemisinin–piperaquine versus artesunate–amodiaquine: superior efficacy and posttreatment prophylaxis against multidrug-resistant Plasmodium falciparum and Plasmodium vivax malaria. Clin Infect Dis. 2007;44:1067–74.
Faranita. Pedoman Tatalaksana Kasus Malaria di Indonesia. 2005
Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial drug resistance: literature review and activities and findings of the ICEMR Network. Am J Trop Med Hyg. 2015;93:57–68.
Sim I-K, Davis TME, Ilett KF. Effects of a high-fat meal on the relative oral bioavailability of piperaquine. Antimicrob Agents Chemother. 2005;49:2407–11.
Dhingra SK, Gabryszewski SJ, Small-Saunders JL, Yeo T, Henrich PP, Nok S, et al. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio. 2019;10:e02731–18.
Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–73.
Witkowski B, Duru V, Khim N, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study [published correction appears in Lancet Infect Dis. 2018;18:829]. Lancet Infect Dis. 2017;17:174–83.
Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria [published correction appears in N Engl J Med. 2009 Oct 22;361:1714]. N Engl J Med. 2009;361:455–67.
Dondorp AM, Fairhurst RM, Slutsker L, Macarthur JR, Breman JG, Guerin PJ, et al. The threat of artemisinin-resistant malaria. N Engl J Med. 2011;365:1073–5.
Ménard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–64.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5.
Fairhurst RM, Dondorp AM. Artemisinin-resistant Plasmodium falciparum malaria. Microbiol Spectr. 2016;4. https://doi.org/10.1128/microbiolspec.EI10-0013-2016.
Asih PBS, Rozi IE, Dewayanti FK, Wangsamuda S, Zulfah S, Robaha M, et al. Efficacy and safety of dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in Northern Papua and Jambi, Indonesia. medRxiv. 2020.
Dalmat R, Naughton B, Kwan-Gett TS, Slyker J, Stuckey EM. Use cases for genetic epidemiology in malaria elimination. Malar J. 2019;18:163.
Daniels RF, Rice BL, Daniels NM, Volkman SK, Hartl DL. The utility of genomic data for Plasmodium vivax population surveillance. Pathog Glob Health. 2015;109:153–61.
Price RN, Hasugian AR, Ratcliff A, Siswantoro H, Purba HL, Kenangalem E, et al. Clinical and pharmacological determinants of the therapeutic response to dihydroartemisinin–piperaquine for drug-resistant malaria. Antimicrob Agents Chemother. 2007;51:4090–7.
Dewi RM. Angka Kegagalan Pengobatan Klorokuin di Daerah dengan Beda Endemisitasnya; Kajian dengan Teknik PCR dan Kovensional. 2002. http://repository.bkpk.kemkes.go.id/1607/
Estiana L. Resistensi fansidar dan klorokuin:: Kajian uji sensitifitas serta faktor-faktor yang berhubungan dengan terjadinya resistensi di Kecamatan Pituruh Kabupaten Purworejo . [Yogyakarta]: Universitas Gadjah Mada; 2000. http://etd.repository.ugm.ac.id/penelitian/detail/5450
Sutanto I, Supriyanto S, Ruckert P, Purnomo, Maguire JD, Bangs MJ. Comparative efficacy of chloroquine and sulfadoxine–pyrimethamine for uncomplicated Plasmodium falciparum malaria and impact on gametocyte carriage rates in the East Nusatenggara province of Indonesia. Am J Trop Med Hyg. 2004;70:467–73.
Arubusman M. Evaluasi hasil guna kombinasi artesunat-amodiakuin dan primakuin pada pengobatan malaria falciparum tanpa komplikasi di Kabupaten Alor Propinsi Nusa Tenggara Timur. [Yogyakarta]: Universitas Gadjah Mada; 2009.
Triglia T, Wang P, Sims PF, Hyde JE, Cowman AF. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 1998;17:3807–15.
Tjitra E, Suprianto S, Currie BJ, Morris PS, Saunders JR, Anstey NM. Therapy of uncomplicated falciparum malaria: a randomized trial comparing artesunate plus sulfadoxine–pyrimethamine versus sulfadoxine–pyrimethamine alone in Irian Jaya, Indonesia. Am J Trop Med Hyg. 2001;65:309–17.
Asih PBS, Rozi IE, Dewayanti FK, Wangsamuda S, Zulfah S, Robaha M, et al. Efficacy and safety of dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in Papua and Sumatra, Indonesia. Malar J. 2022;21:95.
Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, Kosaisavee V, et al. Chloroquine-resistant Plasmodium vivax: in vitro characterization and association with molecular polymorphisms. PLoS ONE. 2007;2: e1089.
Ratcliff A, Siswantoro H, Kenangalem E, et al. Therapeutic response of multidrug-resistant Plasmodium falciparum and P. vivax to chloroquine and sulfadoxine–pyrimethamine in southern Papua, Indonesia. Trans R Soc Trop Med Hyg. 2007;101:351–9.
Poespoprodjo JR, Kenangalem E, Wafom J, Chandrawati F, Puspitasari AM, Ley B, et al. Therapeutic response to dihydroartemisinin–piperaquine for P. falciparum and P. vivax nine years after its introduction in Southern Papua, Indonesia. Am J Trop Med Hyg. 2018;98:677–82.
Fryauff DJ, Soekartono, Tuti S, Leksana B, Suradi, Tandayu S, et al. Survey of resistance in vivo to chloroquine of Plasmodium falciparum and P. vivax in North Sulawesi, Indonesia. Trans R Soc Trop Med Hyg. 1998;92:82–3.
Murphy GS, Basri H, Purnomo, Andersen Em, Bangs MJ, Pount DL, et al. Vivax malaria resistant to treatment and prophylaxis with chloroquine. Lancet. 1993;341:96–100.
Kim ES, Na BK, Park Y-K, Chung M-H, Lee J-S, Cheon S-M, et al. A case of chloroquine-resistant Plasmodium vivax malaria imported from Indonesia. Infect Chemother. 2008;40:52–7.
Tjitra E, Baker J, Suprianto S, Cheng Q, Anstey NM. Therapeutic efficacies of artesunate–sulfadoxine–pyrimethamine and chloroquine–sulfadoxine–pyrimethamine in vivax malaria pilot studies: relationship to Plasmodium vivax dhfr mutations. Antimicrob Agents Chemother. 2002;46:3947–53.
Salwati E, Handayani S, Jekti RP. Identifikasi single nucleotide polymorphism (SNP) Gen pvmdr1 pada Penderita Malaria Vivaks di Minahasa Tenggara (Sulawesi Utara). J Biotek Medisiana Indones. 2014;3:49–57.
Tjitra E, Maladi M, Prasetyorini B, Suprianto S, Harun S, Nurhayati N, et al. Efficacy of chloroquine, chloroquine plus sulphadoxine-pyrimethamine, and amodiaquine for treatment of vivax malaria in Bangka island, Indonesia: a randomized trial. Med J Indones. 2008;96–106
Asih PB, Rozi IE, Herdiana, Pratama NR, Hidayato A, Marantina SS, et al. The baseline distribution of malaria in the initial phase of elimination in Sabang Municipality, Aceh Province, Indonesia. Malar J. 2012;11:291.
Fryauff DJ, Baird JK, Basri H, Sumawinata I, Purnomo, Richie TL, et al. Randomised placebo-controlled trial of primaquine for prophylaxis of falciparum and vivax malaria. Lancet. 1995;346:1190–3.
Baird JK, Basri H, Subianto B, Fryauff DJ, McElroy PD, Leksana B, et al. Treatment of chloroquine-resistant Plasmodium vivax with chloroquine and primaquine or halofantrine. J Infect Dis. 1995;171:1678–82.
Pasaribu AP, Chokejindachai W, Sirivichayakul C, Tanomsing N, Chavez I, Tjitra E, et al. A randomized comparison of dihydroartemisinin–piperaquine and artesunate-amodiaquine combined with primaquine for radical treatment of vivax malaria in Sumatera, Indonesia. J Infect Dis. 2013;208:1906–13.
Baird JK, Wiady I, Fryauff DJ, Sutanihardja MA, Leksana B, Widjaya H, Kysdarmanto. In Vivo resistance to chloroquine by Plasmodium vivax and Plasmodium falciparum at Nabire, Irian Jaya, Indonesia. Am J Trop Med Hyg. 1997;56:627–31.
Sutanto I, Suprijanto S, Nurhayati, Manoempil P, Baird JK. Resistance to chloroquine by Plasmodium vivax at Alor in the Lesser Sundas Archipelago in eastern Indonesia. Am J Trop Med Hyg. 2009;81:338–42.
Spalding MD, Eyase F, Akala HM, Bedno SA, Progge ST, Coldren RL, et al. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu. Kenya Malar J. 2010;9:338.
Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine–pyrimethamine and chlorproguanil–dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8.
Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether–lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–5.
Rachmad B. Isolasi Dan Identifikasi Mutasi Gen Pfk13 (Pf3d7_1343700) Sebagai Penanda Resistensi Artemisinin Pada Isolat Plasmodium falciparum Asal Lampung. Prosiding dalam rangka Rakernas XIV & Temu Ilmiah XXII. 2019. p. 25–40.
Small-Saunders JL, Hagenah LM, Wicht KJ, Dhingra SK, Deni I, Kim J, et al. Evidence for the early emergence of piperaquine-resistant Plasmodium falciparum malaria and modeling strategies to mitigate resistance. PLoS Pathog. 2022;18: e1010278.
Pava Z, Puspitasari AM, Rumaseb A, Handayuni I, Trianty L, Utami R, et al. Molecular surveillance over 14 years confirms reduction of Plasmodium vivax and falciparum transmission after implementation of artemisinin-based combination therapy in Papua, Indonesia. PLoS Negl Trop Dis. 2020;14: e0008295.
Lim P, Chy S, Ariey F, Incardona S, Chim P, Sem R, et al. pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob Agents Chemother. 2003;47:87–94.
Henriques G, Hallett RL, Beshir KB, Gadalla NB, Johnson RE, Burrow R, et al. Directional selection at the pfmdr1, pfcrt, pfubp1, and pfap2mu loci of Plasmodium falciparum in Kenyan children treated with ACT. J Infect Dis. 2014;210:2001–8.
Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–8.
Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–9.
Lederman ER, Maguire JD, Sumawinata IW, Chand K, Elyazar I, Estiana L, et al. Combined chloroquine, sulfadoxine/pyrimethamine and primaquine against Plasmodium falciparum in Central Java, Indonesia. Malar J. 2006;5:108.
Atroosh WM, Al-Mekhlafi HM, Mahdy MA, Surin J. The detection of pfcrt and pfmdr1 point mutations as molecular markers of chloroquine drug resistance, Pahang, Malaysia. Malar J. 2012;11:251.
Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–9.
Roepe PD. Molecular and physiologic basis of quinoline drug resistance in Plasmodium falciparum malaria. Future Microbiol. 2009;4:441–55.
Srimuang K, Miotto O, Lim P, Fairhurst RM, Kwiatkowski DP, Woodrow CJ, et al. Analysis of anti-malarial resistance markers in pfmdr1 and pfcrt across Southeast Asia in the Tracking Resistance to Artemisinin Collaboration [published correction appears in Malar J. 2018;17:325]. Malar J. 2016;15:541.
Bangs MJ, Tjitra E, Kadir A, Ingkokusumo G, Fryauff DJ, Sumawinata I, et al. In vivo responses to antimalarials by Plasmodium falciparum and Plasmodium vivax from isolated Gag Island off northwest Irian Jaya, Indonesia. Am J Trop Med Hyg. 1999;60:542–6.
Sidhu ABS, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–26.
Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;7:11553.
Calçada C, Silva M, Baptista V, Thathy V, Silva-Pedrosa R, Granja D, et al. Expansion of a specific Plasmodium falciparum PfMDR1 haplotype in Southeast Asia with increased substrate transport. mBio. 2020;11:e02093–20.
Marfurt J, Wirjanata G, Prayoga P, Chalfein F, Leonardo L, Sebayang BF, et al. Longitudinal ex vivo and molecular trends of chloroquine and piperaquine activity against Plasmodium falciparum and P. vivax before and after introduction of artemisinin-based combination therapy in Papua, Indonesia. Int J Parasitol Drugs Drug Resist. 2021;17:46–56.
Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–90.
Achieng AO, Muiruri P, Ingasia LA, Opot BH, Juma DW, Yeda R, et al. Temporal trends in prevalence of Plasmodium falciparum molecular markers selected for by artemether–lumefantrine treatment in pre-ACT and post-ACT parasites in western Kenya. Int J Parasitol Drugs Drug Resist. 2015;5:92–9.
Baliraine FN, Rosenthal PJ. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis. 2011;204:1120–4.
Syafruddin D, Asih PBS, Casey GJ, Maguire J, Baird JK, Nagesha HS, et al. Molecular epidemiology of Plasmodium falciparum resistance to antimalarial drugs in Indonesia. Am J Trop Med Hyg. 2005;72:174–81.
Siswantoro H, Hasugian AR, Avrina R, Risniati Y. Efikasi dan Keamanan Dihidroartemisininpiperakuin (Dhp) pada Penderita Malaria Falsiparum Tanpa Komplikasi di Kalimantan Dan Sulawesi. Media Penelitian dan Pengembangan Kesehatan. 2011;21.
Tatura SNN. Efikasi Obat Kloroquine, Kina, Artesunate-SP, Artesunate-Amodiaquine, Artesunate-Lumafentrin pada Anak Malaria Falciparum di BLU RSUP Prof. Dr RD Kandou Manado Sari Pediatri. 2016;10:417.
Ali M, Hidayatullah TA, Alimuddin Z, Sabrina Y. Sequence diversity of pfmdr1 and sequence conserve of pldh in Plasmodium falciparum from Indonesia: its implications on designing a novel antimalarial drug with less prone to resistance. Iran J Parasitol. 2013;8:522–9.
Asih PB, Syafruddin D, Leake J, Sorontou T, Sadikin M, Sauerwein RW, et al. Phenotyping clinical resistance to chloroquine in Plasmodium vivax in northeastern Papua, Indonesia. Int J Parasitol Drugs Drug Resist. 2011;1:28–32.
Sutanto I, Endawati D, Ling LH, Laihad F, Setiabudy R, Baird JK. Evaluation of chloroquine therapy for vivax and falciparum malaria in southern Sumatra, western Indonesia. Malar J. 2010;9:52.
Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, Karyana M, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua. Indonesia PLoS Med. 2008;5: e128.
Brega S, Meslin B, de Monbrison F, Severini C, Gradoni L, Udomsangpetch R, et al. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J Infect Dis. 2005;191:272–7.
Marfurt J, de Monbrison F, Brega S, Barbollat L, Müller I, Sie A, et al. Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine–pyrimethamine: mutations in pvdhfr and pvmdr1. J Infect Dis. 2008;198:409–17.
Sibley CH, Hyde JE, Sims PFG, Plowe C v, Kublin JG, Mberu EK, et al. Pyrimethamine–sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–8.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–13.
Cortese JF, Caraballo A, Contreras CE, Plowe C v. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002;186:999–1006.
Andriantsoanirina V, Durand R, Pradines B, Baret E, Bouchier C, Ratsimbasoa A, et al. In vitro susceptibility to pyrimethamine of DHFR I164L single mutant Plasmodium falciparum. Malar J. 2011;10:283.
Sugaram R, Suwannasin K, Kunasol C, Mathema VB, Day NPJ, Sudathip P, et al. Molecular characterization of Plasmodium falciparum antifolate resistance markers in Thailand between 2008 and 2016. Malar J. 2020;19:107.
Nzila-Mounda A, Mberu EK, Sibley CH, Plowe CV, Winstanley PA, Watkins WM. Kenyan Plasmodium falciparum field isolates: correlation between pyrimethamine and chlorcycloguanil activity in vitro and point mutations in the dihydrofolate reductase domain. Antimicrob Agents Chemother. 1998;42:164–9.
Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8.
Wang P, Lee C-S, Bayoumi R, Djimde A, Doumbo O, Swedberg G, et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol Biochem Parasitol. 1997;89:161–77.
Rahman HA, Zakaria R, Abdullah MS, Nagappan S, Cox-Singh J, Singh B. Short report: differences in dihydrofolate reductase but not dihydropteroate synthase alleles in Plasmodium falciparum isolates from geographically distinct areas in Malaysia. Am J Trop Med Hyg. 2001;64:28–31.
Biswas S, Escalante A, Chaiyaroj S, Angkasekwinai P, Lal AA. Prevalence of point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum isolates from India and Thailand: a molecular epidemiologic study. Trop Med Int Health. 2000;5:737–43.
Jelinek T, Kilian AH, Kabagambe G, von Sonnenburg F. Plasmodium falciparum resistance to sulfadoxine/pyrimethamine in Uganda: correlation with polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes. Am J Trop Med Hyg. 1999;61:463–6.
Sattabongkot J, Lim CS, Kim T-S, Kim K, Chen J-H, Lin K, et al. Mutations in the antifolate-resistance-associated genes dihydrofolate reductase and dihydropteroate synthase in Plasmodium vivax isolates from malaria-endemic countries. Am J Trop Med Hyg. 2010;83:474–9.
Leartsakulpanich U, Imwong M, Pukrittayakamee S, White NJ, Snounou G, Sirawaraporn W, et al. Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol Biochem Parasitol. 2002;119:63–73.
Brega S, de Monbrison F, Severini C, Udomsangpetch R, Sutanto I, Ruckert P, et al. Real-time PCR for dihydrofolate reductase gene single-nucleotide polymorphisms in Plasmodium vivax isolates. Antimicrob Agents Chemother. 2004;48:2581–7.
Hawkins VN, Auliff A, Prajapati SK, Rungsihirunrat K, Hapuarachchi HC, Maestre A, et al. Multiple origins of resistance-conferring mutations in Plasmodium vivax dihydrofolate reductase. Malar J. 2008;7:72.
Imwong M, Pukrittayakamee S, Rénia L, Letourneur F, Charlieu J-P, Leartsakulpanich U, et al. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003;47:1514–21.
Hastings MD, Porter KM, Maguire JD, Susanti I, Kania W, Bamgs MJ, et al. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J Infect Dis. 2004;189:744–50.
Eldin de Pécoulas P, Basco LK, Tahar R, Ouatas T, Mazabraud A. Analysis of the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene sequence. Gene. 1998;211:177–85.
Eldin de Pécoulas P, Tahar R, Ouatas T, Mazabraud A, Basco LK. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol Biochem Parasitol. 1998;92:265–73.
Imwong M, Pukrittakayamee S, Looareesuwan S, Pasvol G, Poirreiz J, White NJ, et al. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine–pyrimethamine: geographical and clinical correlates. Antimicrob Agents Chemother. 2001;45:3122–7.
Risniati Y, Hasugian AR, Siswantoro H, Avrina R, Tjitra E, Delima. Respon klinis dan parasitologis dihidroartemisinin-piperakuin pada subyek malaria falsiparum dan malaria vivaks pada hari ke-3 kunjungan ulang. Media Litbang Kesehatan. 2011;21:57–165.
Lidia K, Deo DA, Pakan PD, Riwu M. Evaluation of therapeutic efficacy and safety of dihydroartemisinin–piperaquine in uncomplicated Plasmodium falciparum monoinfection in Timor Tengah Selatan District, Nusa Tenggara Timur, Indonesia. Southeast Asian J Trop Med Public Health. 2018;49:733–40.
Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lin P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria [published correction appears in N Engl J Med. 2014;371:786]. N Engl J Med. 2014;371:411–23.
Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin–piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–65.
Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, Thuy-Nhien NT, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis. 2019;19:943–51.
Parobek CM, Lin JT, Bailey JA, Lon C, Saunders DL, Juliano JJ, et al. Longitudinal pooled deep sequencing of the Plasmodium vivax K12 Kelch gene in Cambodia reveals a lack of selection by artemisinin. Am J Trop Med Hyg. 2016;95:1409–12.
Loesbanluechai D, Kotanan N, de Cozar C, Kochakarn T, Ansbro MR, Chotivanich K, et al. Overexpression of plasmepsin II and plasmepsin III does not directly cause a reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine, and piperaquine. Int J Parasitol Drugs Drug Resist. 2019;9:16–22.
Gresty K, Anderson K, Pasay C, Waters NC, Cheng Q. Polymorphisms in Plasmodium falciparum Kelch 13 and P. vivax Kelch 12 genes in parasites collected from three South Pacific countries prior to extensive exposure to artemisinin combination therapies. Antimicrob Agents Chemother. 2019;63:e00536–19.
Wang M, Siddiqui FA, Fan Q, Luo E, Cao Y, Cui L. Limited genetic diversity in the PvK12 Kelch protein in Plasmodium vivax isolates from Southeast Asia. Malar J. 2016;15:537.
Duanguppama J, Mathema VB, Tripura R, Day NP, Maxay M, Nguon C, et al. Polymorphisms in Pvkelch12 and gene amplification of Pvplasmepsin4 in Plasmodium vivax from Thailand, Lao PDR and Cambodia. Malar J. 2019;18:114.
WHO. Global report on antimalarial drug efficacy and drug resistance: 2000–2010. Geneva, World Health Organization. Global Malaria Programme. 2010. https://www.who.int/publications/i/item/9789241500470
Lubis IND, Wijaya H, Lubis M, Lubis CP, Beshir KB, Sutherland CJ. Plasmodium falciparum isolates carrying pfk13 polymorphisms harbor the SVMNT allele of pfcrt in Northwestern Indonesia. Antimicrob Agents Chemother. 2020;64:e02539-e2619.
Purworejo Health Report 2019. Available from: http://dinkes.purworejokab.go.id/
Southwest Sumba Health Report 2019. Available from: https://dinkes.sumbabaratkab.go.id/
Kulon Progo Health Report 2016. Available from: https://dinkes.kulonprogokab.go.id/
Gunung Kidul Health Report 2019. Available from: https://gunungkidulkab.go.id/D-7c5c10bd7afb63f86b3f053bd8c45ae7-NR-100-0.html
Wonosobo Health Report 2019. Available from: https://dinkes.wonosobokab.go.id/
Magelang Health Report 2019. Available from: https://dinkes.magelangkab.go.id/
Cilacap Health Report 2019. Available from: https://dinkes.cilacapkab.go.id/
Jepara Health Report 2019. Available from: https://dinkes.jepara.go.id/
Lampung Health Report 2019. Available from: https://dinkes.lampungprov.go.id/
Gorontalo Health Report 2019. Available from: https://dinkes.gorontalokota.go.id/
Makassar district Health Report 2019. Available from: https://makassarkota.go.id/dinas-kesehatan/
Sulawesi Health Report 2019. Available from: https://dinkes.sulselprov.go.id/
Kamelia M, Supargiyono, Wijayanti MA. Study on chloroquine resistance transporter (pfcrt) gene polymorphism of Plasmodium falciparum in malaria patients in Lampung. Trop Med J. 2011;1:1.
Suwandi JF, Asmara W, Kusnanto H, Syafruddin D, Supargiyono S. Efficacy of artemisinin base combination therapy and genetic diversity of Plasmodium falciparum from uncomplicated malaria falciparum patient in district of Pesawaran, Province of Lampung. Indonesia Iran J Parasitol. 2019;14:143–50.
Syafruddin D. Efficacy and safety of dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in 4 sentinel sites in Indonesia. 2016.
Reteng P, Vrisca V, Sukarno I, Djarkoni IH, Jacobs GE, Runtuwene LR, et al. Genetic polymorphisms in Plasmodium falciparum chloroquine resistance genes, pfcrt and pfmdr1, in North Sulawesi, Indonesia. BMC Res Notes. 2017;10:147.
Lamaka B, Arsin A, Alam G, Ishak H, Hatta M, Phihantono P. Plasmodium falciparum gene polymorphisms Pfmdr1 N86Y and drug self-medication in the endemic areas of West Papua Region, Indonesia. Int J Sci Basic Applied Res. 2017;33:103–11.
Suwanarusk R, Chavchich M, Russell B, Jaidee A, Chalfein F, Barends M, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis. 2008;198:1558–64.
Asih PB, Marantina SS, Nababan R, et al. Distribution of Plasmodium vivax pvdhfr and pvdhps alleles and their association with sulfadoxine–pyrimethamine treatment outcomes in Indonesia. Malar J. 2015;14:365.
Acknowledgements
This work was supported by Universitas Muhammadiyah Yogyakarta, Indonesia, for the funding sources and Eijkman Research Centre for Molecular Biology, National Research and Innovation Agency, Jakarta, Indonesia, for all the facilities provided.
Funding
This review was funded by Universitas Muhammadiyah Yogyakarta, Indonesia and Mahidol University, the Thailand Science Research and Innovation (TSRI), Fundamental Fund 2023, RTA6280006, Thailand.This research was funded in whole, or in part, by the Wellcome Trust (220211). For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
FR, PA, FD, and CR chose the articles for inclusion and extracted the data, with disagreements addressed and agreement with other senior authors MI, DS and PC. We considered studies of various designs that looked at molecular profiles of anti-malarial drugs in terms of the extent of related gene polymorphisms in P. falciparum isolates. There were limitations on the date of publication or the language used. All potentially acceptable texts for inclusion were then reread FR, PA, FD, and CR. Data was extracted using a standard form created especially for this review MI, DS, and PC. Furthermore, studies that did not record evidence of P. falciparum and P. vivax infection and uncomplicated infection in Indonesia were omitted (negative PCR or absence of microscopic examination). Figures and tables were created and edited, FD, according the review study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Rahmasari, F.V., Asih, P.B.S., Dewayanti, F.K. et al. Drug resistance of Plasmodium falciparum and Plasmodium vivax isolates in Indonesia. Malar J 21, 354 (2022). https://doi.org/10.1186/s12936-022-04385-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-022-04385-2