Abstract
Background
Drug resistant malaria poses an increasing public health problem in Indonesia, especially eastern Indonesia, where malaria is highly endemic. Widespread chloroquine (CQ) resistance and increasing sulphadoxine-pyrimethamine (SP) resistance prompted Indonesia to adopt artemisinin-based combination therapy (ACT) as first-line therapy in 2004. To help develop a suitable malaria control programme in the district of West Sumba, the seasonal distribution of alleles known to be associated with resistance to CQ and SP among Plasmodium falciparum isolates from the region was investigated.
Methods
Plasmodium falciparum isolates were collected during malariometric surveys in the wet and dry seasons in 2007 using two-stage cluster sampling. Analysis of pfcrt, pfmdr1, pfmdr1 gene copy number, dhfr, and dhps genes were done using protocols described previously.
Results and Discussion
The 76T allele of the pfcrt gene is nearing fixation in this population. Pfmdr1 mutant alleles occurred in 72.8% and 53.3%, predominantly as 1042D and 86Y alleles that are mutually exclusive. The prevalence of amplified pfmdr1 was found 41.9% and 42.8% of isolates in the wet and dry seasons, respectively. The frequency of dhfr mutant alleles was much lower, either as a single 108N mutation or paired with 59R. The 437G allele was the only mutant dhps allele detected and it was only found during dry season.
Conclusion
The findings demonstrate a slighly higher distribution of drug-resistant alleles during the wet season and support the policy of replacing CQ with ACT in this area, but suggest that SP might still be effective either alone or in combination with other anti-malarials.
Similar content being viewed by others
Background
The most widely used anti-malarial drugs, chloroquine (CQ) and sulfadoxine-pyrimethamine (SP), are failing at an accelerating rate in most endemic countries, including Indonesia. In response to this situation the World Health Organization has recommended artemisinin-based combination therapy (ACT), wherein an artemisinin derivative is paired with a second partner anti-malarial drug. This policy, however, has several limitations in resource-poor settings where diagnostic facilities are challenged by availability, cost, compliance and requirements for proper monitoring [1]. In addition, the rapid spread of resistance to currently available anti-malarials is limiting options for ACT partner drugs.
The molecular mechanisms underlying resistance to anti-malarial drugs have been investigated. Several single nucleotide polymorphisms (SNPs) in a number of Plasmodium falciparum genes have been associated with resistance to CQ [2] and SP [3]. In addition, amplification of the Plasmodium falciparum multi-drug resistance 1 (pfmdr1) gene has been associated with mefloquine resistance [4].
In Indonesia CQ, SP and primaquine have been the most widely used anti-malarial drugs. The drugs are readily available but diagnostic facilities are poor, and as a result their use is often inappropriate, leading to the increased spread of resistance. Resistance to CQ was first reported in 1975, resistance to SP appeared in 1978, and by 1997 treatment failures associated with both drugs had been documented in most provinces [5, 6]. Molecular epidemiology studies conducted from 2003 to 2005 in several sentinel sites throughout the Indonesia archipelago indicated widespread distribution of mutant alleles associated with resistance to CQ resistance (pfmdr1 86Y, Plasmodium falciparum chloroquine resistance (pfcrt) 76T) and SP (dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps)) [7–10]. The mutant allele associated with chloroquine resistance, pfcrt 76T, seems to be nearly fixed among the P. falciparum isolates collected, and pfmdr1 1042D alleles were mainly found in the island of Flores [9].
Previous studies of the distribution of drug resistance markers in Indonesia were not based on large-scale systematic sampling. In order to study the distribution of drug resistance markers more systematically, samples from a previously reported district wide study of seasonal malaria prevalence in West Sumba was analyzed [11]. The frequency distributions of the alleles across the villages throughout the district of West Sumba is reported.
Methods
Study site and sampling strategy
Sumba is a member of the Lesser Sunda Archipelago, located in the province of East Nusa Tenggara, Indonesia, at longitude 118.9 - 119.9 East and latitude 9.3 - 9.8 South, with total population of approximately 387,000 in 2007. The study design and sampling strategy have been described elsewhere [11]. Briefly, malariometric surveys were conducted twice, in March 2007 (wet season) and in August 2007 (dry season) using two-stage cluster sampling. The wet season usually occur during November to April whereas the dry season proceeds from May to October. Forty-five clusters (sub-villages) were chosen by probability proportional to size sampling. Within each sub-villages, households were chosen randomly by spinning a pointer, and were included until 100 subjects per cluster had been enrolled. Although the same 45 clusters were used in both surveys, no attempt was made to re-sample the same households; instead a new random selection of households was made within each cluster. The malaria prevalence in West Sumba was seasonal with the parasite rates was 6.83% and 4.95% in the wet and dry season, respectively. The malaria infections were mostly asymptomatic, mainly occurred among children and teenagers, and the geometric mean parasitemia decreased with age [11].
Data collection
In each subject, blood was collected by finger or heel prick for thick and thin films on glass slides and for blood blots on filter paper (Whatman, Schleicher & Schuell, Whatman International Ltd, Maidstone, UK) for malaria diagnosis and parasite genotyping. The study was approved by Eijkman Institute for Molecular Biology Research Ethics Commission, scientific and ethical review boards of the Naval Medical Research Unit #2, and by the Indonesian National Institute of Health Research and Development.
Genomic DNA preparation
DNA was extracted from P. falciparum positive blood samples diagnosed by microscopy as well as 10% of the malaria negative subjects, using chelex-100 ion exchanger (SIGMA, St Louis, USA) according to a procedure described previously [12]. DNA was either used immediately for a polymerase chain reaction (PCR) or stored at -20°C for later analysis.
PCR amplification and genotyping of pfcrt, pfmdr1, dhfr, and dhps codons
Detection of the single nucleotide polymorphisms of pfcrt, pfmdr1, dhfr, and dhps genes was performed using PCR and restriction fragment length polymorphyms (RFLP) as described elsewhere [3, 8, 9, 13, 14].
Pfmdr1 copy number
Pfmdr1 copy number was assessed using a Real Time PCR method [4, 15, 16]. The primers and a FAM-TAMRA (6-carboxyfluorescein 6-carboxy-tetra-methylrhodamine) probe specific to a conserved region of pfmdr1 and the primers and a VIC-TAMRA (chemical structure not released by Applied Biosystems) probe specific to β-tubulin were multiplexed so that both genes could be assayed in the same well. PCR reactions were performed on IQ5 Biorad® and consisted of pfmdr1 probe (150 nmol/L), pfmdr1 primers (300 nmol/L), β-tubulin probe (100 nmol/L), β-tubulin primers (100 nmol/L), IQ Multiplex Powermix (1×), DNA (2-4 μL), and water up to 25 μL. The cycling conditions were: 95°C for 15 minutes for pre denaturation, and 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. The cycle threshold (CT) was calculated with Optical System Biorad software ver. 1.0. DNA from strains 3D7 and W2mef were included on each plate. Pfmdr1 copy number was calculated according to the following formula: copy number = (E βtubulin )CT(βtubulin)/(E pfmdr1 )CT(pfmdr1). The efficiency (E) of β-tubulin, which was higher than that of pfmdr1, was assumed to be 2. Pfmdr1's efficiency, relative to that of β-tubulin, was calculated for each plate by assuming the 3D7 control has one pfmdr1 copy. The additional control, W2mef, was previously determined to have three pfmdr1 copies [17].
Results
Distribution of pfcrt and pfmdr1 gene polymorphisms
PCR yielded 213 and 231 amplicons in 29 and 32 villages in the wet and dry season, respectively. The pfcrt 76T allele was found in 92.9% of the P. falciparum isolates during the wet season and 84.9% during the dry season (Table 1). Analysis of isolates for the pfmdr1 gene revealed the existence of mutually exclusive 86Y and 1042D mutant alleles. The proportion of isolates carrying the 86Y allele was 41.9% in the wet season and 42.8% in the dry season and for the 1042D allele in the proportion were 72.8% and 53.3%. The pfmdr1 mutant alleles 1034C and 1246Y were not found in any of the isolates examined. Allelic combinations of 76T (pfcrt) with 86Y (pfmdr1) were found in 23.6% of the isolates during the wet season and 24.7% in the dry season whereas combinations of 76T (pfcrt) with 1042D (pfmdr1) were found in 6.2% and 5.5% in the wet and dry seasons, respectively (Table 2).
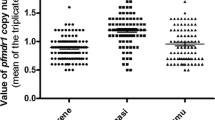
Distribution of pfmdr1 copy number
Analysis of the pfmdr1 gene copy number revealed 10.1% and 25.9% of the P. falciparum isolates in the wet and dry seasons, respectively, harbored more than one copy (Table 1). Of which, 72.2% in the wet and 85.7% dry seasons had more than one pfmdr1 gene copy number combined with the wild type 86N allele. The proportion of isolates that carried more than one copy number of pfmdr1 gene combined with 1042D was 27.8% in the wet and 14.3% in dry season, respectively. The distribution of P. falciparum isolates that carried more than one copy number is shown in Figure 1 in each village. The village that has P. falciparum isolates with the highest pfmdr1 gene copy number is Lamboya Dete.
Distribution of dhfr and dhps mutant alleles
Amplification of the dhfr gene showed that 31.5% of the isolates carried the 108N allele in the wet season whereas in dry season the proportion dropped to 24.7% (Figure 2). The 108T allele was not detected in any of the isolates examined. The 59R allele was found in 25.6% and 25.4% of the P. falciparum isolates in wet and dry seasons respectively, and in most cases was paired with the 108N allele. The other dhfr mutant alleles 16V, 50I, 51I and 164L were not found in any of the isolates examined. Amplification of the dhps gene revealed no mutant alleles in any of the isolates examined during the wet season whereas in the dry season, the 437G was found in 2.2% of the isolates. The other dhps mutant alleles (436A, 540E, 581G and 613S/T) were not detected in any of the isolates examined.
Distribution of parasite haplotypes by village
The pattern of the parasite haplotype found in the wet and dry season is shown in Table 3. The P. falciparum isolates that carried wildtype alleles of the pfcrt, pfmdr1, dhfr and dhps genes, simultaneously were only found in two subjects during dry season in two remote villages of Hoha Wungo (Kodi sub-district) and Lamboya Dete (Lamboya sub-district). On the contrary, due to the very low distribution of mutant alleles of dhps gene, we only found two isolates that carried simoultaneous mutant alleles in the four gene during the dry season.
Discussion
Molecular analysis of the P. falciparum isolates collected throughout the district of West Sumba indicated that the 76T allele of the pfcrt gene, a molecular marker for CQ resistance, has almost gone to fixation in the local population. Furthermore, nearly half of isolates also simultaneously carried pfmdr1 mutant alleles, either as 86Y or 1042D (Figure 2). Five mutant alleles of the pfmdr1 gene have been implicated in CQ resistance: 86Y, 184F, 1032C, 1042D and 1246Y. Of these, 86Y and 1042D are commonly found throughout Asia and Oceania whereas the 1032C and 1246N alleles are typical in P. falciparum isolates of Latin American origin [18, 19]. In Indonesia, the 86Y allele is more prevalent in western provinces whereas the 1042D allele dominates in the east. Interestingly, these alleles were found to be mutually exclusive [9]. These findings endorse abandonment of CQ as the first-line treatment of uncomplicated P. falciparum in the district of West Sumba.
Previous data in Africa suggest an associaton between amodiaquine (AQ) treatment failure and parasite isolates carrying a combination of pfcrt 76T and pfmdr1 86Y alleles [19, 20]. In this study P. falciparum isolates frequently carried the allelic combination of pfcrt 76T and pfmdr1 86Y, but previous study showed artesunate (AS) and AQ combination therapy to be highly effective [10]. This finding suggests that the parasite haplotype 76T and pfmdr 1 86N is still sensitive to AQ treatment, particularly in combination with AS. Otherwise, the high efficacy of AQ might be associated with the absence of 1246Y allele of the pfmdr1 in any of the isolates examined. Previous studies found out the selection of pfmdr1 1246Y alleles following AQ treatment [21–23].
In the present study, the distribution of the Pfcrt 76T, pfmdr1 86Y, dhfr 108N and dhfr 59R alleles were slightly higher in the wet season (Figure 2). It may be suggested that this overrepresentation of the drug-resistant alleles may reflect relatively higher drug pressure during in the wet season. This suggestion is supported by the fact that the wild type P. falciparum isolates that is found during the dry season.
Analysis of the pfmdr1 gene copy number revealed over 10% and 25% of the P. falciparum isolates harbor more than one copy in wet and dry season, respectively. This finding is similar to results in Thailand where the increase in the pfmdr1 gene copy number was associated with resistance to mefloquine and lumefantrine [4]. This finding may indicate that mefloquine and lumefantrine, which are currently used as partner drugs for artesunate and arthemeter, may not be suitable for use in Sumba. Previous studies also reported that increased in copy number of the pfmdr1 gene is more often usually paired with wildtype 86N allele in comparison to the mutant 86Y [4, 19]. This phenomenon is also observed in this study where the 1042D was the most common pfmdr1 allele found. The increase in pfmdr1 copy number was more often found along with either 86N or 1042N in comparison to 86Y of 1042D. The findings indicate that under CQ pressure the parasite may undertake either the pfmdr1 gene amplification or mutations, and that the former usually takes place earlier as indicated in the previous study [24].
The proportion of isolates carrying mutant alleles of the dhfr and dhps gene are relatively small in comparison to the P. falciparum isolates collected from the other parts of Indonesia [7–9]. For the dhfr gene, the majority of isolates carried the double mutation 108N+59R, whereas for dhps only two isolates carried the 437G allele. It follows that the use of SP as an interim alternative option to AS-AQ could be considered in this area, as the majority of the P. falciparum isolates are likely still sensitive to the drug. Treatment failure with SP in Africa is associated with the presence of quintuple mutations in dhfr and dhps genes, respectively [25].
This study shows that the determining mutant allele associated with CQ resistance is nearly fixed in the parasite population found in the district of West Sumba. A high prevalence of pfmdr1 mutant alleles was also found, reinforcing that CQ should no longer be used in this area. Except for the mutant dhps 437G allele which was only detected in two subjects during dry season, the distribution of the other drug-resistant alleles is slightly higher in the wet season. In addition, the increasing number of P. falciparum isolates carrying more than one copy number of pfmdr1 limit candidate partner drugs for artemisinin as this mutation is associated with resistance to mefloquine and lumefantrine, two of the compounds that are currently used in ACT. However, the relatively low prevalence of mutant alleles in the dhfr and dhps genes is encouraging for future ACT formulations in this area.
References
WHO: Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria. 2003, (WHO/HTM/RBM/200350) World Health Organization, Geneva
Djimde A, Doumbo OK, Cortese JF, Kayentao K, Diourte Y, Doumbo S, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV, Coulibaly D: A molecular marker fore chloroquine-resistant falciparum malaria. N Engl J Med. 2001, 344: 257-263. 10.1056/NEJM200101253440403.
Duraisingh MT, Curtis J, Warhurst DC: Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998, 89: 1-8. 10.1006/expr.1998.4274.
Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S: Mefloquine resistance in Plasmodium falciparum and increased pfmdr 1 gene copy number. Lancet. 2004, 364: 438-447. 10.1016/S0140-6736(04)16767-6.
Ebisawa I, Fukuyama T: Choroquine-resistant falciparum malaria from west Irian and East Kalimantan. Ann Trop Med Parasitol. 1975, 69: 275-282.
Tjitra E, Gunawan S, Laihad F, Sulaksono S, Arjono S, Richie TL, Manurung N: [Evaluation of antimalarial drugs in Indonesia 1981-1995]. Buletin Penelitian Kesehatan (in Indonesian). 1997, 25: 27-58.
Syafruddin D, Asih PB, Siregar JE, Tjitra E: Molecular basis of antimalarial drug resistance in Indonesia. Adv Exp Med Biol. 2003, 531: 103-115.
Syafruddin D, Asih PBS, Aggarwal SL, Shankar AH: Frequency distribution of antimalarial drug resistant alleles among the isolates of Plasmodium falciparum in Purworejo District, Central Java Province. Am J Trop Med Hyg. 2003, 33: 325-330.
Syafruddin D, Asih PBS, Casey GJ, Maguire J, Baird JK, Nagesha HS, Cowman AF, Reeder JC: Molecular epidemiology of Plasmodium falciparum resistance to antimalarial drugs in Indonesia. Am J Trop Med Hyg. 2005, 73: 174-181.
Asih PBS, Dewi RM, Tuti S, Sadikin M, Sumarto W, Sinaga B, Ven van der AJAM, Sauerwein , Syafruddin D: Efficacy of artemisinin- based combination therapy for treatment of persons with uncomplicated Plasmodium falciparum malaria in West Sumba District, East Nusa Tenggara Province, Indonesia, and genotypic profiles of parasites. Am J Trop Med Hyg. 2009, 80: 914-918.
Syafruddin D, Krisin , Asih P, Sekartuti , Dewi RM, Coutrier FN, Rozi IS, Susanti AI, Elyazar IR, Sutamiharja A, Rahmat A, Kinzer M, Rogers WO: Seasonal prevalence of malaria in West Sumba district, Indonesia. Malar J. 2009, 8: 8-10.1186/1475-2875-8-8.
Wooden J, Kyes S, Sibley CH: PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993, 9: 303-305. 10.1016/0169-4758(93)90131-X.
Wellems TE, Plowe CV: Chloroquine-Resistant malaria. J Infect Dis. 2001, 184: 770-776. 10.1086/322858.
Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF: Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990, 345: 255-258. 10.1038/345255a0.
Reeder JC, Rieckmann KH, Genton B, Lorry K, Wines B, Cowman AF: Point mutations in the dihydrofolate reductase and dihydropteroate synthase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am J Trop Med Hyg. 1996, 55: 209-213.
Ferreira ID, Rosario VE, Cravo PV: Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006, 5: 1-10.1186/1475-2875-5-1.
Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G: Selection of Plasmodium falciparum pfmdr1 Alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006, 50: 1893-1895. 10.1128/AAC.50.5.1893-1895.2006.
Rogers WO, Sem R, Tero T, Chim P, Lim P, Muth S, Socheat D, Ariey F, Wongsrichanalai C: Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J. 2009, 8: 10-10.1186/1475-2875-8-10.
Babiker HA, Pringle SJ, Abdel-Muchsin A, Mackinnon M, Hunt P, Walliker D: High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene, pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis. 2001, 183: 1535-1538. 10.1086/320195.
Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Bjorkman A: Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol. 2006, 6: 309-314. 10.1016/j.meegid.2005.09.001.
Holmgren G, Hamrin J, Svard J, Martesson A, Gil JP, Bjorkman A: Selection of pfmdr1 mutations after amodiaquine monotheraphy and amodiaquine plus artemisinin combination therapy in East Africa. Infect Genet Evol. 2007, 5: 562-569. 10.1016/j.meegid.2007.03.005.
Humphreys GS, Merinopoulos , Ahmed J, Whitty CJM, Mutabingwa TK, Sutherlands CJ, Hallett RL: Amodiaquine and Artemether-Lumefantrine Select Distict Alleles of the Plasmodium falciparum mdr1 Gene in Tanzanian Children Treated for Uncomplicated Malaria. Antimicrob Agents Chemother. 2007, 51: 991-997. 10.1128/AAC.00875-06.
Ochong EO, Broek van den IV, Keus K, Nzila A: Short report: association between chloroquine and amodiaquine resistance and allelic variation in the Plasmodium falciparum multiple drug resistance 1 gene and the chloroquine resistance transporter gene in isolates from the upper Nile in southern Sudan. Am J Trop Med Hyg. 2003, 69: 184-187.
Nair S, Nash D, Sudimack D, Nosten F, Anderson T: Characterization of segmental amplification of chromosome 5 associated with multidrug resistance in Plasmodium falciparum. Am J Trop Med Hyg. 2005, 93: 197-
Bwijo B, Kaneko A, Takechi M, Zungu IL, Moriyama Y, Lum JK, Tsukahara T, Mita T, Takahashi N, Bergqvist Y, Bjorkman A, Kobayakawa T: High Prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003, 85: 363-373. 10.1016/S0001-706X(02)00264-4.
Acknowledgements
The authors are grateful for the support of the Eijkman Institute, the National Institute of Health Research and Development of Indonesia and USNAMRU-2, Jakarta. The authors wish to thank Michael Kinzer, Iqbal RF Elyazar, Suradi, Suprianto, Sunardi, Faisal Amir of US NAMRU-2, Jakarta, the health professional staff of West Sumba District, and Riyanti Ekowatiningsih of the National Institute of Health Research and Development of Indonesia, Jakarta, for their assistance during the malariometric surveys, Rodiah Nababan of the Eijkman Institute, Jakarta, for her assistance in the malaria laboratory, and the Scientific Writing Course Jakarta, South East Asia Infectious Disease Clinical Research Network for their assistance in manuscript preparation. The opinions expressed are the private opinions of the authors and do not purport to represent those of the U.S. Navy. This study was supported by the U.S. Department of Defence Global Epidemic Information System, a grant-in-aid from the Netherlands Foundation for the advancement of Tropical Research through PRIOR programme, and the publication cost from South East Asia Infectious Diseases Clinical Research Network (SEAICRN).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PBSA, WOR, AIS, AR, EIR, MAK and DS performed field samples collection, molecular assays, data analysis, and the manuscripts writing. K, ST, RMD, FNC, and AS collected field samples and performed data analysis. AJAM and RWS contributed to data analysis, and the manuscript writing. DS, WOR, AJAM and RWS design the study and were responsible for management and fund raising for this study. All authors read and approved the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Asih, P.B., Rogers, W.O., Susanti, A.I. et al. Seasonal distribution of anti-malarial drug resistance alleles on the island of Sumba, Indonesia. Malar J 8, 222 (2009). https://doi.org/10.1186/1475-2875-8-222
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-8-222